Abstract
Candida albicans often resides in the oral cavity of healthy humans as a harmless commensal organism. This opportunistic fungus can cause significant disease in critically ill patients, such as those undergoing mechanical ventilation in the intensive care unit (ICU) having compromised local airway defense mechanisms. The goal of this study was to determine the intra- and inter-patient genetic relationship between strains of C. albicans recovered from dental plaque, tracheal secretions, and the lower airway by bronchoalveolar lavage of patients undergoing mechanical ventilation. Three pulsed-field gel electrophoresis (PFGE) typing methods were used to determine the genetic relatedness of the C. albicans strains, including electrophoretic karyotyping (EK) and restriction endonuclease analysis of the genome using SfiI (REAG-S) and BssHII (REAG-B). The C. albicans isolates from dental plaque and tracheo-bronchial sites from the same patient were genetically indistinguishable and retained over time, whereas strains from different patients usually separated into different genotypes. Among the three methods, REAG-B proved to be the most discriminatory method to differentiate isolates. The finding of genetically similar strains from the oral and tracheo-bronchial sites from the same patient supports the notion that the oral cavity may serve as an important source for C. albicans spread to the trachea and lung of mechanically ventilated patients.
Keywords: yeast, pulsed-field gel electrophoresis (PFGE), molecular epidemiology, mechanical ventilation, oral cavity
Candida albicans is an ubiquitous, dimorphic commensal yeast (1) that resides in the oral cavities of most healthy humans (2, 3). However, in critically ill patients with compromised local airway defense mechanisms C. albicans may act as an opportunistic fungal pathogen that can cause infections resulting in considerable morbidity and mortality (4–9). Risk factors predisposing an ICU patient to increased mucosal colonization by Candida species include the use of broad-spectrum antibiotics and corticosteroids, which upset the homeostatic balance of the commensal flora. In addition, intravascular catheters, complex surgical procedures, and acute renal failure increase the risk of Candida species infections (8, 10, 11).
For patients undergoing mechanical ventilation, the endotracheal tube can serve as a conduit for pathogen transmission to the lower airway (12, 13). While respiratory colonization by the yeast form of C. albicans in the airway is not a good marker for Candida pneumonia in critically ill patients, it may be a marker for increased risk of hospital morbidity and mortality (14–18). Indeed, it was shown that increased mortality of suspected ventilator-associated pneumonia (VAP) patients colonized only by Candida species in respiratory secretions was associated with increased inflammatory markers, rather than the presence of Candida (18). C. albicans colonization of the tracheo-bronchial tract has been associated with longer intensive care unit (ICU) and hospital stays and higher costs (14, 19, 20). In addition, several studies have documented the interaction of C. albicans with Pseudomonas aeruginosa, where C. albicans can undergo transformation to the filamentous form, allowing P. aeruginosa to form biofilms (14, 21–23). Consequently, these patients appear to have a greater risk of Pseudomonas VAP, though by itself C. albicans rarely causes VAP in non-immunocompromised patients (15, 16, 24, 25).
Molecular epidemiology has been used to analyze genetic relationships and identify the route of transmission for C. albicans (26). Methods such as pulsed-field gel electrophoresis (PFGE), Ca3 fingerprinting, and multilocus sequencing typing (MLST) offer high-resolution, greater stability, and have proved to be more discriminatory than phenotypic methods (27). Contour-clamped homogenous electrophoresis (CHEF), the most commonly used PFGE system, has been successfully applied as a tool for typing Candida strains (28–32). The PFGE system has been used to electrophoretically karyotype (EK) yeast through the separation of intact chromosomal DNA (33), but it has limitations in its ability to differentiate C. albicans isolates when compared to restriction analysis (34). To further delineate strains of Candida species, restriction endonuclease analysis of the genome (REAG) using either BssHII or SfiI as the restriction enzyme has proven to be a reliable method with high discriminatory abilities (34, 35).
We previously used PFGE to document the genetic relationships between bacterial respiratory isolates recovered from oral and tracheo-bronchial sites from mechanically ventilated patients in the ICU (13). These patients that were part of a clinical trial assessing the efficacy of 0.12% chlorhexidine oral rinse in reducing VAP (36), which postulated that reduction of potential respiratory pathogens in the mouth would reduce the incidence VAP. Molecular epidemiology studies were performed on samples from a subset of these patients suspected of having VAP who underwent bronchoalveolar lavage (13). Bacterial isolates from dental plaque were genetically identical to those from bronchoscopic cultures done at the time of suspected pneumonia. In the current study, a molecular epidemiology investigation was performed on C. albicans isolates recovered from the same oral and tracheo-bronchial samples. The aim of this study was to assess the clonal relatedness of intra- and inter-patient C. albicans strains from these patients, to document the route of transmission to the lower airway, and to determine an endogenous or exogenous origin of the strains. Three PFGE-based typing methods, electrophoretic karyotyping (EK), restriction endonuclease analysis of genome using SfiI (REAG-S), and restriction endonuclease analysis of genome using BssHII (REAG-B) were used to genetically analyze each C. albicans isolate recovered from the supragingival dental plaque (SG), tracheal secretions (TS), and bronchoalveolar lavage (BL) fluids from patients in the ICU undergoing mechanical ventilation.
Materials and Methods
Study population
The subjects were recruited for a randomized clinical trial (ClinicalTrials.gov identifier: NCT00123123) that tested the effect of 0.12% chlorhexidine gluconate oral rinse on oral colonization by respiratory pathogens (13, 37). The University at Buffalo Human Subjects Institutional Review Board approved the study protocol. Patients admitted to Erie County Medical Center (ECMC) trauma ICU during 14 February 2005 to 15 May 2006 who were intubated and mechanically ventilated within 48 h of admission were recruited to be subjects in this study. All participants or their surrogates provided informed consent. Exclusion criteria for the study, Acute Physiology and Chronic Health Evaluation II score, pneumonia diagnosis criteria, BL sampling, and oral examination protocols have been previously described (13, 37).
Oral, tracheal, and lung microbial sampling
Samples were collected from SG and TS every 48 h, and BL samples were collected as described previously (13). Samples were immediately transported on ice to the clinical microbiology laboratory where they were diluted, plated, and incubated for 72 h at 37°C in 5% carbon dioxide. Growth of target VAP pathogens and C. albicans was assessed on sheep blood agar and MacConkey agar, as previously described (13). C. albicans was identified using standard microbiological methods; the API 20C (bioMerieux, France) was used for further speciation as required.
In-gel cell lysis
Pulsed-field gel electrophoresis was performed according to the protocol described previously (38) with minor modifications. C. albicans isolates were subcultured from stock cultures to DifcoTM Sabouraud dextrose agar (Becton Dickinson [BD], Sparks, MD) and incubated for 48 h at 37°C. A single colony was transferred to 4 ml of YPD broth (1% yeast extract [BD], 1% BactoTM peptone [BD], and 2% dextrose) and incubated overnight with constant shaking at 37°C. After 14 h of incubation, cells were pelleted by centrifugation, resuspended in 4 ml of 50 mM EDTA, pH 8.0, and the optical density measured at 600 nm. The cell suspension was adjusted to 1.5 at 600 nm with additional 50 mM EDTA to ensure a similar number of cells per isolate for each lane of the PFGE gel. Cells were collected from a 1.5 ml aliquot by centrifugation at top speed for 30 sec. Each cell pellet was resuspended in 1 ml of 50 mM EDTA, pH 8.0, followed by centrifugation. After aspiration of supernatant, cells were resuspended in 100 µl of 50 mM EDTA, pH 8.0. Lyticase solution was prepared fresh just prior to use by mixing 1 ml of SCE (1 M sorbitol, 0.1 M sodium citrate, and 60 mM EDTA) solution with 4 µl lyticase (3 mg/ml; Sigma, St. Louis, MO), and 50 µl β-mercaptoethanol (Sigma). Lyticase solution (50 µl) was added to the cell suspension and mixed immediately with 250 µl of melted 1% (weight-to-volume) low-melting-point (LMP) InCert agarose (Cambrex Bio Science Rockland, Rockland, ME) in 0.125 M EDTA, pH 7.0 equilibrated to 55°C. Each sample was mixed and applied to five wells of a disposable plug mold (Bio-Rad, Hercules, CA), as per manufacturer's instructions. After the plugs were set by placing them at –20°C for 5 min, each set of plugs were extruded into a single 15 ml tube of freshly prepared EDTA-Tris-β-mercaptoethanol (ETB) solution (for 10.5 ml ETB: 9 ml 0.5 M EDTA, pH 8.0; 1 ml 1 M Tris-HCl, pH 8.0; and 0.5 ml β-mercaptoethanol). Tubes were incubated horizontally at 37°C with gentle rotation for a minimum of 4 h. ETB solution was removed and the plugs were rinsed twice with 5 ml of 50 mM EDTA. To each tube of five plugs, 2.5 ml proteinase K solution (for 10 ml proteinase K solution: 9 ml 0.5 M EDTA, pH 8.0; 1 ml 10% laurylsarcosine in 0.5 M EDTA; 0.1 mg RNase A, 10 µl of 10 mg/ml RNase solution); and 32 µg (32 µl of 1 mg/ml proteinase K [Sigma] solution) were added. The tubes were placed horizontally and rotated at 37°C for at least 6 h. After removing the proteinase solution, the plugs were rinsed with 5 ml of 50 mM EDTA, and incubated in 3–4 ml 1X TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) buffer for 1 h with gentle agitation. The plugs were used immediately or stored in 1 ml of storage solution (for 10 ml: 1 ml 1 M Tris-HCl and 9 ml 0.5 M EDTA, pH 8.0) at 4°C. For EK, plugs were used as is without restriction enzyme digestion.
In-plug restriction enzyme digestion
If the plugs containing genomic DNA were in storage solution, one plug was removed and placed in a microcentrifuge tube and washed twice by adding 1 ml of 1X TE buffer, and incubating for 30 min at room temperature. For SfiI digestion, the plug was transferred to a microcentrifuge tube containing 500 µL of buffer 2 (50 mM NaCl, 10 mMTris-HCl, 10 mM MgCl2, 1 mM dithiothreitol), and incubated on ice for 30 min. After aspirating the buffer, the plug was placed in 160 µL (4X plug volume) of buffer 2 containing 20 U SfiI and 2 µL BSA (New England BioLabs, Ipswich, MA), and incubated overnight at 50°C. For BssHII digestion, the plug was transferred to a microcentrifuge tube containing 500 µl of buffer 3 (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol), and incubated on ice for 30 min. After aspirating buffer, the plug was placed in 160 µl of buffer 3 containing 4 U of BssHII (New England BioLabs), and incubated overnight at 50°C. Following aspiration, each plug was soaked for 1 h at room temperature in 1 ml 1X TE to remove buffer salts. The plugs were then placed into the agarose gel wells for electrophoresis.
PFGE
DNA was recovered from each C. albicans isolate once and subjected to each of the PFGE methods described. Electrophoresis was performed with contour-clamped homogenous electric field apparatus (CHEF DR-II; Bio-Rad). PFGE analysis was performed as described (38) with minor modifications. For electrophoretic karyotyping (EK) the following parameters were used: switch time 90–325 sec, temperature 14°C, 4 V/cm in 1% agarose gel (SeaKem GTG agarose; FMC) with 0.5X TBE (1:20 dilution of 10X TBE stock, 890 mM Tris base, 890 mM boric acid, 20 mM EDTA, pH 8) for 48 h. For restriction endonuclease analysis of genomic DNA using SfiI (REAG-S) and BssHII (REAG-B), the switch time was 5–50 sec, temperature 14°C, 6 V/cm in 1% agarose gel (SeaKem GTG agarose; Cambrex Bio Science Rockland) with 0.5X TBE for 20 h. After electrophoresis, gels were stained in ethidium bromide solution for 30 min and destained with distilled water. Gels were photographed using AlphaEase FC and Alpha-Imager (Alpha Innotech, San Leandro, CA). For all EK gels, Saccharomyces cerevisiae DNA was included as a size marker. For all REAG-S and REAG-B, λ DNA ladder (Bio-Rad) was included as the size marker. C. albicans SC 5314 (ATCC MYA-2876), used as a reference strain, was provided by Dr. Mira Edgerton.
Band pattern analysis and interpretation
Dendrogram analysis was performed with BioNumerics software (Applied Maths, Austin, TX). The software assigned bands automatically. Each gel was normalized to respective size markers (S. cerevisiae for EK, λ DNA ladder for REAGs). The position tolerance and optimization was 1 and 3%, respectively, as used in previous study (35). The unweighted pair group method using arithmetic averages (UPGMA) was used to construct the phylogenetic trees. Dice coefficients were used to calculate the similarity percentage of the band patterns. The isolates were grouped into three categories: genetically identical when the number and size of the bands matched perfectly (similarity percentage 100%), genetically related (genetically similar) when the bands had minor differences (similarity percentage ≥95%, but less than 100%), and genetically unrelated (genetically different) when the bands differed (similarity percentage less than 95%) (34, 39).
Results
Colonization of patients
Sixty-two of 96 (65.6%) subjects studied were colonized by Candida species in at least one site; 54 (87.1%) of isolates were identified as C. albicans. In colonized patients who were sampled up to day 10, detection of Candida species increased from 34.4% on day 0 to 84% by day 10 or later. The number of days after ICU admission that C. albicans was first isolated ranged from day zero (day of the admission) to 18 days after admission.
This study focused on C. albicans strains isolated from a subset of patients with suspected pneumonia who underwent BL. Of the 30 patients undergoing BL, 20 patients harbored C. albicans in at least one site: 19 (63.3%) were colonized orally, 16 (53.3%) colonized in the trachea, and 2 (6.7%) were colonized in the BL. Two additional mechanically ventilated patients (ID nos. 45 and 74), also colonized with C. albicans, but not undergoing BL were also screened for strain comparison. A total of 50 isolates from these 22 patients were analyzed by PFGE. The source of the isolates was as follows: 27 from SG, 21 from TS, and 2 from BL. Among the 22 patients, C. albicans was recovered from one site in five patients, two sites in 15 patients, and three sites in two patients. Of the 17 patients who harbored isolates from more than one site, C. albicans was recovered concurrently from SG and TS in nine patients, from SG and subsequently in TS in seven patients, and from TS prior to SG and BL from patient four only. In patients who harbored C. albicans at all three sites, VAP bacterial pathogens (Escherichia coli, Staphylococcus aureus, and Klebsiella pneumoniae) were co-cultured in the BL from patient 41, and normal oral flora (alpha hemolytic streptococci, diphtheroids, and coagulase negative staphylococcus) from patient four. The same microbial flora was recovered from all three sites in these patients, suggesting that the route of transmission was from the mouth to the lower airway.
PFGE analysis of the isolates
Regardless of the PFGE method used, the band pattern of the SG isolate from patient 76 was very different from the TS isolate, which was very similar to the other isolates. It was later discovered that this SG isolate was actually Candida glabrata identified by carbohydrate assimilation assays (API 20C). Another 76 SG isolate from the same day, identified as C. albicans, was not analyzed. Since the focus of this study was C. albicans, this isolate was excluded from subsequent analyses.
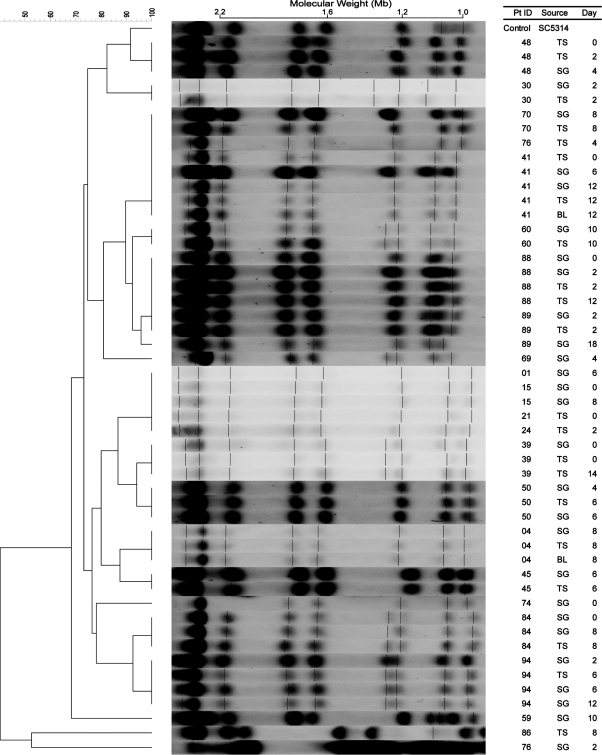
Electrophoretic karyotyping (EK) of chromosomal DNA (Fig. 1) from the C. albicans isolates generated 6 to 10 bands (mean, 8) from the isolates and 8 bands from the control strain SC5314. Analysis of the EK patterns yielded 17 karyotypes from the 49 isolates obtained from 22 patients, ranging in similarity from 47.5 to 100%.
Fig. 1.
Electrophoretic karyotypes (EK) with dendrogram for Candida albicans isolates. A genetic similarity percentage is shown above the dendrogram. Patient identification (Pt ID), sample site (Source), and number of days after admission to the intensive care unit that the strain was isolated (Day) are included along each PFGE lane. Saccharomyces cerevisiae DNA concatemers were used as the molecular size marker. Sizes are measured in megabases (Mb). C. albicans strain SC5314 (ATCC MYA-2876) was used as the control strain. Abbreviations: SG, supragingival dental plaque; TS, tracheal secretion; BL, bronchoalveolar lavage.
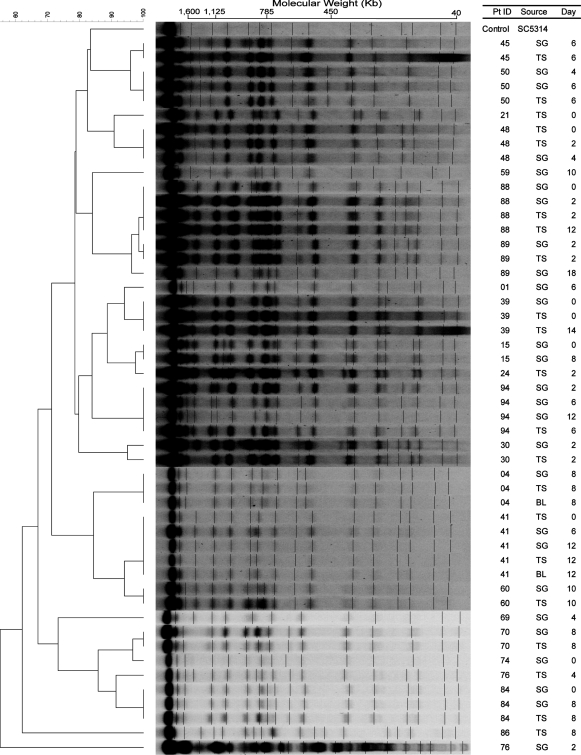
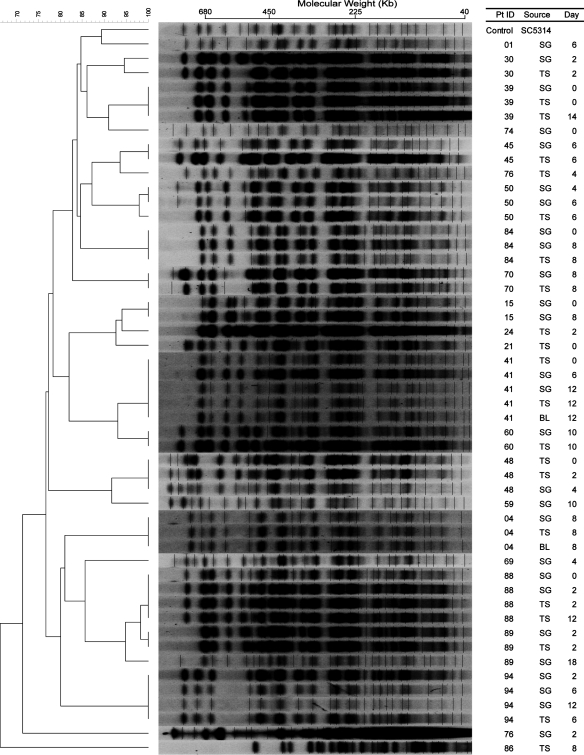
In comparison, restriction endonuclease analysis of the genome (REAG) from the 49 isolates of 22 patients yielded 24 patterns after digestion with SfiI (Fig. 2) and 25 patterns after digestion with BssHII (Fig. 3). The similarity percentage ranged from 62 to 100% for REAG-S and 66 to 100% for REAG-B. The enzyme SfiI cuts in all Major Repeat Sequences (MRS) and the 13 base pair restriction site (5′-GGCCNNNNNGGCC-3). The enzyme BssHII cuts DNA in the six base pair restriction site (5′-GCGCGC-3′). Digesting an isolate with REAG-B generally generated more bands than REAG-S, thus increasing discriminatory power.
Fig. 2.
Pulsed-field gel electrophoresis (PFGE) patterns of SfiI restriction endonuclease analysis of genomic DNA (REAG-S) with dendrogram for Candida albicans isolates. A genetic similarity percentage is shown above the dendrogram. Patient identification (Pt ID), sample site (Source), and number of days after admission to the intensive care unit that the strain was isolated (Day) are included along each PFGE lane. Saccharomyces cerevisiae DNA concatemers and λ DNA ladder were used as size markers. Sizes are measured in kilobases (Kb). C. albicans strain SC5314 (ATCC MYA-2876) was used as the control strain. Abbreviations: SG, supragingival dental plaque; TS, tracheal secretion; BL, bronchoalveolar lavage.
Fig. 3.
Pulsed-field gel electrophoresis (PFGE) patterns of BssHII restriction endonuclease analysis of genomic DNA (REAG-B) with dendrogram for Candida albicans isolates. A genetic similarity percentage is shown above the dendrogram. Patient identification (Pt ID), sample site (Source), and number of days after admission to the intensive care unit that the strain was isolated (Day) are included along each PFGE lane. Saccharomyces cerevisiae DNA concatemers and λ DNA ladder were used as size markers. Sizes are measured in kilobases (Kb). C. albicans strain SC5314 (ATCC MYA-2876) was used as the control strain. Abbreviations: SG, supragingival dental plaque; TS, tracheal secretion; BL, bronchoalveolar lavage.
Intra-patient PFGE analysis
By the EK method (Fig. 1), concurrent or sequential C. albicans isolates, i.e. those from different sites on the same day and/or different days, were identical in 13 of 14 (92.9%). For patient 89, the similarity percentage was 96%, indicating that though the isolates may not be a perfect match they are considered genetically related. Similar results were obtained by the REAG-S method (Fig. 2), i.e. isolates from 12 patients were identical and the isolates from patients 30 and 89 though not identical were genetically related. However, more diversity was observed using the REAG-B method (Fig.3). Multiple concurrent or sequential isolates from 11 of 14 (78.6%) individual patients had identical REAG-B band patterns. Nonetheless, the similarity percentages of the remaining isolates were >95%, and thus considered to be genetically related. These data reveal that intra-patient isolates were clonal in all 14 patients. Additionally, nine patients with C. albicans isolates recovered on multiple days retained the same genotype.
Inter-patient PFGE analysis
Strains from most patients separated into different EK types as expected, but the same three EK patterns were also displayed in multiple patients. Strains from patients 1, 15, 21, and 24 showed the same EK pattern and were grouped into one type. Patients 41, 70, and 76 (TS only) harbored strains with the same EK pattern and were grouped into another type. Strains from patients 88 and 89 were grouped into another EK type.
However, further examination of the strains with REAG-S and REAG-B indicated that these groupings were only partially accurate. EK was correct to group the strains from patients 15 and 24 together, as well as strains from patients 88 and 89. The high percentage of similarity values of both REAG-S and REAG-B for these strains supported these groupings (Table 1). A plausible explanation for these two instances of different patients having a high percentage of similarity values would be patient-to-patient transmission or environment-to-patient contamination. Potentially epidemiologically related patients 15 and 24 were in the ICU within the same month and there was an overlap in the ICU stay for patients 88 and 89.
Table 1.
Inter-patient comparisons
| Methoda | EKb (%) | REAG-Sb (%) | REAG-Bb (%) |
|---|---|---|---|
| Patients 1 vs. 21 | 100 | 79 | 78 |
| Patients 1 vs.15 | 100 | 89 | 78 |
| Patients 15 vs. 21 | 100 | 79 | 93 |
| Patients 15 vs. 24 | 100 | 98 | 94 |
| Patients 41 vs. 70 | 100 | 67 | 78 |
| Patients 41 vs. 76 | 100 | 67 | 78 |
| Patient 70 vs. 76 | 100 | 86 | 83 |
| Patients 88 vs. 89 (day 2) | 100 | 99 | 98 |
| Patients 88 vs. 89 (day 18) | 96 | 96 | 95 |
Electrophorectic karyotype (EK); restriction endonuclease analysis of the genome using SfiI (REAG-S); restriction endonuclease analysis of the genome using BssHII (REAG-B).
Genetically identical (similarity value of 100%), genetically similar (similarity value of >¯95% and less than 100%), genetically different (similarity value <95%).
On the other hand, strains of patients 1, 15, and 21 should not have been grouped together, as well as those from patients 41, 70, and 76 (TS) (Table 1). These incorrect groupings occurred due to the low number of bands (6 to 10) resolved by the EK method in this study. The discriminatory ability of the EK method is insufficient when PFGE resolves less than 10 distinct bands (40). REAG-S and REAG-B methods were able to discriminate the differences in these strains, as the average number of bands resolved by the two methods was 17 and 26, respectively. The discriminatory abilities of REAG-S and REAG-B supported our finding that most strains from different patients separated into different groups, except for the two instances where possible patient-to-patient transmission or environment-to-patient contamination are suspected.
Discussion
C. albicans, a commensal organism, can also act as an opportunistic pathogen in high-risk individuals such as the ICU patient. In the present study, C. albicans colonization was examined in 22 of 96 mechanically ventilated ICU patients. This yeast was harbored in the mouth of 19 patients, in the trachea of 17 patients, and in the lung of 2 patients. In the present and previous studies (35), the genotype of most isolates was clonal and patient-specific. Among the 22 patients examined in this study, 12 patients harbored the yeast in two sites, while 2 patients had the yeast in all three sites. Of the 14 patients analyzed with isolates from two or more sites, 12 patients had genetically identical isolates and one patient had closely related isolates.
The endotracheal tube in mechanically ventilated ICU patients impairs local defense mechanisms and facilitates the passage of C. albicans from the mouth or stomach into the lower airway. As a normal constituent of the normal oral flora in many healthy humans, C. albicans may be commonly found in the mouth and the trachea of patients. But the low frequency of this species in the lung (2/20 BL or 10% patients in this study) indicates that host defense mechanisms limit its colonization of the lower airway. The artificial surface of the catheter bypasses anatomic barriers and facilitates the transport of the pathogen from the oral cavity to the trachea and to the lungs. A previous study (12) found that the endotracheal tube can serve as a reservoir for pathogens in VAP patients and can facilitate the transport of pathogens to the lungs. Concordant with these findings, in patients 4 and 41, the bacterial strains co-cultured with the C. albicans from the bronchoalveolar lavage were also present in the mouth and tracheal secretions. While we cannot rule out the stomach as the source of the C. albicans isolates, based on our molecular epidemiology data, the tracheo-bronchial isolates of the same genotype are also found in the mouth. This is finding concurs with several previous investigations of microbial colonization patterns in mechanically ventilated patients (41–43).
Patient isolates recovered from different sites and/or from multiple days usually shared the same clonal ancestry, yet may not be perfect matches. In this study, in 3 of 22 patients (13.6%) minor differences in the band patterns were observed, despite the same apparent clonal origin of the strains. These differences can be attributed to microevolution (44, 45). This low rate of microevolution may be ascribed to the duration of colonization and the genetic stability of the strain in the patient's environment. In a previous study from Shin and colleagues (44), a similar low rate of microevolution of C. albicans in ICU patients was described, which was attributed to insufficient time for strains to diversify. However, a more discriminatory method such as MLST may be necessary to better clarify microevolution of individual isolates, which is beyond the scope of this paper.
Comparison of the strains from different patients indicates that there may be a possibility of acquisition of the organism from the environment after admission to the hospital. As in previous studies (34, 46), PFGE typing methods may be used to detect nosocomial transmission. In the current study, most strains were patient-specific, indicating an endogenous origin of the strains. But in some cases, strains may have been transmitted exogenously through patient-to-patient transmission or environment-to-patient contamination. Further study using a more discriminatory molecular typing method, along with environmental samples, may be required to validate exogenous transmission.
Among the three PFGE typing methods, electrophoretic karyotyping (EK) was the least discriminating method. Strains from different individuals appear to share identical EK profiles, which upon examination using other methods revealed otherwise. On the other hand, REAG-B proved to be the most discriminatory method, as in other studies using PFGE (35, 46). Despite the most discriminatory nature of this method, REAG-B proved to be the most time consuming in underscoring the bands. Other alternative methods, such as MLST have been shown to be more discriminatory for C. albicans, but not other Candida species (47–49). However, we have shown that PFGE could easily demonstrate inter-patient strain diversity.
In summary, these data indicate that dental plaque can be an important source of C. albicans and that mechanical ventilation may facilitate C. albicans colonization of the lower airway. While there is the possibility of nosocomial transmission in some cases, most oral and airway isolates are genetically indistinguishable within a patient and are retained over time.
Acknowledgements
We thank Dr. Daniel Amsterdam of the Department of Laboratory Medicine, Erie County Medical Center Healthcare Network (Buffalo, NY) and his staff for isolation and identification of yeast strains; Drs. Timothy F. Murphy and Alan J. Lesse of the Division of Infectious Diseases, Veterans Affairs Western New York Healthcare System (Buffalo, NY), for their technical and bionumeric support; and study nurses Angela Vacanti and Noreen Frawley, for enrolling patients and sample collection. This work was supported by the National Institute of Dental and Craniofacial Research grant DE014685. S.M. Heo and R.S. Sung contributed equally to this work.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
References
- 1.Vazquez JA, Beckley A, Sobel JD, Zervos MJ. Comparison of restriction enzyme analysis and pulsed-field gradient gel electrophoresis as typing systems for Candida albicans. J Clin Microbiol. 1991;29:962–7. doi: 10.1128/jcm.29.5.962-967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hellstein J, Vawter-Hugart H, Fotos P, Schmid J, Soll DR. Genetic similarity and phenotypic diversity of commensal and pathogenic strains of Candida albicans isolated from the oral cavity. J Clin Microbiol. 1993;31:3190–9. doi: 10.1128/jcm.31.12.3190-3199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutler JE. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 4.McCullough MJ, Ross BC, Dwyer BD, Reade PC. Genotype and phenotype of oral Candida albicans from patients infected with the human immunodeficiency virus. Microbiology. 1994;140:1195–202. doi: 10.1099/13500872-140-5-1195. [DOI] [PubMed] [Google Scholar]
- 5.Millon L, Manteaux A, Reboux G, Drobacheff C, Monod M, Barale T, et al. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J Clin Microbiol. 1994;32:1115–8. doi: 10.1128/jcm.32.4.1115-1118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, Rhine-Chalberg J, Redding SW, Smith J, Farinacci G, Fothergill AW, et al. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J Clin Microbiol. 1994;32:59–64. doi: 10.1128/jcm.32.1.59-64.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doi M, Homma M, Iwaguchi S, Horibe K, Tanaka K. Strain relatedness of Candida albicans strains isolated from children with leukemia and their bedside parents. J Clin Microbiol. 1994;32:2253–9. doi: 10.1128/jcm.32.9.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voss A, le Noble JL, Verduyn Lunel FM, Foudraine NA, Meis JF. Candidemia in intensive care unit patients: risk factors for mortality. Infection. 1997;25:8–11. doi: 10.1007/BF02113499. [DOI] [PubMed] [Google Scholar]
- 9.Wright WL, Wenzel RP. Nosocomial Candida. Epidemiology, transmission, and prevention. Infect Dis Clin North Am. 1997;11:411–25. doi: 10.1016/s0891-5520(05)70363-9. [DOI] [PubMed] [Google Scholar]
- 10.Bouza E, Munoz P. Epidemiology of candidemia in intensive care units. Int J Antimicrob Agents. 2008;32:S87–91. doi: 10.1016/S0924-8579(08)70006-2. [DOI] [PubMed] [Google Scholar]
- 11.Adiguzel N, Karakurt Z, Gungor G, Yazicioglu Mocin O, Acarturk E, Sogukpinar O, et al. Mortality rates and risk factors associated with nosocomial Candida infection in a respiratory intensive care unit. Tuberk Toraks. 2010;58:35–43. [PubMed] [Google Scholar]
- 12.Adair CG, Gorman SP, Feron BM, Byers LM, Jones DS, Goldsmith CE, et al. Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med. 1999;25:1072–6. doi: 10.1007/s001340051014. [DOI] [PubMed] [Google Scholar]
- 13.Heo SM, Haase EM, Lesse AJ, Gill SR, Scannapieco FA. Genetic relationships between respiratory pathogens isolated from dental plaque and bronchoalveolar lavage fluid from patients in the intensive care unit undergoing mechanical ventilation. Clin Infect Dis. 2008;47:1562–70. doi: 10.1086/593193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azoulay E, Timsit JF, Tafflet M, de Lassence A, Darmon M, Zahar JR, et al. Candida colonization of the respiratory tract and subsequent pseudomonas ventilator-associated pneumonia. Chest. 2006;129:110–7. doi: 10.1378/chest.129.1.110. [DOI] [PubMed] [Google Scholar]
- 15.El-Ebiary M, Torres A, Fabregas N, de la Bellacasa JP, Gonzalez J, Ramirez J, et al. Significance of the isolation of Candida species from respiratory samples in critically ill, non-neutropenic patients. An immediate postmortem histologic study. Am J Respir Crit Care Med. 1997;156:583–90. doi: 10.1164/ajrccm.156.2.9612023. [DOI] [PubMed] [Google Scholar]
- 16.Rello J, Esandi ME, Diaz E, Mariscal D, Gallego M, Valles J. The role of Candida sp, isolated from bronchoscopic samples in nonneutropenic patients. Chest. 1998;114:146–9. doi: 10.1378/chest.114.1.146. [DOI] [PubMed] [Google Scholar]
- 17.Wood GC, Mueller EW, Croce MA, Boucher BA, Fabian TC. Candida sp. isolated from bronchoalveolar lavage: clinical significance in critically ill trauma patients. Intensive Care Med. 2006;32:599–603. doi: 10.1007/s00134-005-0065-6. [DOI] [PubMed] [Google Scholar]
- 18.Williamson DR, Albert M, Perreault MM, Delisle MS, Muscedere J, Rotstein C, et al. The relationship between Candida species cultured from the respiratory tract and systemic inflammation in critically ill patients with ventilator-associated pneumonia. Can J Anaesth. 2011;58:275–84. doi: 10.1007/s12630-010-9439-5. [DOI] [PubMed] [Google Scholar]
- 19.Delisle MS, Williamson DR, Perreault MM, Albert M, Jiang X, Heyland DK. The clinical significance of Candida colonization of respiratory tract secretions in critically ill patients. J Crit Care. 2008;23:11–17. doi: 10.1016/j.jcrc.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Olaechea PM, Palomar M, Leon-Gil C, Alvarez-Lerma F, Jorda R, Nolla-Salas J, et al. Economic impact of Candida colonization and Candida infection in the critically ill patient. Eur J Clin Microbiol Infect Dis. 2004;23:323–30. doi: 10.1007/s10096-004-1104-x. [DOI] [PubMed] [Google Scholar]
- 21.Ader F, Faure K, Guery B, Nseir S. Pseudomonas aeruginosa and Candida albicans interaction in the respiratory tract: from pathophysiology to a therapeutic perspective. Pathol Biol (Paris) 2008;56:164–9. doi: 10.1016/j.patbio.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–32. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 23.Nseir S, Jozefowicz E, Cavestri B, Sendid B, Di Pompeo C, Dewavrin F, et al. Impact of antifungal treatment on Candida-Pseudomonas interaction: a preliminary retrospective case-control study. Intensive Care Med. 2007;33:137–42. doi: 10.1007/s00134-006-0422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meersseman W, Lagrou K, Spriet I, Maertens J, Verbeken E, Peetermans WE, et al. Significance of the isolation of Candida species from airway samples in critically ill patients: a prospective, autopsy study. Intensive Care Med. 2009;35:1526–31. doi: 10.1007/s00134-009-1482-8. [DOI] [PubMed] [Google Scholar]
- 25.Masur H, Rosen PP, Armstrong D. Pulmonary disease caused by Candida species. Am J Med. 1977;63:914–25. doi: 10.1016/0002-9343(77)90546-0. [DOI] [PubMed] [Google Scholar]
- 26.Leung WK, Dassanayake RS, Yau JY, Jin LJ, Yam WC, Samaranayake LP. Oral colonization, phenotypic, and genotypic profiles of Candida species in irradiated, dentate, xerostomic nasopharyngeal carcinoma survivors. J Clin Microbiol. 2000;38:2219–26. doi: 10.1128/jcm.38.6.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McEllistrem MC, Pass M, Elliott JA, Whitney CG, Harrison LH. Clonal groups of penicillin-nonsusceptible Streptococcus pneumoniae in Baltimore, Maryland: a population-based, molecular epidemiologic study. J Clin Microbiol. 2000;38:4367–72. doi: 10.1128/jcm.38.12.4367-4372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sangeorzan JA, Zervos MJ, Donabedian S, Kauffman CA. Validity of contour-clamped homogeneous electric field electrophoresis as a typing system for Candida albicans. Mycoses. 1995;38:29–36. doi: 10.1111/j.1439-0507.1995.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 29.Carruba G, Pontieri E, De Bernardis F, Martino P, Cassone A. DNA fingerprinting and electrophoretic karyotype of environmental and clinical isolates of Candida parapsilosis. J Clin Microbiol. 1991;29:916–22. doi: 10.1128/jcm.29.5.916-922.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doebbeling BN, Lehmann PF, Hollis RJ, Wu LC, Widmer AF, Voss A, et al. Comparison of pulsed-field gel electrophoresis with isoenzyme profiles as a typing system for Candida tropicalis. Clin Infect Dis. 1993;16:377–83. doi: 10.1093/clind/16.3.377. [DOI] [PubMed] [Google Scholar]
- 31.Essayag SM, Baily GG, Denning DW, Burnie JP. Karyotyping of fluconazole-resistant yeasts with phenotype reported as Candida krusei or Candida inconspicua. Int J Syst Bacteriol. 1996;46:35–40. doi: 10.1099/00207713-46-1-35. [DOI] [PubMed] [Google Scholar]
- 32.Dib JC, Dube M, Kelly C, Rinaldi MG, Patterson JE. Evaluation of pulsed-field gel electrophoresis as a typing system for Candida rugosa: comparison of karyotype and restriction fragment length polymorphisms. J Clin Microbiol. 1996;34:1494–6. doi: 10.1128/jcm.34.6.1494-1496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 34.Shin JH, Og YG, Cho D, Kee SJ, Shin MG, Suh SP, et al. Molecular epidemiological analysis of bloodstream isolates of Candida albicans from a university hospital over a five-year period. J Microbiol. 2005;43:546–54. [PubMed] [Google Scholar]
- 35.Chen KW, Lo HJ, Lin YH, Li SY. Comparison of four molecular typing methods to assess genetic relatedness of Candida albicans clinical isolates in Taiwan. J Med Microbiol. 2005;54:249–58. doi: 10.1099/jmm.0.45829-0. [DOI] [PubMed] [Google Scholar]
- 36.Scannapieco FA, Yu J, Raghavendran K, Vacanti A, Owens SI, Wood K, et al. A randomized trial of chlorhexidine gluconate on oral bacterial pathogens in mechanically ventilated patients. Crit Care. 2009;13:R117. doi: 10.1186/cc7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818–29. [PubMed] [Google Scholar]
- 38.Maringele L, Lydall D. Pulsed-field gel electrophoresis of budding yeast chromosomes. Methods Mol Biol. 2006;313:65–73. doi: 10.1385/1-59259-958-3:065. [DOI] [PubMed] [Google Scholar]
- 39.Voss A, Hollis RJ, Pfaller MA, Wenzel RP, Doebbeling BN. Investigation of the sequence of colonization and candidemia in nonneutropenic patients. J Clin Microbiol. 1994;32:975–80. doi: 10.1128/jcm.32.4.975-980.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonten MJ, Gaillard CA, van Tiel FH, Smeets HG, van der Geest S, Stobberingh EE. The stomach is not a source for colonization of the upper respiratory tract and pneumonia in ICU patients. Chest. 1994;105:878–84. doi: 10.1378/chest.105.3.878. [DOI] [PubMed] [Google Scholar]
- 42.Ewig S, Torres A, El-Ebiary M, Fabregas N, Hernandez C, Gonzalez J, et al. Bacterial colonization patterns in mechanically ventilated patients with traumatic and medical head injury. Incidence, risk factors, and association with ventilator-associated pneumonia. Am J Respir Crit Care Med. 1999;159:188–98. doi: 10.1164/ajrccm.159.1.9803097. [DOI] [PubMed] [Google Scholar]
- 43.George DL, Falk PS, Wunderink RG, Leeper KV, Jr, Meduri GU, Steere EL, et al. Epidemiology of ventilator-acquired pneumonia based on protected bronchoscopic sampling. Am J Respir Crit Care Med. 1998;158:1839–47. doi: 10.1164/ajrccm.158.6.9610069. [DOI] [PubMed] [Google Scholar]
- 44.Shin JH, Park MR, Song JW, Shin DH, Jung SI, Cho D, et al. Microevolution of Candida albicans strains during catheter-related candidemia. J Clin Microbiol. 2004;42:4025–31. doi: 10.1128/JCM.42.9.4025-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lockhart SR, Fritch JJ, Meier AS, Schroppel K, Srikantha T, Galask R, et al. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol. 1995;33:1501–9. doi: 10.1128/jcm.33.6.1501-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voss A, Pfaller MA, Hollis RJ, Rhine-Chalberg J, Doebbeling BN. Investigation of Candida albicans transmission in a surgical intensive care unit cluster by using genomic DNA typing methods. J Clin Microbiol. 1995;33:576–80. doi: 10.1128/jcm.33.3.576-580.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen KW, Chen YC, Lin YH, Chou HH, Li SY. The molecular epidemiology of serial Candida tropicalis isolates from ICU patients as revealed by multilocus sequence typing and pulsed-field gel electrophoresis. Infect Genet Evol. 2009;9:912–20. doi: 10.1016/j.meegid.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Chen KW, Chen YC, Lo HJ, Odds FC, Wang TH, Lin CY, et al. Multilocus sequence typing for analyses of clonality of Candida albicans strains in Taiwan. J Clin Microbiol. 2006;44:2172–8. doi: 10.1128/JCM.00320-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin CY, Chen YC, Lo HJ, Chen KW, Li SY. Assessment of Candida glabrata strain relatedness by pulsed-field gel electrophoresis and multilocus sequence typing. J Clin Microbiol. 2007;45:2452–9. doi: 10.1128/JCM.00699-07. [DOI] [PMC free article] [PubMed] [Google Scholar]