Abstract
First discovered in Drosophila, the Hippo signaling pathway is a conserved regulator of organ size. Central to this pathway is a kinase cascade leading from the tumor suppressor Hippo (Mst1 and Mst2 in mammals) to the oncoprotein Yki (YAP and TAZ in mammals), a transcriptional coactivator of target genes involved in cell proliferation and survival. Here, I review recent progress in elucidating the molecular mechanism and physiological function of Hippo signaling in Drosophila and mammals. These studies suggest that the core Hippo kinase cascade integrates multiple upstream inputs, enabling dynamic regulation of tissue homeostasis in animal development and physiology.
Introduction
The control of organ size is a long-standing puzzle in developmental biology. Classic embryological studies suggest that many organs possess intrinsic information about their final size. For example, when two-thirds of a mouse liver is surgically removed, the remaining one-third regenerates its original mass within 7–10 days and then ceases growth (reviewed in Michalopoulos and DeFrances, 1997). Similarly, when imaginal discs from newly hatched larvae are transplanted into adult flies, they grow to a final size characteristic of that seen in situ (Bryant and Simpson, 1984). The molecular mechanisms that stop organ growth at the appropriate point during development or regeneration remain poorly understood today.
The discovery of the Hippo signaling pathway provides an important entry point to addressing these long-standing questions. The first four components of the Hippo pathway, including the NDR family protein kinase Warts (Wts) (Justice et al., 1995; Xu et al., 1995), the WW domain-containing protein Salvador (Sav) (Tapon et al., 2002; Kango-Singh et al., 2002), the Ste20-like protein kinase Hippo (Hpo) (Wu et al., 2003; Udan et al., 2003; Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003) and the adaptor protein Mob-as-tumor-suppressor (Mats) (Lai et al., 2005), were discovered in genetic screens in Drosophila for tumor suppressor genes. Loss-of-function mutant clones for any of these four genes lead to a strong tissue overgrowth phenotype characterized by increased proliferation and diminished cell death. Biochemically, these four tumor suppressors form a kinase cascade in which the Hpo-Sav kinase complex phosphorylates and activates the Wts-Mats kinase complex (Wu et al., 2003; Wei et al., 2007). The prime target of this kinase cascade in growth regulation is transcriptional coactivator Yorkie (Yki), which was isolated as a Hippo pathway component in a yeast two-hybrid screen for Wts-binding proteins (Huang et al., 2005). Yki functions as an oncogene and its overexpression phenocopies loss of Hippo signaling. Genetic analysis placed yki downstream of hpo, sav or wts, and biochemical studies demonstrated that Wts directly phosphorylates and inactivates Yki in a Hpo-regulated manner (Huang et al., 2005). Thus, from these pioneering studies, a kinase cascade leading from Hpo to Yki phosphorylation emerged (reviewed in Edgar, 2006; Pan, 2007).
The elucidation of the Hippo kinase cascade in Drosophila has stimulated intense research into the molecular mechanism and the physiological function of this emerging pathway in both flies and vertebrates. While still a relatively young field, research on Hippo signaling is escalating rapidly. Here, I review current understanding of the Hippo signaling network in Drosophila and mammals, its diverse roles in multiple physiological contexts, and its involvement in cancer development, highlighting important progress in the past 3 years.
The Hippo Signaling Network in Drosophila
Following the discovery of the four tumor suppressors that constitute the core kinase cassette, candidate gene-based approaches and forward genetic screens have implicated at least seven additional tumor suppressors whose activities converge on Hpo and/or Wts. These include the FERM domain proteins Merlin (Mer) and Expanded (Ex), the protocadherins Fat (Ft) and Dachsous (Ds), the CK1 family kinase Disc overgrown (Dco), the WW and C2 domain-containing protein Kibra, and the apical transmembrane protein Crumbs (Crb) (Table 1). While the mechanisms by which these upstream regulators converge on the Hippo kinase cascade are complex and in some instances unresolved, a notable feature is that mutations in each of these genes lead to a relatively mild overgrowth phenotype, suggesting that these upstream components function in a combinatorial or additive manner to regulate the Hippo kinase cassette.
Table 1.
Hippo Pathway Components in Drosophila and Human
| Drosophila | Human | Conserved Domains and Motifs |
|---|---|---|
| The Core Pathway | ||
| Hippo | Mst1-2 | Ste20 Ser/Thr kinase and SARAH domains |
| Salvador | Sav1/WW45 | WW and SARAH domain |
| Warts | Lats1-2 | NDR Ser/Thr kinase domain, PPXY motif |
| Mats | MOBKL1A-B | Mob1/phocein domain |
| Yorkie | YAP, TAZ | WW and TEAD-binding domains |
| Scalloped | TEAD1-4 | TEA/ATTS and Yki/YAP- binding domains |
| Upstream Apical Complex | ||
| Kibra | KIBRA/WWC1,WWC2 | WW and C2 domains |
| Expanded | FRMD6/Ex1,FRMD1/Ex2 | FERM domain |
| Merlin | NF2/Merlin | FERM domain |
| Fat Effectors and Regulators | ||
| Fat | Fat4/Fat-j | EGF-like, Laminin G and Cadherin repeat domains |
| Dachsous | Dchs1-2 | Cadherin repeat domain |
| Four-jointed | Fjx1 | Golgi Ser/Thr kinase |
| Dachs | ? | Myosin motor domain |
| Discs overgrown | CK1δ, CK1ε | Ser/Thr kinase domain |
| Approximated | ZDHHC14 | DHHC zinc finger domain |
| Lowfat | Lix1, Lix1L | unknown conserved domain |
| Apical-Basal Polarity | ||
| Crumbs | Crb1-3 | EGF-like and Laminin G domains, PDZ- and FERM-binding motifs |
| Lgl | Lgl1-2 | LLGL2 domain |
| aPKC | aPKCλ, aPKCζ | PKC kinase, PB1 and C1 domains |
| Other Modulators | ||
| dRASSF | RASSF1-6 | Ras association and SARAH domains |
| dSTRIPAK PP2A | STRIPAK PP2A | PP2A Ser/Thr phosphatase complex |
| dJuba | Ajuba, LIMD1, WTIP | LIM domain |
An Apical Protein Complex Containing Kibra, Ex, and Merlin
The initial discovery of the Hippo kinase cascade posed a major question: how does this intracellular signaling module respond to extracellular cues? The first advance in addressing this question was provided by Hamaratoglu et al. (2006), who implicated Merlin (Mer) and Expanded (Ex)—two members of the “4.1, Ezrin, Radixin, Moesin” (FERM) domain-containing family of proteins—as potential upstream regulators of Hpo. Mer and Ex bind to each other and colocalize to the apical domain of epithelial cells (McCartney et al., 2000). Genetically, mer and ex function in a partially redundant manner to regulate the Hippo pathway, as double mutant clones display a much stronger overgrowth phenotype than mutants of either gene (Hamaratoglu et al., 2006). Consistent with the genetic analysis, overexpression of Mer and Ex was shown to promote Wts phosphorylation in a cooperative manner. Given the general role of FERM proteins as adaptors that bridge transmembrane proteins to the cytoskeleton, Mer and Ex are uniquely positioned to link the Hippo kinase cascade to potential transmembrane receptors.
Three recent reports identified the WW and C2 domain-containing protein Kibra as another apically localized tumor suppressor that functions together with Mer and Ex to stimulate Hippo signaling (Yu et al., 2010; Baumgartner et al., 2010; Genevet et al., 2010). Kibra, Ex and Mer cooperatively promote Wts phosphorylation, and accordingly, double mutant combinations among the three genes revealed stronger Hippo signaling defects than any single mutants. The Kibra-Ex-Mer protein complex physically associates with the Hpo-Sav complex and is required for membrane association of Hpo (Yu et al., 2010). Interaction between Kibra and Wts was also reported (Genevet et al., 2010). Thus, the Kibra-Ex-Mer complex appears to interact with the Hippo kinase cascade through multiple protein-protein interactions, and these physical interactions are likely critical for bringing the Hippo kinase cassette to plasma membrane for activation. Indeed, a recent study provided direct evidence demonstrating that the tumor suppressor Mats is activated at the cell membrane (Ho et al., 2010). Interestingly, the expression of these upstream apical regulators is negatively regulated by Hippo signaling, as hpo, sav, or wts mutant clones have been reported to show increased transcript levels of kibra and ex, as well as elevated protein levels of Kibra, Ex, and Mer (Genevet et al., 2010; Hamaratoglu et al., 2006).
Unlike protein complexes, such as TSC1-TSC2, in which each component is obligatory to the function of the whole complex, components of the Kibra-Ex-Mer complex appear to function in a partially redundant manner to regulate Hippo signaling. Furthermore, the extent to which each component contributes to Hippo pathway regulation varies according to the tissue type. For example, diap1 regulation is primarily dependent on ex in imaginal discs (Pellock et al., 2006), while mer and kibra are more strongly required in the developing egg chamber for the proper maturation of posterior follicle cells and anterior-posterior polarity of the underlying oocyte (Yu et al., 2008, 2010; Polesello and Tapon, 2007; Meignin et al., 2007). Along the same line, it was suggested that Ex is preferentially required for Hippo signaling in the larval eye whereas Mer is preferentially required in the pupal eye (Milton et al., 2010). Thus, the Kibra-Ex-Mer complex may function as a dynamic integrator of upstream signals in a temporally and spatially regulated manner.
Transmembrane Protein Fat: Regulation and Function
The atypical cadherin Fat (Ft) was the first transmembrane protein shown to impact Hippo signaling (Bennett and Harvey, 2006; Silva et al., 2006; Willecke et al., 2006; Cho et al., 2006; Tyler and Baker, 2007). Ft is constitutively cleaved by an unknown protease(s); the N-terminal 450 kDa extracellular domain forms a stable heterodimer with the C-terminal 110 kDa transmembrane fragment (Sopko et al., 2009; Feng and Irvine, 2009). Besides its role as a growth regulator, Ft also affects planar cell polarity (PCP) (Yang et al., 2002; Casal et al., 2006), a process by which cells in the plane of an epithelium orient themselves along an axis orthogonal to the apical-basal axis, as seen in the coordinated orientation of wing hairs in Drosophila or sensory hair cells in the mouse inner ear. So far, two distinct mechanisms have been proposed to link Ft to Hippo signaling. In one model, it was proposed that Ft, Ex, and Hpo function in a linear pathway whereby Ft is required for apical membrane localization and/or stability of Ex (Bennett and Harvey, 2006; Silva et al., 2006; Willecke et al., 2006). In an alternative model, it was proposed that Ft and Ex function in parallel with each other, with Ft and Ex influencing the protein level and the phosphorylation of Wts protein, respectively (Cho et al., 2006; Feng and Irvine, 2007). Further studies are required to resolve these models, and the possibility exists that Ft may regulate Hippo signaling through both Ex-dependent and -independent mechanisms.
The key downstream mediator of Ft action on the Hippo pathway is the unconventional myosin Dachs, which functions antagonistically to Ft and upstream of Wts (Cho et al., 2006). Dachs associates with Wts when overexpressed in Drosophila cell cultures and may function by promoting Wts proteolysis. Interestingly, Ft activity regulates preferential accumulation of the Dachs protein on the distal side of the wing imaginal disc epithelial cells (Rogulja et al., 2008). Another protein that impacts Dachs activity is the palmitoyltransferase Approximated (App), which antagonizes Ft signaling by regulating the levels and subcellular localization of Dachs (Matakatsu and Blair, 2008). Whether this function is mediated by palmitoylation of Dachs or an unknown regulator of Dachs remains to be determined.
Ft functions by binding to another atypical cadherin called Dachsous (Ds) (Matakatsu and Blair, 2006), a process that is modulated by Four-jointed (Fj), a Golgi-resident kinase that phosphorylates the cadherin domains of Ft and Ds as they transit through the Golgi network (Ishikawa et al., 2008). Interestingly, Fj and Ds are expressed in opposing gradients in many developing tissues, and their expression, in turn, is regulated by morphogens such as Decapentaplegic (Dpp, a BMP), Wingless (a Wnt), and Hedgehog (Yang et al., 2002; Casal et al., 2006; Rogulja et al., 2008), providing a potential mechanism to coordinate tissue size (growth) with tissue shape (patterning). Fj-mediated phosphorylation enhances the ability of Ft to bind Ds while inhibiting the ability of Ds to bind Ft, which suggests a molecular mechanism for how the graded expression of Fj across many Drosophila tissues can be translated to polarized Ft activity within every cell as reflected by the polarized localization of Dachs (Simon et al., 2010; Brittle et al., 2010). Another modulator of Ft is the casein kinase Discs overgrown (Dco), which can phosphorylate the intracellular domain of Ft in a Ds-regulated manner (Sopko et al., 2009; Feng and Irvine, 2009). A recently identified regulator of Ft is Lowfat (Lft), a conserved cytoplasmic protein that binds to the cytoplasmic domains of Ft and Ds and influences the stability of the latter (Mao et al., 2009). Conversely, Ft and Ds control the sub-apical localization of Lft (Mao et al., 2009).
The Ds-Ft system has several unusual characteristics that distinguish it from conventional receptor-ligand interactions. First, Ft signaling is regulated not simply by the amount of Ds, but rather by the steepness of the Ds gradient (Rogulja et al., 2008; Willecke et al., 2008). A steep gradient of Ds inactivates Ft, whereas a shallow gradient activates Ft. Second, Ds is required in both signal-sending cells as well as signal-responding cells, suggesting that Ds has both ligand- and receptor-like activity (Willecke et al., 2008; Casal et al., 2006). Lastly, Ds and Fj have opposite effects on Ft-dependent PCP signaling (and are indeed expressed in opposing gradients in many tissues) (Casal et al., 2002; Strutt and Strutt, 2002; Yang et al., 2002) but have similar effects on Ft-dependent Hippo signaling (Rogulja et al., 2008; Willecke et al., 2008). These contrasting effects can be explained by a model in which PCP is regulated by the directionality of Ft polarization within a cell, whereas Hippo signaling is regulated by the magnitude of the differential polarization when one compares opposite sides of the cell (Rogulja et al., 2008; Willecke et al., 2008).
Transmembrane Protein Crumbs: A Link to Apical-Basal Polarity
Despite a clear influence of Ft on Hippo signaling in imaginal discs, several lines of evidence argue against a simple model that positions Ft as the sole cell-surface receptor for the Hippo pathway. Loss of Ft generally results in a weaker phenotype than mutations of the Hippo kinase cassette. For example, hpo or wts mutant clones in the pupal retina are characterized by a signature “extra interommatidial cell” phenotype with >40 extra cells per ommatidium. In contrast, ft mutant clones contain an average of 3.3 extra cells per ommatidium (Silva et al., 2006). An even greater dichotomy is seen in the developing egg chamber in which ft, but not the core kinase cassette or the Kibra-Ex-Mer complex, is dispensable for posterior follicle cell differentiation and oocyte polarity (Polesello and Tapon, 2007; Meignin et al., 2007; Yu et al., 2008). Thus, depending on the tissue context, the Kibra-Ex-Mer complex is regulated by signals in addition to or instead of Ft. Consistent with this view, ft mer double mutant clones show a much stronger “extra interommatidial cell” phenotype in pupal retina compared to single mutant clones, suggesting that Mer may transduce a signal from an unknown cell surface receptor (Silva et al., 2006; Willecke et al., 2006). Thus, there has been intense interest in identifying additional cell surface proteins that signal to the Hippo pathway.
Crumbs (Crb) is an apical transmembrane protein best known for its role in organizing apical-basal polarity in Drosophila embryos (Tepass et al., 1990). It contains 28 EGF-like and four laminin AG-like repeats in its extracellular domain and a short intracellular domain including a juxtamembrane FERM-binding motif (FBM) and a C-terminal PDZ-binding motif (PBM). Crb forms a protein complex with the apical polarity proteins Stardust (Sdt) and Patj through its PBM (Bachmann et al., 2001; Hong et al., 2001; Bhat et al., 1999), but the molecular function of Crb’s FBM has been less clear. Several recent studies reported that the FBM affects Ex stability and localization (Robinson et al., 2010; Ling et al., 2010; Chen et al., 2010a). Indeed, the FBM of Crb binds to the FERM domain of Ex and is required for Ex apical localization, and double mutant analysis revealed that Crb functions with Ex in a linear pathway, but in parallel with Mer and Kibra (Ling et al., 2010). Conversely, Crb overexpression induces FBM-dependent tissue overgrowth and depletion of apical Ex protein likely from a dominant-negative effect (Robinson et al., 2010; Chen et al., 2010a). Like other upstream regulators of the Hippo pathway such as kibra and ex, transcription of crb is negatively regulated by Hippo signaling, although to a lesser extent (Genevet et al., 2009).
Are there other links between apical-basal polarity and Hippo signaling? Loss of Lgl (a basolateral protein) or activation of aPKC (a component of a different apical protein complex) can both mislocalize Hpo and upregulate selected Hippo pathway target genes (Grzeschik et al., 2010). Further studies are required to fully elucidate which manifestations of polarity impact the Hippo signaling pathway and how this coupling works at a biochemical level.
All Roads Lead to Rome: Regulation of Yki Activity
While the regulation of Hippo signaling by upstream components seems dauntingly complex, these upstream components all converge to and signal through a common downstream growth regulatory effector, the transcriptional coactivator Yki. Indeed, Wts directly phosphorylates Ser168 of Yki, creating a binding site for 14-3-3 proteins, which promote nuclear exclusion and cytoplasmic accumulation of Yki (Dong et al., 2007; Zhao et al., 2007; Oh and Irvine, 2008). Consistent with this model, loss of Hippo signaling (Dong et al., 2007; Oh and Irvine, 2008), knockdown of 14-3-3 (Ren et al., 2010), or mutation of the 14-3-3 binding motif (Zhao et al., 2007) all lead to nuclear accumulation and/or aberrant activation of endogenous Yki. Wts can also influence the activity and nuclear localization of overexpressed Yki via two additional sites (at S111 and S250), albeit to a lesser extent than S168, and independently of 14-3-3 (Oh and Irvine, 2009; Ren et al., 2010). It remains to be determined what contribution and mechanism of action S111 and S250 phosphorylation might have in the context of endogenous Yki activity.
Two recent studies have also shown that direct binding between the WW domains of Yki and the PPxY motifs of Ex, Wts and Hpo may sequester Yki (Badouel et al., 2009; Oh et al., 2009). This repression was suggested to be phosphorylation-independent, based on the observation that overexpressed Ex and/or Wts/Hpo can repress the activity of a Yki transgene that eliminates all three possible Wts phosphorylation sites (Oh et al., 2009). Further study is needed to assess the contribution of this mode of regulation under physiological conditions, e.g., at endogenous levels of Ex/Wts/Hpo and Yki. Because the PPxY motifs implicated in Ex-Yki and Hpo-Yki binding are absent in the mammalian homologs of Hpo and Ex, these mammalian homologs are unlikely to engage a similar phosphorylation-independent mechanism.
Transcriptional Regulation by Yki: Partners and Targets
As a transcriptional coactivator, Yki does not bind to DNA directly but, rather, partners with DNA-binding transcription factors, such as the TEAD/TEF family transcription factor Scalloped (Sd), which mediates Yki-induced tissue overgrowth (Wu et al., 2008; Zhang et al., 2008; Goulev et al., 2008). Sd directly binds to a minimal 26 bp Hippo response element (HRE) that confers Hippo-dependent transcriptional regulation of diap1, a well characterized Hippo target gene (Wu et al., 2008). That said, Sd, but not Yki, is largely dispensable for the normal growth of imaginal discs, suggesting that other DNA-binding transcription factors regulate target gene expression in response to basal levels of Yki activity. Indeed, the minimal diap1 HRE contains non-Sd-binding sequence that is indispensable for activity (Wu et al., 2008). A relevant transcription factor has not yet been identified, but other DNA-binding partners for Yki have been reported recently. In one study, Homothorax (Hth) was reported as a Yki partner that promotes cell survival and proliferation anterior to the morphogenetic furrow in the eye imaginal disc (Peng et al., 2009). In this cellular context, Hth has minimal influence on the expression of diap1 but, instead, regulates the expression of another Yki target, the microRNA bantam. In another study, Smad proteins were reported to interact with Yki/YAP (Alarcón et al., 2009). CDK8/9, components of transcriptional mediator and elongation complexes, phosphorylate the Smad linker region, enabling its binding to the WW domains of Yki. This interaction is believed to potentiate the transcriptional response to BMP/TGF-β signaling, highlighting a possible crosstalk between Hippo and BMP/TGF-β pathways. Because the WW domains are required for endogenous Yki activity (Wu et al., 2008), it will be important to determine whether the basal levels of Yki activity are mediated by Smad or other unknown WW domain-binding transcription factors.
Loss of Hippo signaling (or increased Yki activity) can drive the expression of at least three classes of genes. The first class corresponds to genes with a cell-autonomous role in cell proliferation or survival, such as the cell death inhibitor diap1 (Wu et al., 2003), the microRNA bantam (Thompson and Cohen, 2006; Nolo et al., 2006), and the cell-cycle regulators cyclin E (Tapon et al., 2002) and E2F1 (Goulev et al., 2008). However, Yki targets with direct roles in cell growth have remained elusive. In this issue of Developmental Cell, Johnston and colleagues have considerably filled this gap by identifying dMyc, a potent regulator of ribosome biogenesis and cell growth, as a transcriptional target of Yki. Interestingly, not only does Yki positively regulate the expression of dMyc, dMyc also negatively regulates Yki expression and activity. The negative feedback loop between dMyc and Yki may suggest a homeostatic mechanism that integrates the activity of two key mediators of size control and offers a potential molecular explanation for Myc’s well-documented proapoptotic activity (reviewed in Green and Evan, 2002).
The second class of genes encode upstream regulators of the Hippo pathway, such as Kibra, Ex, Crb, and Fj (Genevet et al., 2010; Hamaratoglu et al., 2006; Genevet et al., 2009; Cho et al., 2006). Feedback regulation of upstream pathway components appears to be a common feature of many signaling pathways, which has been suggested to play important roles in maintaining steady-state levels of signaling strength (reviewed in Stelling et al., 2004).
The third class of genes represent potential means of crosstalk to other modes of cell-cell interaction and cell signaling, such as E-Cadherin (Genevet et al., 2009), Serrate (a Notch ligand) and Wingless (Cho et al., 2006), Vein (an EGFR ligand) (Zhang et al., 2009b), and the heparan sulfate proteoglycans Dally and Dally-like (Baena-Lopez et al., 2008), both of which are cell surface proteins implicated in regulating the extracellular diffusion and signal efficiency of secreted morphogens, including Hedgehog, Dpp, and Wingless. This complex crosstalk between pathways regulating tissue growth and patterning may ensure their coordination during animal development. Of note, with the exception of diap1, Hippo responsive elements (HREs) have not been molecularly defined for the known Hippo target genes. A comprehensive understanding of the transcriptional landscape in response to Hippo signaling will require functional dissection of HREs in multiple Hippo target genes.
Future Directions for the Hippo Signaling Network in Drosophila
It is conspicuous that the kinase cascade leading from Hpo to Yki phosphorylation appears to be invariant and strictly required, while the upstream inputs that influence kinase activity are much more diverse and, indeed, partially redundant (Figure 1). A major question in the field is to identify what features of cell adhesion and polarity are the primary “signals” activating the Hippo pathway and to understand how these signals are interpreted biochemically by the Hippo kinase cascade.
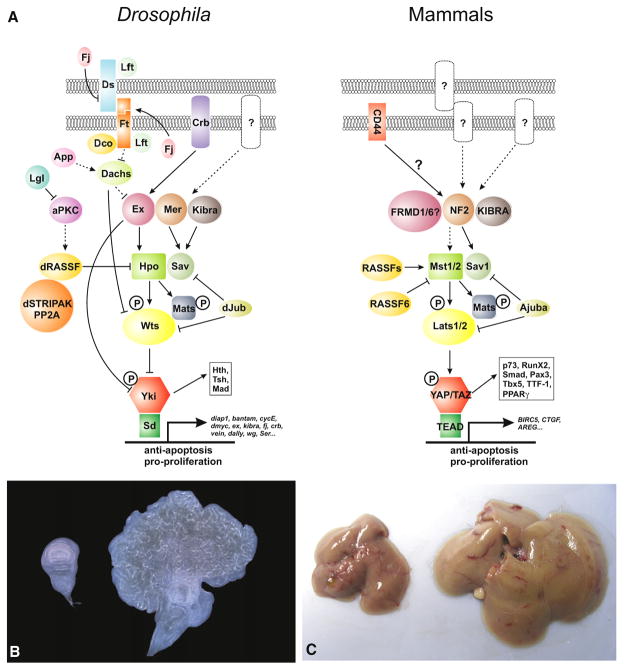
Figure 1. The Hippo Signaling Network in Drosophila and Mammals.
(A) Signaling diagram. Corresponding proteins in Drosophila and mammals are indicated by matching colors and shapes. Direct biochemical interactions are indicated by solid lines or drawn as proteins in direct contact with each other. Dashed lines indicate genetic interactions for which no direct protein-protein interactions have been reported. Arrowed or blunted ends indicate activation or inhibition, respectively. Also shown are selected target genes. Yki- or YAP/TAZ-interacting transcription factors other than Sd (Drosophila) or TEAD (mammals) are collectively listed in a box.
(B) A normal (left) and a yki-overexpressing (right) Drosophila wing imaginal disc. Image adapted from Huang et al. (2005).
(C) A normal (left) and a YAP-overexpressing (right) mouse liver. Image adapted from Dong et al. (2007).
The dramatic increase in organ size induced by Yki/YAP overexpression illustrates the potent growth-regulatory activity of Hippo signaling in Drosophila and mammals.
Given its direct interactions with both upstream receptors and downstream kinase cassette, the apical Kibra-Ex-Mer complex is uniquely positioned as a platform for signal integration. Besides Ft and Crb, which have been linked to this complex via their effect on Ex localization and/or stability, it is conceivable that additional cell-surface proteins may regulate this complex through their influence on Kibra and Mer. While accumulating evidence suggests that the Hippo kinase cascade is activated at the plasma membrane, the biochemical mechanism of kinase activation at the plasma membrane remains to be elucidated. Also unknown is whether the Hippo kinase cassette can be activated in additional subcellular compartments such as the nucleus, where some components of the mammalian Hippo kinase cassette have been implicated in controlling mitotic exit and centrosome duplication (Nishiyama et al., 1999; Bothos et al., 2005; Guo et al., 2007; Hergovich et al., 2009).
Besides positive upstream regulators of Hippo signaling, additional nodes of signal integration may be provided by proteins that negatively modulate the Hippo kinase cascade. Such proteins include the aforementioned myosin-like protein Dachs (Mao et al., 2006), the Drosophila Ras association family protein dRASSF (Polesello et al., 2006), and the LIM family protein Ajuba (Das Thakur et al., 2010). Like its mammalian counterparts, dRASSF directly binds to Hpo, an interaction that is mediated by the C-terminal SARAH domain of each protein (Polesello et al., 2006). However, unlike its tumor suppressor counterparts in mammals (see below), dRASSF functions as a positive regulator of growth by competing with Sav for binding to Hpo (Polesello et al., 2006) and recruiting a Hpo-inactivating PP2A complex called dSTRIPAK (Ribeiro et al., 2010). The Ajuba proteins inhibit Hippo signaling through physical interactions with Wts and Sav (Das Thakur et al., 2010). While the precise mechanism by which the Ajuba proteins influence the Hippo kinase cascade remains to be determined, it is notable that the Ajuba proteins are components of the adherens junctions (Marie et al., 2003) and, thus, are ideally positioned to link the apical upstream regulators to the downstream Hippo kinase cassette. Overall, the Hippo kinase cascade appears to be regulated by myriad upstream inputs, many of which are yet to be identified.
The Hippo Signaling Network in Mammals: Conservation, Complexity, and Divergence
The discovery of the founding members of the Hippo signaling pathway in Drosophila has provided a paradigm for investigating Hippo signaling in mammals. While recent molecular and genetic studies have experimentally validated some conserved aspects of Hippo signaling in mammals, such as the composition of the core kinase cascade and its role in organ size control, these studies also revealed additional complexity and, in some cases, divergence from Drosophila (Figure 1).
The Hippo Kinase Cascade in Mammals and Divergent Mechanisms of YAP/TAZ Inactivation
Given its central position in Hippo signaling, earlier studies in mammals have focused on the core kinase cascade. The mammalian genome contains two Hpo homologs (Mst1 and Mst2), one Sav homolog (WW45 or Sav1), two Wts homologs (Lats1 and Lats2), and two Mats homologs (MOBKL1A and MOBKL1B, often collectively referred to as Mob1). These proteins form a conserved kinase cassette (Callus et al., 2006; Praskova et al., 2008; Chan et al., 2005) that phosphorylates and inactivates the mammalian Yki homologs YAP (Dong et al., 2007; Zhao et al., 2007; Lei et al., 2008) and TAZ (Lei et al., 2008) on multiple HxRxxS motifs in response to cell density, of which only one (YAP S127 and TAZ S89, like Yki S168) serves as a 14-3-3-binding site and plays the most critical role in regulating nuclear-cytoplasmic translocation. This cell density-dependent activation of Hippo signaling is required for contact inhibition of cultured mammalian cells (Zhao et al., 2007). Thus, a highly analogous Hippo kinase cascade appears to exist in mammalian cells.
The physiological relevance of the conserved Hippo kinase cascade was supported by studies of transgenic and knockout mice. Transgenic overexpression of YAP (Dong et al., 2007; Camargo et al., 2007) or liver-specific knockout of Mst1/2 or Sav1 (Zhou et al., 2009; Lu et al., 2010; Song et al., 2010; Lee et al., 2010) each expanded the liver size and ultimately induced hepatocellular carcinoma (HCC), revealing a conserved role for the Hippo pathway in regulating organ size in mammals. Examination of Lats1/2 and YAP phosphorylation in Mst1/2- or Sav1-deficient livers, however, has produced inconsistent results concerning the specific requirement for Mst1/2 in Lats1/2 phosphorylation (Zhou et al., 2009; Lu et al., 2010), as well as the requirement for Sav1 in YAP phosphorylation (Lu et al., 2010; Lee et al., 2010). There also appear to be tissue-specific requirements for pathway components, as in contrast to the murine liver, Mst1/2 are not required for YAP phosphorylation in immortalized mouse embryonic fibroblasts (Zhou et al., 2009). Such complexity may suggest the existence of non-canonical components that replace certain canonical components of the Hippo kinase cascade. Along this line, it was suggested that an Mst1/2-regulated kinase distinct from Lats1/2 may be responsible for YAP phosphorylation in the murine liver (Zhou et al., 2009). These recent findings highlight the complexity and possible tissue-specific organization of the Hippo kinase cascade in mammals.
Another unanticipated aspect of the mammalian Hippo kinase cascade concerns the mechanism of YAP/TAZ inactivation. While previous studies have focused on YAP nuclear-cytoplasmic translocation as a major mechanism of YAP inactivation, a recent report described an additional mode of regulation in which phosphorylation of a specific HxRxxS motif at S381 by Lats1/2 primes YAP for subsequent phosphorylation by CK1 delta/epsilon, followed by the recruitment of the E3 ubiquitin ligase SCFβ-TRCP and ubiquitin-mediated degradation of YAP (Zhao et al., 2010). Both S381-mediated YAP degradation and the previously described S127-mediated nuclear-cytoplasmic translocation are critical for YAP inactivation because both phosphorylation sites must be mutated before YAP can transform NIH 3T3 cells (Zhao et al., 2010). Interestingly, the S381 phosphodegron of YAP is conserved in TAZ, but not Yki, revealing an important divergence between Drosophila and mammals. Because the analysis of Lats1/2 phosphorylation sites in YAP/TAZ has been carried out using overexpressed constructs, it will be important to define the contribution of individual phosphorylation site or their combinations to the regulation of endogenous YAP/TAZ activity.
Transcriptional Regulation by YAP/TAZ: Partners and Targets
Studies in mammalian cells have revealed multiple DNA-binding transcription factor partners for YAP, such as the p53 family member p73 (Strano et al., 2001), the Runt family member Runx2 (Yagi et al., 1999), and the TEAD/TEF family transcription factors (Vassilev et al., 2001). Likewise, TAZ has been reported to interact with multiple DNA-binding transcription factors, such as peroxisome proliferator-activated receptor γ (Hong et al., 2005), thyroid transcription factor-1 (Park et al., 2004), Pax3 (Murakami et al., 2006), Tbx5 (Murakami et al., 2005), Runx2 (Cui et al., 2003; Hong et al., 2005), TEAD1 (Mahoney et al., 2005), and Smad2/3/4 (Varelas et al., 2008). Among these YAP/TAZ-interacting proteins, the TEAD/TEF family transcription factors, which represent homologs of the Drosophila Sd protein, have emerged as the prime mediators of YAP/TAZ function in Hippo signaling (Zhao et al., 2008; Ota and Sasaki, 2008; Zhang et al., 2009a; Chan et al., 2009). The mammalian genome contains four highly homologous TEAD/TEF family members (TEAD1 to -4), which are expressed in diverse tissues from preimplantation embryos to adult tissues (Kaneko and DePamphilis, 1998). Most adult tissues express at least one TEAD member, and different TEAD members appear to function in a redundant manner (Sawada et al., 2008). Consistent with the view that the TEAD transcription factors are the major mediators of YAP/TAZ function, TEAD1/TEAD2 and YAP were shown to regulate largely overlapping sets of target genes (Zhao et al., 2008; Ota and Sasaki, 2008), and RNAi of TEAD factors, or mutations in YAP/TAZ that disrupt their interactions with TEAD factors, diminished the ability of YAP/TAZ in promoting anchorage-independent growth and epithelial-mesenchymal transition (EMT) (Zhao et al., 2008; Zhang et al., 2009a; Chan et al., 2009).
The list of transcriptional partners discussed above, and the shared transactivation of TEAD factors in particular, suggests that YAP and TAZ have partially separable functions but may also be redundant in some crucial ways. Consistent with this view, YAP null mice die at embryonic day 8.5 (Morin-Kensicki et al., 2006) and TAZ null mice are viable (Hossain et al., 2007; Tian et al., 2007; Makita et al., 2008), but mice lacking both YAP and TAZ die extremely early, before the formation of 16-cell morula embryos (Nishioka et al., 2009). Parallel analysis of gene expression profiles revealed common as well as distinct target genes regulated by YAP and TAZ in human MCF10A mammary epithelial cells (Zhang et al., 2009a). This raises the question of whether target gene specificity is merely dependent on the utilization of distinct DNA-binding partners or might actually be guided by YAP and TAZ themselves. A precedent for coactivator-dictated target specificity has been reported in Drosophila, where Sd-Yki and Sd-Vg complexes can regulate distinct target genes (Wu et al., 2008).
Comprehensive expression profiling has revealed a large number of genes that are induced by YAP in mouse livers and different cell lines in a context-dependent manner (Dong et al., 2007; Zhao et al., 2007; Hao et al., 2008; Ota and Sasaki, 2008; Lu et al., 2010). Some of these genes, such as the IAP family member BIRC5 (Dong et al., 2007), the secreted Cystein-rich protein connective tissue growth factor (CTGF) (Zhao et al., 2007; Zhang et al., 2009a), and the EGF family member amphiregulin (AREG) (Zhang et al., 2009b), have been subjected to secondary studies and shown to contribute to YAP-activated growth in cell culture. Indeed, CTGF is a direct target of both YAP and TAZ (Zhao et al., 2007; Zhang et al., 2009a), and AREG is a direct target of YAP (Zhang et al., 2009b). In the future, it will be important to examine whether these reported YAP/TAZ targets contribute to Hippo signaling under physiological conditions such as within intact mammalian tissues.
Upstream of the Mammalian Hippo Kinase Cascade
With the exception of Dachs, the mammalian genome contains homologs for all the reported upstream regulators of the Hippo pathway, including two Kibra homologs (KIBRA/WWC1 and WWC2), two possible Ex homologs (FRMD6/Ex1 and FRMD1/Ex2), one Mer homolog (NF2/Mer), one Ft homolog (Fat4/Fat-j), two Ds homologs (Dchs1 and Dchs2), one Fj homolog (Fjx1), two Lft homologs (Lix1 and Lix1-like), and three Crb homologs (Crb1, Crb2 and Crb3) (Table 1). Loss-of-function mutations are not available for most of these mammalian homologs, thus preventing a rigorous dissection of their physiological functions. However, genetic characterization of the few genes for which loss-of-function mutations are available has revealed important insights into upstream regulation of the mammalian Hippo pathway.
Among the mammalian genes whose Drosophila counterparts have been implicated as upstream regulators of Hippo signaling, NF2/Mer has been most intensively studied (reviewed in McClatchey and Giovannini, 2005; Okada et al., 2007). Mer/NF2 was discovered nearly two decades ago as a tumor suppressor whose mutations cause Neurofibromatosis 2, an autosomal dominant disorder characterized by the development of benign schwannomas and other Schwann-cell-derived tumors associated with the nervous system (Rouleau et al., 1993; Trofatter et al., 1993). Since then, NF2/Mer has been linked to a wide spectrum of effector pathways and cellular functions in mammalian cells, such as Ras, Rac, STAT, or PI3K signaling, as well as endocytosis/degradation of membrane receptors, such as EGFR (reviewed in McClatchey and Giovannini, 2005; Okada et al., 2007), or inhibition of the E3 ubiquitin ligase CRL4DCAF1 in the nucleus (Li et al., 2010a). A particularly important aspect of NF2 function from the standpoint of this review is its prominent role in contact inhibition mediated by cell-surface receptors (e.g., the hyaluronic acid receptor CD44; Morrison et al., 2001) or adherens junctions. However, the relative contribution of effector pathways to NF2 function has not been defined genetically in intact tissues. Recently, it was shown that inactivation of Nf2 led to hepatocellular carcinoma (HCC) and bile duct tumors (Zhang et al., 2010; Benhamouche et al., 2010). Intriguingly, mutations in core Hippo pathway components, such as Mst1/2 (Yu et al., 2010; Lu et al., 2010) and Sav1 (Lee et al., 2010), reveal similar dual tumor phenotypes, and it has been proposed that this dual effect on hepatocyte and cholangiocyte proliferation may reflect the expansion of oval cells, putative bipotential liver progenitor cells (Benhamouche et al., 2010; Lee et al., 2010; Yu et al., 2010; Lu et al., 2010). That said, cholangiocyte tumors were not observed in studies of YAP-transgenic livers (Dong et al., 2007; Camargo et al., 2007), and further work is needed to evaluate the apparent discrepancy as well as the (patho)physiological nature of oval cell lineages in vivo.
Strikingly, loss of Yap leads to loss of hepatocytes and biliary epithelial cells, whereas heterozygous deletion of Yap, which results in no phenotypical manifestation by itself, significantly suppressed the Nf2-deficient liver phenotypes as well as cataract formation caused by Nf2-deficiency in the lens epithelium (Zhang et al., 2010). These findings provide strong genetic evidence placing NF2/Mer upstream of the mammalian Hippo signaling pathway in a physiological setting. This relationship may not be entirely straightforward, however, as NF2/Mer also controls liver homeostasis through EGFR signaling (Benhamouche et al., 2010), and conflicting evidence for Hippo pathway regulation by NF2 was reported (Zhang et al., 2010; Benhamouche et al., 2010). It is worth noting in this context that overexpressed human homologs of some components of the Drosophila Kibra-Ex-Mer complex can physically interact with each other (Genevet et al., 2010; Yu et al., 2010) or with canonical Hippo pathway components (Yu et al., 2010) and can stimulate Lats1/2 phosphorylation (Yu et al., 2010) or inhibit YAP activity (Zhao et al., 2007) in mammalian cell cultures. Of note, the Ex homolog FRMD6 does not contain the extended C-terminal region, which Ex requires for its growth-inhibitory activity (Boedigheimer et al., 1997) and KIBRA binding (Genevet et al., 2010). Future studies will be needed to fully assess whether a homologous Kibra-FRMD-Mer complex regulates Hippo signaling in mammals.
To date, there is little evidence implicating mammalian homologs of Ft or Crb in Hippo signaling. Of the four atypical mammalian cadherins with extracellular homology to Ft, only Fat4 seems to possess a cytoplasmic tail similar to that of Drosophila Ft. Analysis of knockout mice revealed an essential role for Fat4 in controlling PCP in inner hair cell orientation and neural tube elongation; Fat4 mutants also developed cystic kidney disease characterized by a loss of orientated cell division (Saburi et al., 2008). None of these PCP-related phenotypes were reported in mice lacking essential components of the Hippo kinase cascade, such as Mst1/2 or Sav1 (Zhou et al., 2009; Lu et al., 2010; Song et al., 2010; Lee et al., 2010; Lee et al., 2008). Conversely, the phenotypes characteristic of mouse knockouts of Sav1 or Mst1/2, such as hepatomegaly, were not reported in Fat4 mutant mice. It remains unclear whether Fat4 may participate in Hippo signaling in yet-to-be-defined cellular contexts. Likewise, the importance of Crb family proteins in mammalian apicobasal polarity (reviewed in Bazellieres et al., 2009) and pathophysiology is illustrated by the fact that mutations in human Crb1 lead to early-onset retinal degenerative diseases, including retinitis pigmentosa and Leber congenital amaurosis (den Hollander et al., 1999). But these Crb-deficient phenotypes are not obviously related to Hippo signaling. Interestingly, Fat4 and Crb3 have been implicated as potential tumor suppressors in breast cancers (Qi et al., 2009; Karp et al., 2008), suggesting a possible crosstalk between these proteins in certain tissue contexts.
Complexity of Mst1/2 Regulation and Substrates in Mammals
Mst1/2 proteins have long been the subject of intense study because of their involvement in stress-induced apoptosis (reviewed in Radu and Chernoff, 2009). In this context, Mst1/2 are activated by autophosphorylation and caspase-dependent cleavage (Graves et al., 1998, 2001; Glantschnig et al., 2002; Lee and Yonehara, 2002), which liberates the ~35 KDa N-terminal kinase domain from a C-terminal autoinhibitory domain; active kinase then translocates the nucleus and promotes apoptosis by phosphorylating relevant substrates such as Histone H2B (Cheung et al., 2003). However, the caspase cleavage site of Mst1/2 is not conserved in Hpo, and mutant alleles of hpo that encode an analogous ~35 KDa product lead to loss- rather than gain-of-function mutations in Drosophila (Wu et al., 2003; Harvey et al., 2003). Indeed, the 35 KDa Mst1/2 cleavage products lack the C-terminal SARAH domain that is required for Mst/Hpo-WW45/Sav binding in Drosophila (Wu et al., 2003; Udan et al., 2003; Callus et al., 2006). It, therefore, appears that at least in some contexts, Mst1/2 are regulated differently and connect to their downstream effectors via different mechanisms compared to their Drosophila counterpart. Indeed, studies in mouse livers indicate that Mst1/2 cleavage correlates with YAP phosphorylation and inactivation via a Lats1/2-independent mechanism, and this cleavage is frequently lost in human HCCs (Zhou et al., 2009).
Mst1/2 activity is also regulated by heterodimerization with RASSF family proteins (reviewed in Richter et al., 2009; Avruch et al., 2009). The human genome encodes six distinct RASSF family members (RASSF1 to -6), each of which contains a Ras association (RA) domain and a C-terminal SARAH domain. Most RASSF family members have been implicated as tumor suppressors, and their expression is frequently inactivated through epigenetic silencing in human cancers (reviewed in Richter et al., 2009). Their tumor suppressor activity can often be attributed, at least in part, to SARAH domain-mediated heter-odimerization with Mst1/2. This interaction may activate Mst1/2 by targeting Mst1/2 to their endogenous activators and substrates (Praskova et al., 2004) or by liberating Mst1/2 from their inhibitors (such as the kinase Raf-1) (Matallanas et al., 2007). An exception is RASSF6, which has been reported to inhibit Mst2 (Ikeda et al., 2009). The molecular relationship between the Mst/Hpo-RASSF complex and the Mst/Hpo-WW45/Sav complex remains to be defined: whereas a trimeric complex of Mst2, WW45, and RASSF1A (or RASSF6) has been reported in mammalian cells (Guo et al., 2007; Ikeda et al., 2009), dRASSF and Sav bind to Hpo in a mutually exclusive manner in Drosophila (Polesello et al., 2006). Another emerging theme concerns the regulation of RASSF proteins by small GTPases. In mammalian cell cultures, endogenous Nore1/RASSF5 was reported to bind active Ras upon serum stimulation (Vavvas et al., 1998), and further studies implicated the Nore1-Mst1 complex as a Ras effector that mediates the proapoptotic effect of activated Ras (Praskova et al., 2004). Another small GTPase, Rap1, has been implicated as a regulator of the Nore1-Mst1 complex in lymphocyte trafficking and motility (Katagiri et al., 2003).
Accompanying the complex mechanisms of Mst1/2 regulation is the complexity of known Mst1/2 substrates. Besides the core Hippo pathway components WW45, Mob1 and Lats1/2, Mst1/2 have been shown to phosphorylate a diverse array of substrates, such as Histone H2B (Cheung et al., 2003), transcription factors FOXO1 and FOXO3 (Lehtinen et al., 2006; Jang et al., 2007), and the Lats1/2-related kinases Ndr1/Ndr2 (Vichalkovski et al., 2008), mostly in response to apoptotic stimuli. It will be important to define the contribution of these pathways to the overall tumor suppressor function of Mst1/2, especially in the context of intact mammalian tissues.
Future Directions for the Hippo Signaling Network in Mammals
A notable feature of the mammalian Hippo pathway is the large number of RASSF family proteins compared to Drosophila. While the underlying reason for this difference is unknown at present, one can speculate that the mammalian RASSF proteins may have evolved to link Mst1/2 to a greater diversity of upstream signals in mammals. Such complexity reinforces the emerging view from genetic dissection of Hippo signaling in Drosophila that the core Hippo kinase cascade may be viewed as a signaling module that integrates diverse upstream inputs, possibly in a context-dependent manner. In light of this, it is perhaps more accurate to consider Hippo signaling as a complex network rather than a discrete linear pathway. A major challenge in the future is to identify the complete repertoire of cell surface inputs into the Hippo signaling network in both Drosophila and mammals.
As the nuclear effectors of the Hippo signaling pathway, YAP and TAZ represent attractive points of crosstalk. Indeed, Sonic hedgehog signaling is known to upregulate YAP transcription in cerebellar granule neural precursors (Fernandez-L et al., 2009). Conversely, YAP and TAZ can control BMP/TGF-β-dependent Smad signaling (Alarcón et al., 2009; Varelas et al., 2008). In a recent study, Varelas et al. reported crosstalk between the Hippo and the Wnt signaling pathways as well, such that Lats1/2-phosphorylated and cytoplasmic-localized TAZ protein inhibits Wnt signaling through direct physical interaction with the Dishevelled protein (Varelas et al., 2010). Since Lats1/2-phosphorylated TAZ is inactive as a transcriptional coactivator, this study revealed a hitherto unknown cytoplasmic and growth-inhibitory function for TAZ. Thus, the Hippo signaling pathway may restrict tissue growth not only by inactivating YAP/TAZ’s oncogenic activity in the nucleus, but also by enhancing its growth-suppressive activity in the cytoplasm. Future studies will likely reveal additional modes of crosstalk between Hippo and other signaling pathways, which may shed light on how different developmental pathways are integrated to specify organs of characteristic size and shape during animal development.
Physiological Function of Hippo Signaling in Animal Development
Although the Hippo pathway was first discovered for its pivotal role in restricting imaginal disc growth by promoting cell-cycle exit and apoptosis, more recent studies in Drosophila have expanded the function of this pathway into other developmental contexts, such as the mitotic-to-endocycle switch of the posterior follicle cells in adult egg chambers (Polesello and Tapon, 2007; Meignin et al., 2007; Yu et al., 2008), neuroepithelial cell differentiation in larval optic lobe (Reddy et al., 2010), photore-ceptor R8 subtype specification in pupal retina (Mikeladze-Dvali et al., 2005), and dendrite morphogenesis of larval sensory neurons (Emoto et al., 2006). These studies emphasize two general physiological functions for Hippo signaling: coordinating a timely transition from cell proliferation to cellular quiescence and ensuring proper cellular differentiation. These two processes are intimately linked in many development contexts.
Studies of Hippo signaling in vertebrates have reinforced these general themes. For example, overexpression of YAP in murine intestine or chick neural tubes resulted in expansion of progenitor cells and concomitant loss of differentiated cells (Camargo et al., 2007; Cao et al., 2008). Likewise, loss of WW45 led to progenitor cell hyperplasia and defective terminal differentiation in skin, intestine, and lung epithelia (Lee et al., 2008). In cell culture models of myogenesis and keratinocyte differentiation, it was shown that Hippo signaling is activated at the onset of cell differentiation and that YAP hyperactivation led to failure of cell-cycle exit and terminal differentiation (Watt et al., 2010; Lee et al., 2008). The loss of differentiated cell types upon YAP hyperactivation may explain, at least in part, why ubiquitous inactivation of Hippo signaling resulted in increased organ size in only selected tissues (Lee et al., 2008; Song et al., 2010).
In some developmental contexts, however, the often coupled roles for Hippo signaling in promoting cellular quiescence and differentiation may be separated from each other. For example, Hippo signaling is required to maintain the terminally differentiated hepatocytes of mammalian livers in a quiescent state. Overexpression of YAP or loss of Mst1/2 leads to ectopic proliferation of the differentiated hepatocytes (Dong et al., 2007; Camargo et al., 2007; Zhou et al., 2009; Lu et al., 2010). In this context, Hippo signaling regulates cell proliferation without an obvious effect on hepatocyte differentiation. There are also contexts in which the opposite is true. In blastocyst stage mouse embryos, differential Hippo signaling activity leads to cytoplasmic and nuclear YAP localization in the presumptive inner cell mass (ICM) (inside the blastocyst) and the presumptive trophectoderm lining the exterior of the blastocyst, respectively (Nishioka et al., 2009). This position-dependent Hippo signaling activity may derive in part from the difference in the degree of cell-cell contacts between the inside and the outside cells. In this context, cell-contact-mediated Hippo signaling regulates cell-fate specification without inducing proliferation arrest. This dedicated role in cell differentiation is reminiscent of the requirement for Hippo signaling in photoreceptor R8 subtype specification and dendrite morphogenesis in Drosophila, although the latter involve postmitotic rather than proliferating cells (Mikeladze-Dvali et al., 2005; Emoto et al., 2006). Thus, Hippo signaling may regulate distinct cellular outcomes in different contexts.
Hippo Signaling in Human Diseases and Cancers
Consistent with the critical roles of Hippo signaling in mammalian physiology, mutations in Hippo pathway components have been linked to human diseases. Besides Mer/NF2 as the tumor suppressor underlying Neurofibromatosis 2, a heterozygous missense mutation in TEAD1, Y421H, was identified in two independent pedigrees as the cause of Sveinsson’s chorioretinal atrophy (SCRA) (also known as helicoid peripapillary chorioretinal degeneration), a rare autosomal dominant disease characterized by progressive lesions radiating from the optic disc involving the retina and the choroid (Fossdal et al., 2004). In the recently resolved three dimensional structure of the TEAD1-YAP or the TEAD4-YAP complex, Y421 was shown to be engaged in hydrogen bond and hydrophobic interaction with YAP (Chen et al., 2010b; Li et al., 2010b). Accordingly, TEAD1/2/4 proteins carrying the disease-mimicking mutation disrupted TEAD1/2/4-YAP/TAZ binding (Kitagawa, 2007; Chen et al., 2010b; Li et al., 2010b; Tian et al., 2010). It is unknown at present whether SCRA lesions result from loss of heterozygosity or haploinsufficiency of the dominant Y421G allele. Identifying the relevant target genes should shed light on the pathophysiological mechanism of SCRA.
Other than NF2/Mer, DNA mutations in tumor suppressor components of the Hippo pathway are rare in human cancers (http://www.sanger.ac.uk/genetics/CGP/cosmic/). Deletion of WW45 was reported in two renal cancer cell lines (Tapon et al., 2002), and mutations in Mob1 were found in two cDNAs derived from a human melanoma and a mouse mammary carcinoma, respectively (Lai et al., 2005). In contrast, there is increasing evidence implicating epigenetic silencing as a prevalent mechanism of inactivating Hippo pathway tumor suppressor genes. Besides the frequent hypermethylation of RASSF family genes in human cancers (Richter et al., 2009), hypermethylation of Mst1/2 (in soft tissue sarcoma; Seidel et al., 2007) and Lats1/2 (in astrocytoma and breast cancers; Jiang et al., 2006; Takahashi et al., 2005) has also been reported. More generally, decreased expression of Mst1/2 (in colorectal and prostate cancers; Minoo et al., 2007; Cinar et al., 2007) and Mob1 (in colorectal and lung cancers; Kosaka et al., 2007; Sasaki et al., 2007) may be functionally significant, irrespective of methylation status. miRNA-mediated silencing of Hippo pathway tumor suppressors in human cancers has also emerged, as exemplified by the suppression of Lats2 expression by miR372 and miR373, two related oncogenic miRNAs in testicular germ cell tumors (Voorhoeve et al., 2006).
The YAP/TAZ-TEAD transcription factor complex represents a common target of oncogenic transformation. Amplification of the YAP gene locus has been reported at varying frequencies in a wide spectrum of human and murine tumors, such as medulloblastomas, oral squamous-cell carcinomas, and carcinomas of the lung, pancreas, esophagus, liver, and mammary gland (Fernandez-L et al., 2009; Dai et al., 2003; Snijders et al., 2005; Bashyam et al., 2005; Imoto et al., 2001; Zender et al., 2006; Overholtzer et al., 2006). Interestingly, in nearly all cases, the YAP amplicon also contains cIAP2, a mammalian homolog of diap1, suggesting a potential cooperation between YAP and cIAP2 in tumorigenesis. Consistently, comprehensive survey of the most common solid cancer types revealed widespread and frequent YAP overexpression in lung, ovarian, pancreatic, colorectal, hepatocellular, and prostate carcinomas (Dong et al., 2007; Steinhardt et al., 2008), and YAP was shown to be an independent prognostic marker for disease-free survival and overall survival of HCC patients (Xu et al., 2009). A comprehensive survey of TAZ protein expression across multiple tumor types is unavailable at present. In one study focusing on mammary tumors, TAZ overexpression was detected in 21% of primary breast cancers (Chan et al., 2008). This study further suggested that TAZ may govern the invasiveness of breast cancer cells. Consistent with these findings, overexpression of YAP or TAZ can induce anchorage-independent growth and EMT of immortalized mammary and pancreatic epithelial cells in vitro (Overholtzer et al., 2006; Zhao et al., 2008; Zhang et al., 2009a; Chan et al., 2009; Dong et al., 2007). Interestingly, the YAP/TAZ partner TEAD4 has also been reported to be amplified in various cancers (Nowee et al., 2007; Skotheim et al., 2006; Adélaïde et al., 2007), and TEAD4 alone promoted anchorage-independent growth of MCF10A cells in vitro (Chan et al., 2009). Whether other TEAD homologs are amplified or overexpressed in human cancers remains to be determined. It is worth noting that besides its role as a potent oncogene, YAP has also been implicated as a potential tumor suppressor by potentiating p73-mediated apoptosis (Strano et al., 2001; Basu et al., 2003; Matallanas et al., 2007; Oka et al., 2008; Yuan et al., 2008). Paradoxically, this proapoptotic activity of YAP was reported to be activated (Matallanas et al., 2007) or inhibited (Oka et al., 2008) by Lats1-mediated phosphorylation. Future studies are required to determine whether the seemingly opposing function of YAP as an oncoprotein versus a tumor suppressor is dictated by cell contexts.
Concluding Remarks
Research in the past several years has greatly advanced our understanding of the molecular mechanism and the physiological function of the Hippo signaling pathway. These studies have firmly established the Hippo signaling pathway as a central mechanism that regulates organ size and tissue homeostasis in species spanning from Drosophila to mammals. The fundamental importance of this pathway is further solidified by the realization that dysregulation of Hippo signaling underlies various human diseases including cancer.
Despite recent progress, our knowledge about this important growth regulatory pathway remains incomplete. First and foremost, while a conserved Hippo kinase cascade has been established, potentially important variations on this cascade await characterization in mammals, and many of the upstream inputs remain to be defined (Figure 1). A major challenge in the future is to elucidate the molecular nature of these upstream signals, the physiological contexts of their action, and the molecular mechanism by which they regulate the Kibra-Ex-Mer complex and/or the core kinase cassette. Conversely, the outputs of the pathway remain incompletely defined, especially in intact mammalian tissues. Another challenge is to understand how the growth-regulatory Hippo pathway is integrated with other developmental pathways involved in pattern formation, cell growth, survival, and differentiation to coordinately define the characteristic size, shape, and cellular composition of a given organ during animal development. Likewise, investigation of how dysregulation of Hippo and other pathways cooperate to drive tumorigenesis and other forms of human disease is likely to be an active and exciting topic. Finally, small molecule modulators of Hippo signaling may be exploited for tissue engineering and regenerative medicine, as well as therapeutic intervention of relevant human cancers.
Acknowledgments
Given the extensive and ever-expanding literature on Hippo signaling, I apologize to colleagues whose work could not be cited in this review because of space limitations. I am particularly grateful for Dr. Yonggang Zheng for help with the table and the figure. Research in the Pan laboratory is supported in part by grants from National Institutes of Health (EY015708) and Department of Defense (NF093145). D.P. is an investigator of the Howard Hughes Medical Institute.
References
- Adélaïde J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J, Xavier R, Bardeesy N, Zhang XF, Praskova M, Zhou D, Xia F. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284:11001–11005. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann A, Schneider M, Theilenberg E, Grawe F, Knust E. Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature. 2001;414:638–643. doi: 10.1038/414638a. [DOI] [PubMed] [Google Scholar]
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Baena-Lopez LA, Rodríguez I, Baonza A. The tumor suppressor genes dachsous and fat modulate different signalling pathways by regulating dally and dally-like. Proc Natl Acad Sci USA. 2008;105:9645–9650. doi: 10.1073/pnas.0803747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, Tibshirani R, Maitra A, Pollack JR. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Bazellieres E, Assemat E, Arsanto JP, Le Bivic A, Massey-Harroche D. Crumbs proteins in epithelial morphogenesis. Front Biosci. 2009;14:2149–2169. doi: 10.2741/3368. [DOI] [PubMed] [Google Scholar]
- Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, McClatchey AI. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Bhat MA, Izaddoost S, Lu Y, Cho KO, Choi KW, Bellen HJ. Discs Lost, a novel multi-PDZ domain protein, establishes and maintains epithelial polarity. Cell. 1999;96:833–845. doi: 10.1016/s0092-8674(00)80593-0. [DOI] [PubMed] [Google Scholar]
- Boedigheimer MJ, Nguyen KP, Bryant PJ. Expanded functions in the apical cell domain to regulate the growth rate of imaginal discs. Dev Genet. 1997;20:103–110. doi: 10.1002/(SICI)1520-6408(1997)20:2<103::AID-DVG3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005;65:6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- Brittle AL, Repiso A, Casal J, Lawrence PA, Strutt D. Four-jointed modulates growth and planar polarity by reducing the affinity of dachsous for fat. Curr Biol. 2010;20:803–810. doi: 10.1016/j.cub.2010.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant PJ, Simpson P. Intrinsic and extrinsic control of growth in developing organs. Q Rev Biol. 1984;59:387–415. doi: 10.1086/414040. [DOI] [PubMed] [Google Scholar]
- Callus BA, Verhagen AM, Vaux DL. Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. FEBS J. 2006;273:4264–4276. doi: 10.1111/j.1742-4658.2006.05427.x. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J, Struhl G, Lawrence PA. Developmental compartments and planar polarity in Drosophila. Curr Biol. 2002;12:1189–1198. doi: 10.1016/s0960-9822(02)00974-0. [DOI] [PubMed] [Google Scholar]
- Casal J, Lawrence PA, Struhl G. Two separate molecular systems, Dachsous/Fat and Starry night/Frizzled, act independently to confer planar cell polarity. Development. 2006;133:4561–4572. doi: 10.1242/dev.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schäfer A, Nigg EA, Silljé HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–14358. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA. 2010a;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, Song H. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010b;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Cinar B, Fang PK, Lutchman M, Di Vizio D, Adam RM, Pavlova N, Rubin MA, Yelick PC, Freeman MR. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J. 2007;26:4523–4534. doi: 10.1038/sj.emboj.7601872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui CB, Cooper LF, Yang X, Karsenty G, Aukhil I. Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ. Mol Cell Biol. 2003;23:1004–1013. doi: 10.1128/MCB.23.3.1004-1013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A, Wu YZ, Rush LJ, Ross P, Molina JR, et al. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum Mol Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, ten Brink JB, de Kok YJ, van Soest S, van den Born LI, van Driel MA, van de Pol DJ, Payne AM, Bhattacharya SS, Kellner U, et al. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12) Nat Genet. 1999;23:217–221. doi: 10.1038/13848. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci USA. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Processing and phosphorylation of the Fat receptor. Proc Natl Acad Sci USA. 2009;106:11989–11994. doi: 10.1073/pnas.0811540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-L A, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, Taylor MD, Kenney AM. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossdal R, Jonasson F, Kristjansdottir GT, Kong A, Stefansson H, Gosh S, Gulcher JR, Stefansson K. A novel TEAD1 mutation is the causative allele in Sveinsson’s chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum Mol Genet. 2004;13:975–981. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- Genevet A, Polesello C, Blight K, Robertson F, Collinson LM, Pichaud F, Tapon N. The Hippo pathway regulates apical-domain size independently of its growth-control function. J Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantschnig H, Rodan GA, Reszka AA. Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J Biol Chem. 2002;277:42987–42996. doi: 10.1074/jbc.M208538200. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Graves JD, Gotoh Y, Draves KE, Ambrose D, Han DK, Wright M, Chernoff J, Clark EA, Krebs EG. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 1998;17:2224–2234. doi: 10.1093/emboj/17.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JD, Draves KE, Gotoh Y, Krebs EG, Clark EA. Both phosphorylation and caspase-mediated cleavage contribute to regulation of the Ste20-like protein kinase Mst1 during CD95/Fas-induced apoptosis. J Biol Chem. 2001;276:14909–14915. doi: 10.1074/jbc.M010905200. [DOI] [PubMed] [Google Scholar]
- Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17:700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr Biol. 2009;19:1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Ho LL, Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated at the cell membrane to control tissue growth and organ size in Drosophila. Dev Biol. 2010;337:274–283. doi: 10.1016/j.ydbio.2009.10.042. [DOI] [PubMed] [Google Scholar]
- Hong Y, Stronach B, Perrimon N, Jan LY, Jan YN. Drosophila Stardust interacts with Crumbs to control polarity of epithelia but not neuroblasts. Nature. 2001;414:634–638. doi: 10.1038/414634a. [DOI] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, Qi Z, Ponniah S, Hong W, Hunziker W. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci USA. 2007;104:1631–1636. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Kawata A, Nishikawa M, Tateishi Y, Yamaguchi M, Nakagawa K, Hirabayashi S, Bao Y, Hidaka S, Hirata Y, Hata Y. Hippo pathway-dependent and -independent roles of RASSF6. Sci Signal. 2009;2:ra59. doi: 10.1126/scisignal.2000300. [DOI] [PubMed] [Google Scholar]
- Imoto I, Yang ZQ, Pimkhaokham A, Tsuda H, Shimada Y, Imamura M, Ohki M, Inazawa J. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- Ishikawa HO, Takeuchi H, Haltiwanger RS, Irvine KD. Four-jointed is a Golgi kinase that phosphorylates a subset of cadherin domains. Science. 2008;321:401–404. doi: 10.1126/science.1158159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SW, Yang SJ, Srinivasan S, Ye K. Akt phosphorylates MstI and prevents its proteolytic activation, blocking FOXO3 phosphorylation and nuclear translocation. J Biol Chem. 2007;282:30836–30844. doi: 10.1074/jbc.M704542200. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Li X, Hu J, Zhou W, Jiang Y, Li G, Lu D. Promoter hypermethylation-mediated down-regulation of LATS1 and LATS2 in human astrocytoma. Neurosci Res. 2006;56:450–458. doi: 10.1016/j.neures.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev Genet. 1998;22:43–55. doi: 10.1002/(SICI)1520-6408(1998)22:1<43::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Karp CM, Tan TT, Mathew R, Nelson D, Mukherjee C, Degenhardt K, Karantza-Wadsworth V, White E. Role of the polarity determinant crumbs in suppressing mammalian epithelial tumor progression. Cancer Res. 2008;68:4105–4115. doi: 10.1158/0008-5472.CAN-07-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol. 2003;4:741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- Kitagawa M. A Sveinsson’s chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem Biophys Res Commun. 2007;361:1022–1026. doi: 10.1016/j.bbrc.2007.07.129. [DOI] [PubMed] [Google Scholar]
- Kosaka Y, Mimori K, Tanaka F, Inoue H, Watanabe M, Mori M. Clinical significance of the loss of MATS1 mRNA expression in colorectal cancer. Int J Oncol. 2007;31:333–338. [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lee KK, Yonehara S. Phosphorylation and dimerization regulate nucleocytoplasmic shuttling of mammalian STE20-like kinase (MST) J Biol Chem. 2002;277:12351–12358. doi: 10.1074/jbc.M108138200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim TS, Yang TH, Koo BK, Oh SP, Lee KP, Oh HJ, Lee SH, Kong YY, Kim JM, Lim DS. A crucial role of WW45 in developing epithelial tissues in the mouse. EMBO J. 2008;27:1231–1242. doi: 10.1038/emboj.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, Lim DS. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villén J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, You L, Cooper J, Schiavon G, Pepe-Caprio A, Zhou L, Ishii R, Giovannini M, Hanemann CO, Long SB, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010a;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y. Structural insights into the YAP and TEAD complex. Genes Dev. 2010b;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci USA. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, Johnson RL. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney WM, Jr, Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388:217–225. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol. 2008;294:F542–F553. doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed] [Google Scholar]
- Mao Y, Rauskolb C, Cho E, Hu WL, Hayter H, Minihan G, Katz FN, Irvine KD. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development. 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- Mao Y, Kucuk B, Irvine KD. Drosophila lowfat, a novel modulator of Fat signaling. Development. 2009;136:3223–3233. doi: 10.1242/dev.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, Moss SJ, Troyanovsky S, Attwell D, Longmore GD, Braga VM. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278:1220–1228. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. The DHHC palmitoyltransferase approximated regulates Fat signaling and Dachs localization and activity. Curr Biol. 2008;18:1390–1395. doi: 10.1016/j.cub.2008.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O’neill E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, Kulikauskas RM, LaJeunesse DR, Fehon RG. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- McClatchey AI, Giovannini M. Membrane organization and tumorigenesis–the NF2 tumor suppressor Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- Meignin C, Alvarez-Garcia I, Davis I, Palacios IM. The salvador-warts-hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr Biol. 2007;17:1871–1878. doi: 10.1016/j.cub.2007.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Milton CC, Zhang X, Albanese NO, Harvey KF. Differential requirement of Salvador-Warts-Hippo pathway members for organ size control in Drosophila melanogaster. Development. 2010;137:735–743. doi: 10.1242/dev.042309. [DOI] [PubMed] [Google Scholar]
- Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR, Lugli A. Prognostic significance of mammalian sterile20-like kinase 1 in colorectal cancer. Mod Pathol. 2007;20:331–338. doi: 10.1038/modpathol.3800740. [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O’Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, Gutmann DH, Ponta H, Herrlich P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Nakagawa M, Olson EN, Nakagawa O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc Natl Acad Sci USA. 2005;102:18034–18039. doi: 10.1073/pnas.0509109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Tominaga J, Makita R, Uchijima Y, Kurihara Y, Nakagawa O, Asano T, Kurihara H. Transcriptional activity of Pax3 is co-activated by TAZ. Biochem Biophys Res Commun. 2006;339:533–539. doi: 10.1016/j.bbrc.2005.10.214. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Hirota T, Morisaki T, Hara T, Marumoto T, Iida S, Makino K, Yamamoto H, Hiraoka T, Kitamura N, Saya H. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett. 1999;459:159–165. doi: 10.1016/s0014-5793(99)01224-7. [DOI] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Nowee ME, Snijders AM, Rockx DA, de Wit RM, Kosma VM, Hämäläinen K, Schouten JP, Verheijen RH, van Diest PJ, Albertson DG, Dorsman JC. DNA profiling of primary serous ovarian and fallopian tube carcinomas with array comparative genomic hybridization and multiplex ligation-dependent probe amplification. J Pathol. 2007;213:46–55. doi: 10.1002/path.2217. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- Okada T, You L, Giancotti FG. Shedding light on Merlin’s wizardry. Trends Cell Biol. 2007;17:222–229. doi: 10.1016/j.tcb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Léopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Park KS, Whitsett JA, Di Palma T, Hong JH, Yaffe MB, Zannini M. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J Biol Chem. 2004;279:17384–17390. doi: 10.1074/jbc.M312569200. [DOI] [PubMed] [Google Scholar]
- Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev Biol. 2006;304:102–115. doi: 10.1016/j.ydbio.2006.12.021. Published online December 15, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesello C, Tapon N. Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr Biol. 2007;17:1864–1870. doi: 10.1016/j.cub.2007.09.049. [DOI] [PubMed] [Google Scholar]
- Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Curr Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Khoklatchev A, Ortiz-Vega S, Avruch J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem J. 2004;381:453–462. doi: 10.1042/BJ20040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C, Zhu YT, Hu L, Zhu YJ. Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int J Cancer. 2009;124:793–798. doi: 10.1002/ijc.23775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu M, Chernoff J. The DeMSTification of mammalian Ste20 kinases. Curr Biol. 2009;19:R421–R425. doi: 10.1016/j.cub.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Reddy BV, Rauskolb C, Irvine KD. Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Zhang L, Jiang J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol. 2010;337:303–312. doi: 10.1016/j.ydbio.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro PS, Josué F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- Saburi S, Hester I, Fischer E, Pontoglio M, Eremina V, Gessler M, Quaggin SE, Harrison R, Mount R, McNeill H. Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat Genet. 2008;40:1010–1015. doi: 10.1038/ng.179. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Kawano O, Endo K, Suzuki E, Yukiue H, Kobayashi Y, Yano M, Fujii Y. Human MOB1 expression in non-small-cell lung cancer. Clin Lung Cancer. 2007;8:273–276. doi: 10.3816/CLC.2007.n.006. [DOI] [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel C, Schagdarsurengin U, Blümke K, Würl P, Pfeifer GP, Hauptmann S, Taubert H, Dammann R. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46:865–871. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Simon MA, Xu A, Ishikawa HO, Irvine KD. Modulation of fat: dachsous binding by the cadherin domain kinase four-jointed. Curr Biol. 2010;20:811–817. doi: 10.1016/j.cub.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim RI, Autio R, Lind GE, Kraggerud SM, Andrews PW, Monni O, Kallioniemi O, Lothe RA. Novel genomic aberrations in testicular germ cell tumors by array-CGH, and associated gene expression changes. Cell Oncol. 2006;28:315–326. doi: 10.1155/2006/219786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, Albertson DG. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Silva E, Clayton L, Gardano L, Barrios-Rodiles M, Wrana J, Varelas X, Arbouzova NI, Shaw S, Saburi S, et al. Phosphorylation of the tumor suppressor fat is regulated by its ligand Dachsous and the kinase discs overgrown. Curr Biol. 2009;19:1112–1117. doi: 10.1016/j.cub.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J, Sauer U, Szallasi Z, Doyle FJ, III, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D. Nonautonomous planar polarity patterning in Drosophila: dishevelled-independent functions of frizzled. Dev Cell. 2002;3:851–863. doi: 10.1016/s1534-5807(02)00363-5. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, Noguchi S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Tepass U, Theres C, Knust E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 1990;61:787–799. doi: 10.1016/0092-8674(90)90189-l. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Tian Y, Kolb R, Hong JH, Carroll J, Li D, You J, Bronson R, Yaffe MB, Zhou J, Benjamin T. TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell Biol. 2007;27:6383–6395. doi: 10.1128/MCB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Yu J, Tomchick DR, Pan D, Luo X. Structural and functional analysis of the YAP-binding domain of human TEAD2. Proc Natl Acad Sci USA. 2010;107:7293–7298. doi: 10.1073/pnas.1000293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavvas D, Li X, Avruch J, Zhang XF. Identification of Nore1 as a potential Ras effector. J Biol Chem. 1998;273:5439–5442. doi: 10.1074/jbc.273.10.5439. [DOI] [PubMed] [Google Scholar]
- Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008;18:1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Watt KI, Judson R, Medlow P, Reid K, Kurth TB, Burniston JG, Ratkevicius A, De Bari C, Wackerhage H. Yap is a novel regulator of C2C12 myogenesis. Biochem Biophys Res Commun. 2010;393:619–624. doi: 10.1016/j.bbrc.2010.02.034. [DOI] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci USA. 2008;105:14897–14902. doi: 10.1073/pnas.0805201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT, Zender L, Lowe SW, Poon RT, Luk JM. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–4585. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Axelrod JD, Simon MA. Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell. 2002;108:675–688. doi: 10.1016/s0092-8674(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Yu J, Poulton J, Huang YC, Deng WM. The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS ONE. 2008;3:e1761. doi: 10.1371/journal.pone.0001761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P, Danovi SA, et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008;15:1752–1759. doi: 10.1038/cdd.2008.108. [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009a;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009b;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF (beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]