Abstract
Keratinocytes undergo a process of terminal cell differentiation that results in the construction of a multilayered epithelium designed to produce a structure that functions to protect the body from dehydration, abrasion and infection. These protective properties are due to the production of a crosslinked layer of protein called the cornified envelope. Type I transglutaminase (TG1), an enzyme that catalyzes the formation of ε-(γ-glutamyl)lysine bonds, is the key protein responsible for generation of the crosslinks. The mechanisms that lead to activation of transglutaminase during terminal differentiation are not well understood. We have identified a protein that interacts with TG1 and regulates its activity. This protein, tazarotene-induced gene 3 (TIG3), is expressed in the differentiated layers of the epidermis and its expression is associated with transglutaminase activation and cornified envelope formation. We describe a novel mechanism whereby TIG3 regulates TG1 activity.
Keywords: Transglutaminase, TIG3, Keratinocyte differentiation, Calcium, RIG1, H-rev 107 tumor suppressor
Introduction
Transglutaminases comprise a family of proteins that catalyze the formation of protein–protein crosslinks to ensure tissue stability. This property arises from these proteins catalyzing the formation of protein–protein ε-(γ-glutamyl)lysine isopeptide bonds (Ahvazi and Steinert 2003; Nemes and Steinert 1999). A major area of research is identification of the mechanisms that lead to transgluta-minase activation. This is particularly important in epidermis, where the level of the type I transglutaminase (TG1) isoform and its activation state are important controllers of the terminal events in keratinocyte differentiation. Inadequate TG1 level or activity leads to abnormal differentiation associated with lamellar ichthyosis (Petit et al. 1997; Yang et al. 2001). It has been known for many years that calcium is an important cofactor that is required for activation of TG1 (Lorand and Graham 2003). However, there is an ongoing effort to identify other factors, especially proteins, which influence TG1 activation. In pursuing this effort, we identified a novel protein tazarotene-induced gene 3 (TIG3) that interacts with TG1 and facilitates its activation. This review will focus on the role of TG1 in catalyzing crosslink formation in surface epithelia, and our current knowledge regarding the role of TIG3 as a regulator of TG1 activity.
Epithelial differentiation
Surface epithelia are designed to provide protection from the environment. The epidermis, which is among the least permeable of these epithelia, is comprised of multiple cell layers, each of which has a specific function. This property is the result of a unique form of regulated cell death (i.e., cell differentiation) that leads to the formation of a multilayered epithelium (Eckert et al. 1997). In this process, cells in the basal layer (the innermost layer) undergo regulated proliferation to give rise to the daughter cells that occupy the upper epidermal layers. Ultimately these cells are released from the epidermal surface during the final stages of the differentiation process. The basal layer is located at the dermal/epidermal junction. These cells give rise to non-dividing cells that occupy the spinous layer which is characterized by an extensive network of desmosomes. As the cells move into the next layer, the granular layer, they produce intracellular membrane-bound structures that enclose protein and lipid products which are subsequently released and deposited onto the cell surface during terminal differentiation (Bickenbach et al. 1995). Above the granular layer, the cell death process accelerates, resulting in the complete destruction of intracellular structures. The outermost layer, the cornified layer, consists of completely differentiated cells called corneocytes (Eckert et al. 1997). Millions of corneocytes cover the epidermal surface to assure structural integrity and provide protection.
A major component of the corneocyte is the cornified envelope, a dense protein/lipid structure that forms beneath the plasma membrane during terminal keratinocyte differentiation (Green 1980; Kim et al. 1992, 1995; Nemes and Steinert 1999; Steven and Steinert 1994). Assembly of this structure is gradual—it is first visible in the late spinous and granular layers and assembly is completed in the transition zone cells (Eckert et al. 1993; Nemes and Steinert 1999). The envelope is constructed from a variety of soluble (e.g., involucrin and small proline-rich proteins) and insoluble (e.g., loricrin, periplakin, envoplakin) proteins (Chakravarty and Rice 1989; Eckert et al. 1993; Nemes and Steinert 1999; Phillips et al. 1990, 1992; Rice et al. 1992) that are glued together by isopeptide bonds formed by the action of transglutaminases (Eckert et al. 1997; Jarnik et al. 1998). Several transglutaminases are present in the epidermis and in keratinocytes and participate to varying degrees in formation of this structure (Eckert et al. 1997; Huber et al. 1995; Polakowska et al. 1991; Simon and Green 1984).
Transglutaminases in epidermis
Transglutaminases are calcium-activated enzymes that catalyze isopeptide bond formation (Aeschlimann et al. 1998; Grenard et al. 2001; Kim et al. 1991; Phillips et al. 1990). Among these, Factor XIII catalyzes fibrin clot formation (Lorand and Graham 2003; Siefring Jr et al. 1978), TG4 forms clots in prostate seminal fluid (Dubbink et al. 1998), Band 4.2 is inactive and is present in erythroblasts and erythrocytes (Lorand et al. 1987), and TG2 is involved in multiple processes (Fesus and Piacentini 2002; Griffin et al. 2002).
Type I transglutaminase (Phillips et al. 1990; Polakowska et al. 1991), TG3 (Kim et al. 1994) and TG5 (Candi et al. 2001, 2002, 2004; Grenard et al. 2001) are present in the keratinocytes in surface epithelia. Each is located at a specific subcellular location, suggesting specific roles in the keratinocyte terminal differentiation program (Candi et al. 1995, 1999; Tarcsa et al. 1997, 1998). The contention that they have particular functions is supported by the observation that specific patterns of protein crosslinking are observed (Candi et al. 1995). For example, TG3 promotes the formation of intrachain crosslinks that utilize specific lysine and glutamine residues in loricrin. In contrast, TG1 uses multiple glutamine and lysine residues to link loricrin monomers into multimers. This preference is reflected in vivo, as the loricrin residues selected for in vitro crosslink formation by the combined action of TG1 and TG3 mirror the residues utilized in vivo (Candi et al. 1995). This suggests that these enzymes cooperate to produce the in vivo product. Specific crosslinking is also apparent for another family of precursor proteins, the SPR proteins. TG3 forms crosslinks at the SPR1 head A-domain, while TG1 cross-links the SPR1 head B-domain (Candi et al. 1999).
The most important transglutaminase isoform in the context of keratinocyte cornified envelope formation is TG1. TG1 first appears in the spinous layer, and expression is maintained in the granular and cornified layers (Michel et al. 1992). TG1 is a unique transglutaminase in that it is anchored to the inner surface of the keratinocyte plasma membrane via myristyl and palmityl anchors present at the amino terminus (Phillips et al. 1993; Rice et al. 1990). The sites of anchor addition are clusters of five N-terminal cysteine residues (Cys47, Cys48, Cys50, Cys51 and Cys53) (Phillips et al. 1993).
Calcium is considered a necessary cofactor for TG1 activation and TG1 activity is increased at high intracellular calcium concentrations. This calcium sensitivity is consistent with the pattern of TG1 activity in epidermis. TG1 activity is increased in the suprabasal epidermis in layers; cells in these layers contain increased free calcium (Menon et al. 1985). Calcium also has a role in cultured cells. For example, shifting cultured keratinocytes to cell culture medium containing ≥0.3 mM calcium triggers differentiation that is associated with increased TG1 activity and enhanced cornified envelope formation (Eckert et al. 1997; Green 1980). Thus, calcium is an important regulator of TG1 activity. Until recently, little information was available regarding other factors that may regulate TG1 activity; however, recent studies suggest that TIG3 is a protein regulator of TG1 function (Sturniolo et al. 2003, 2005).
TIG3—member of a family of tumor suppressors
Tazarotene-induced gene 3 is a 164 amino acid protein member of the H-rev family of class II tumor suppressors (DiSepio et al. 1998; Huang et al. 2000) and also a member of the NlpC/P60 superfamily (Anantharaman and Aravind 2003; Xue and Rando 2004). TIG3 was initially discovered in human keratinocytes as part of a search to identify genes regulated by the synthetic retinoid, tazarotene (DiSepio et al. 1998). This study and several additional studies indicate that TIG3 is retinoid-regulated (Higuchi et al. 2003; Huang et al. 2000; Jiang et al. 2005). Several findings suggested that TIG3 may be involved in growth regulation and cell survival. First, TIG3 is expressed at reduced levels in the hyperproliferative keratinocytes present in epidermal psoriatic lesions (DiSepio et al. 1998; Duvic et al. 2000, 2003). Second, TIG3 displays homology to various known class II tumor suppressors, including H-rev 10, which are known to suppress cell survival (Deucher et al. 2000; DiSepio et al. 1998; Sers et al. 1997). Third, TIG3 expression is reduced in a host of tumor cell types (Higuchi et al. 2003; Lotz et al. 2005; Tsai et al. 2006). Fourth, treatment of cultures of primary human keratinocytes or psoriatic lesions with vitamin A-derivative results in an increase in TIG3 level that is associated with disease normalization (DiSepio et al. 1998). These findings, which suggest the importance of TIG3 in cell survival regulation, led to biological studies designed to understand its function. TIG3 has been reported to influence activity in several signal transduction cascades (Ou et al. 2008; Tsai et al. 2006, 2007), affect function of subcellular organelles (Tsai et al. 2007) and regulate transglutaminase activity (Sturniolo et al. 2003).
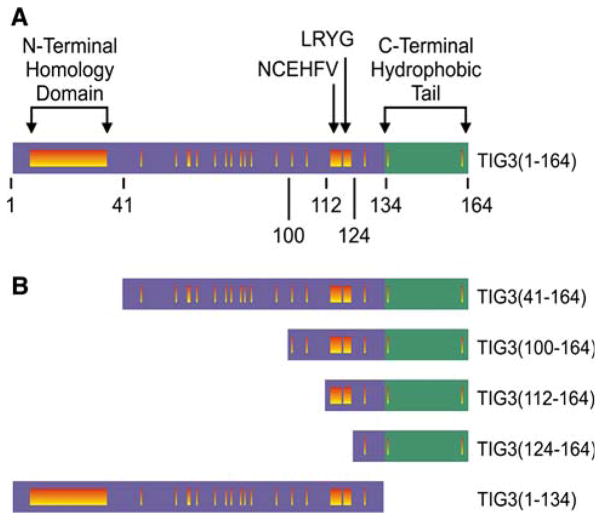
Tazarotene-induced gene 3 (DiSepio et al. 1998), also called RIG1 (Jiang et al. 2005) and H-rev 107–2 (Lotz et al. 2005), displays homology to the human, mouse and rat forms of H-rev 107. The overall structure of these proteins includes an N-terminal hydrophilic region and a hydrophobic C-terminal domain. Although not conserved at the amino acid sequence level, the hydrophobic property of the C-terminal domain is conserved among these proteins. In TIG3 the hydrophilic region includes amino acids 1–134, and the hydrophobic C-terminal domain is contained within amino acids 135–164 (Fig. 1a). The N-terminal homology domain and the sequences NCEHFV and LRYG are motifs that are conserved among these proteins (Xue and Rando 2004). Based on homology with lecithin retinol acyltransferase, it has been suggested that these regions may form part of an enzymatic catalytic site (Anantharaman and Aravind 2003; Bok et al. 2003; Jahng et al. 2003; Xue and Rando 2004)—a possibility that has yet to be explored.
Fig. 1.
Structure of TIG3 and TIG3 mutants. a Structural features of full-length TIG3. The hydrophilic N-terminal region (amino acids 1–134), hydrophobic membrane-anchoring domain (amino acids 134–164) and the N-terminal homology domain, LRYG and NCEHFV motifs are shown (Deucher et al. 2000). Regions of sequence homology shared by TIG3 and other members of the H-rev 107 family are represented as vertical bars. b Structure of TIG3 mutants. Individual mutants were constructed in a systematic manner to eliminate specific homologous elements from the N-terminal hydrophilic domain or to eliminate the membrane-anchoring domain
TIG3—a regulator of keratinocyte survival
The biology of TIG3 action in keratinocytes has recently been examined. These studies show that delivery of TIG3 to immortalized keratinocytes (HaCaT cells) causes a reduction in cell survival that is associated with a peri-nuclear distribution of the expressed protein (Deucher et al. 2000). However, the mechanism responsible for this reduction in cell survival was not understood. This has been studied further in normal human epidermal keratinocytes. As is the case with most normal cells, non-differentiated normal human keratinocytes in monolayer culture produce barely detectable levels of TIG3 (Sturniolo et al. 2005). It is interesting that vector-based expression of TIG3 in these cultures results in a substantial reduction in cell viability. The fact that monolayer cultures of keratinocytes cannot survive in the presence of TIG3 suggests that it is a potent regulator of cell viability. Reduced survival coincides with expression of TIG3, and is observed within 24 h following infection with a TIG3-encoding adenovirus (Sturniolo et al. 2003). Within 48–60 h post-infection cell survival is reduced to 10% of cells. Dose–response studies reveal a direct relationship between the amount of TIG3 present in the cells and the reduction in cell survival (Sturniolo et al. 2003, 2005).
The advent of TIG3 expression is also associated with a change in cell morphology. This includes the appearance in TIG3-expressing cells of structures that resemble the terminal product of keratinocyte differentiation—the cornified envelop (Sturniolo et al. 2003). These structures survive boiling in detergent and reducing agent—properties shared by cornified envelopes that form in vivo (Sturniolo et al. 2003). Since transglutaminase activity is required for cornified envelope assembly, we hypothesized that TIG3 may associate with and trigger TG1 activation.
TIG3—a novel regulator of type I transglutaminase activity
We initiated studies to examine this possibility by assaying for co-localization of TIG3 and TG1 by fluorescence microscopy. Indirect immunofluorescence studies provided strong evidence that TG1 and TIG3 co-localize within cells (Jans et al. 2008; Sturniolo et al. 2003, 2005). To assess whether this interaction is associated with TG1 activation, keratinocytes were equilibrated with fluorescein cadaverine (FC), a cell permeable fluorescent transglutaminase substrate. FC is covalently linked to proteins at sites of transglutaminase activation. These studies show extensive FC incorporation at sites of TIG3 localization, suggesting that TIG3 co-localization with TG1 is associated with increased TG1 activity. There is complete correspondence between TIG3 and TG1 co-localization and TG1 activity. These findings strongly suggest that TIG3 interacts with and activates TG1. This finding is also consistent with the fact that both TIG3 and TG1 are membrane-anchored proteins, suggesting that membrane association may be required for interaction. To assess this possibility, we tested that ability of a mutant, TIG31–134, which lacks the membrane-anchor domain, to promote cell death and activate transglutaminase (Sturniolo et al. 2003). Consistent with a role for membrane localization, expression of TIG31–134 in keratinocytes does not cause cell death or TG1 activation. Moreover, TIG31–134 displays a diffuse cytoplasmic staining pattern that differs markedly from the membrane-associated pattern observed for TIG31–164 (Sturniolo et al. 2003). As noted above, TG1 (Phillips et al. 1990; Polakowska et al. 1991), TG3 (Kim et al. 1994), and TG5 (Candi et al. 2001, 2002, 2004; Grenard et al. 2001) are expressed in human keratinocytes. Thus, TIG3 could be interacting with any one of these forms. However, the fact that both TIG3 and TG1 associate with membranes suggests that TG1 is the primary responsive isoform. Furthermore, TIG31–134, which cannot associate with membranes, does not activate crosslinking.
Since overexpression of TG1 is associated with enhanced activity (unpublished), expression of TIG3 could act by either increasing TG1 level or activity, or a combination of these mechanisms. To address this issue, we assessed the impact of vector-mediated TIG3 expression on TG1 levels and activity. These studies reveal that TIG3 does not change cellular TG1 levels, as measured by mRNA content. However, TG1 activity, assayed by trans-glutaminase substrate incorporation by whole cell extracts prepared from TIG3-expressing and non-expressing cells, is markedly increased by TIG3 (Sturniolo et al. 2005). As with all protein expression experiments, the cell responses to TIG3 expression (reduced viability, increased cornified envelope formation) could be due to a combination of events—some of which may be more important than others. To assess the relative importance of TG1 activation on these responses, TIG3-expressing cells were treated with monodansylcadaverine, a substrate competitive inhibitor of transglutaminase action. Treatment with this agent results in a near-complete reversal of the pro-differentiation morphological changes associated with TIG3 expression. The fact that a relatively specific transglutaminase inhibitor can inhibit most of the biology suggests a central role for transglutaminase in TIG3-mediated cell death (Sturniolo et al. 2005).
The fact that TIG3 and TG1 co-localize at the microscopic level suggests that they might associate within the cell. To assess this possibility, a FLAG epitope-labeled form of TG1 (FLAG-TG1) was co-expressed in keratinocytes with TIG3, and cell extracts were prepared for co-immunoprecipitation analysis. These studies demonstrate that TIG3 co-immunoprecipitates with TG1-FLAG, suggesting that TIG3 associates with TG1, either directly or indirectly. A striking additional finding is that the amino terminal truncated form of TIG3, TIG31–134, does not co-immunoprecipitate with TG1. This finding is consistent with functional data showing that TIG31–134 does not activate TG1 and does not cause cell death. Thus, it appears that TG1/TIG3 interaction at membranes is required for TIG3-mediated functional responses (Sturniolo et al. 2005).
TIG3 is a transglutaminase substrate
Most proteins that come into proximity with TG1 are captured as transglutaminase substrates. Classical examples of transglutaminase substrates in keratinocytes are cornified envelope-associated proteins—e.g., involucrin, S100 proteins, SPR proteins and loricrin (Eckert et al. 2005; Ruse et al. 2001). All of these proteins serve as components of the cornified envelope and not as regulators of transglutaminase activity. Since TIG3 is the first protein identified in keratinocytes that functions as a TG1 regulator, we assessed whether it also serves as a trans-glutaminase substrate. These studies indicate that TIG3 is a transglutaminase substrate, and that the TG1-dependent crosslinking of TIG3 is specifically inhibited in the presence of monodansylcadaverine (Sturniolo et al. 2005). Moreover, TIG3 serves as an amine acceptor in this reaction, indicating that specific glutamine residues present in TIG3 function as transglutaminase substrates. This evidence, measured as ability of TG1 to attach a primary amine substrate, biotin-x-cadaverine, to TIG3 (Sturniolo et al. 2005), is consistent with the immunoprecipitation and fluorescence microscopy data showing that TIG3 and TG1 are in close proximity. In contrast, TIG31–134, which does not associate with membranes or TG1, is not a substrate (Sturniolo et al. 2005).
Identification of TIG3 domain that interacts with TG1
The above studies strongly suggest that TIG3 interaction with TG1 leads to transglutaminase activation and protein crosslink formation, which ultimately leads to cornified envelope assembly and reduced cell survival. These data further suggest that a key step in this regulation is TIG3 interaction with TG1. Regardless of whether the interaction that leads to TG1 activation is direct or indirect, it must require specific TIG3 domains. To examine this question, a series of TIG3 mutants were constructed and expressed in keratinocytes, and each protein’s intracellular localization, pro-differentiation activity and ability to activate trans-glutaminase was monitored. The mutants are shown in Fig. 1b. These results are interesting in that all of the N-terminal deletion mutants interacted with TG1. This is evidenced by TIG3 co-localization and co-immunoprecipitation with TG1, ability to promote FC incorporation in cells, and ability to serve as TG1 substrates. Thus, all of the N-terminal TIG3 deletion mutants interact with and activate TG1, suggesting that the TIG3 protein segment required for interaction with TG1 is present in the segment that spans amino acids 124–164 (Fig. 1B). In contrast, the TIG31–134 mutant, which lacks the C-terminal membrane-anchoring domain, does not interact with TG1. These findings suggest that TIG3 association with TG1 requires the C-terminal hydrophobic tail (membrane-anchoring domain) and several additional amino acids derived from the hydrophilic domain. The precise mapping of the interaction domain is a focus of ongoing studies.
A surprising finding was that the TIG3 truncation mutants cause apoptosis (Jans et al. 2008). This is in marked contrast to the pro-differentiation function of the full-length protein (Sturniolo et al. 2003, 2005). We observed mitochondria-dependent apoptosis, characterized by release of cytochrome c, procaspase-3 activation and PARP cleavage. This is particularly evident for the most severely truncated mutants, TIG3112–164 and TIG3124–164 (Jans et al. 2008). Instead of producing cornified envelopes, as is the case for full-length TIG3, these mutants cause the cells to produce apoptosis-associated (Efimova et al. 2004) bleb structures (Jans et al. 2008). An additional property of these mutants is enhanced association with punctuate membrane-associated structures, a feature that may be important for the response. This enhanced membrane association can be attributed to the increased relative hydrophobicity of the mutants as compared to the intact protein. We do not presently know whether these truncated forms are produced or have biological activity in vivo. In stress conditions, such as exposure to ultraviolet irradiation, keratinocytes undergo apoptotic cell death, which is characterized by caspase activation and PARP cleavage (Jans et al. 2008). The above findings suggest that truncation of TIG3 converts it from a pro-differentiation to a pro-apoptosis protein. Thus, it is possible that a cleaved form of TIG3 may be produced during and may mediate stress-associated apoptosis.
Summary and proposed mechanism of action
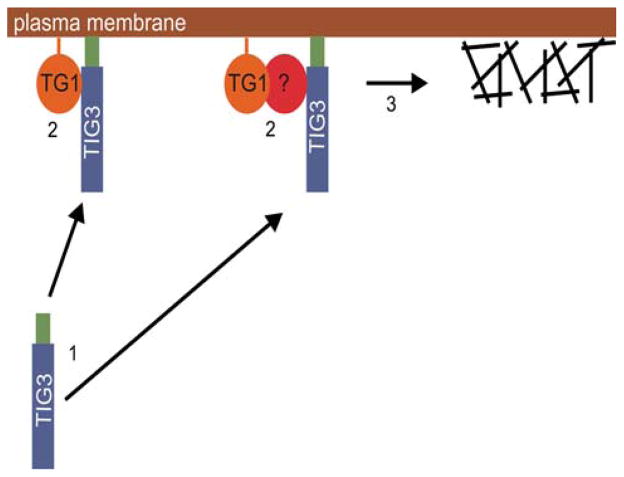
Identifying factors that regulate TG1 level and activity is an important goal, as TG1 activity is absolutely essential for cornified envelope assembly and maintenance of barrier function, and inappropriate TG1 function leads to skin disease (Huber et al. 1995). Calcium is thought to be required for TG1 function and previous studies indicate that TG1 activity is increased when cell culture medium is supplemented with increased calcium (Kruszewski et al. 1991). We now describe TIG3 as a novel regulator that increases TG1 activity in a manner that does not require supplementation of cell culture medium with increased calcium (Sturniolo et al. 2003, 2005) and is not associated with changes in intracellular calcium concentration (unpublished). Based on these findings, we propose the model shown in Fig. 2. In this model TIG3 is produced in cells in the suprabasal epidermis and then associates with the inner surface of the plasma membrane via the TIG3 C-terminal anchor domain. Once positioned along the internal membrane surface, TIG3 then moves in the plane of the membrane until it locates TG1 or another protein that bridges its interaction with TG1. The result of this interaction is activation of TG1 crosslinking function which leads to the formation of intracellular crosslinks in regions of the cell adjacent to the plasma membrane inner surface. It is possible that TIG3 activates TG1 by shifting the TG1 structure to an active conformation. We further propose that the TIG3 domain required for interaction with TG1 is located near its C-terminus (Jans et al. 2008). Studies are underway that are designed to test this model.
Fig. 2.
Proposed mechanism of TIG3 action in keratinocytes. TIG3 is a 164 amino acid protein that is expressed in suprabasal keratinocytes. Upon expression TIG3 associates with the inner surface of the plasma membrane via its C-terminal membrane-anchoring domain (step 1). TIG3 then moves in the plane of the membrane to interact directly or indirectly via an intervening bridge protein (step 2). This TIG3 site required for interaction with TG1 is located near the TIG3 C-terminus. The TIG3/TG1 interaction serves to activate TG1 cross-linking activity and TG1 then assembles crosslinked structures in the vicinity of the plasma membrane (step 3). This ultimately leads to cell death
Acknowledgments
This work was supported by a grant to RLE from the National Institutes of Health (AR49713).
Abbreviations
- TG1
Transglutaminase type 1
- TIG3
Tazarotene-induced gene 3
- FC
Fluorescein cadaverine
Contributor Information
Richard L. Eckert, Email: reckert@umaryland.edu, Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA. Department of Dermatology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Greenebaum Cancer Center, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA
Michael T. Sturniolo, Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Department of Dermatology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Greenebaum Cancer Center, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA
Ralph Jans, Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Department of Dermatology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Greenebaum Cancer Center, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA.
Catherine A. Kraft, Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Department of Dermatology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Greenebaum Cancer Center, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA
Haibing Jiang, Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Department of Dermatology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Greenebaum Cancer Center, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA.
Ellen A. Rorke, Department of Dermatology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Greenebaum Cancer Center, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA, Department of Microbiology and Immunology, University of Maryland School of Medicine, 108 N. Greene Street, Baltimore, MD 21201, USA
References
- Aeschlimann D, Koeller MK, Allen-Hoffmann BL, Mosher DF. Isolation of a cDNA encoding a novel member of the transglutaminase gene family from human keratinocytes. Detection and identification of transglutaminase gene products based on reverse transcription-polymerase chain reaction with degenerate primers. J Biol Chem. 1998;273:3452–3460. doi: 10.1074/jbc.273.6.3452. [DOI] [PubMed] [Google Scholar]
- Ahvazi B, Steinert PM. A model for the reaction mechanism of the transglutaminase 3 enzyme. Exp Mol Med. 2003;35:228–242. doi: 10.1038/emm.2003.31. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickenbach JR, Greer JM, Bundman DS, Rothnagel JA, Roop DR. Loricrin expression is coordinated with other epidermal proteins and the appearance of lipid lamellar granules in development. J Invest Dermatol. 1995;104:405–410. doi: 10.1111/1523-1747.ep12665896. [DOI] [PubMed] [Google Scholar]
- Bok D, Ruiz A, Yaron O, Jahng WJ, Ray A, Xue L, Rando RR. Purification and characterization of a transmembrane domain-deleted form of lecithin retinol acyltransferase. Biochemistry. 2003;42:6090–6098. doi: 10.1021/bi0342416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Melino G, Mei G, Tarcsa E, Chung SI, Marekov LN, Steinert PM. Biochemical, structural, and transglutaminase substrate properties of human loricrin, the major epidermal cornified cell envelope protein. J Biol Chem. 1995;270:26382–26390. doi: 10.1074/jbc.270.44.26382. [DOI] [PubMed] [Google Scholar]
- Candi E, Tarcsa E, Idler WW, Kartasova T, Marekov LN, Steinert PM. Transglutaminase cross-linking properties of the small proline-rich 1 family of cornified cell envelope proteins. Integration with loricrin. J Biol Chem. 1999;274:7226–7237. doi: 10.1074/jbc.274.11.7226. [DOI] [PubMed] [Google Scholar]
- Candi E, Oddi S, Terrinoni A, Paradisi A, Ranalli M, Finazzi-Agro A, Melino G. Transglutaminase 5 cross-links loricrin, involucrin, and small proline-rich proteins in vitro. J Biol Chem. 2001;276:35014–35023. doi: 10.1074/jbc.M010157200. [DOI] [PubMed] [Google Scholar]
- Candi E, Oddi S, Paradisi A, Terrinoni A, Ranalli M, Teofoli P, Citro G, Scarpato S, Puddu P, Melino G. Expression of transglutaminase 5 in normal and pathologic human epidermis. J Invest Dermatol. 2002;119:670–677. doi: 10.1046/j.1523-1747.2002.01853.x. [DOI] [PubMed] [Google Scholar]
- Candi E, Paradisi A, Terrinoni A, Pietroni V, Oddi S, Cadot B, Jogini V, Meiyappan M, Clardy J, Finazzi-Agro’ A, Melino G. Transglutaminase 5 is regulated by guanine/adenine nucleotides. Biochem J. 2004;381:313–319. doi: 10.1042/BJ20031474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty R, Rice RH. Acylation of keratinocyte transglutaminase by palmitic and myristic acids in the membrane Anchorage region. J Biol Chem. 1989;264:625–629. [PubMed] [Google Scholar]
- Deucher A, Nagpal S, Chandraratna RA, Di Sepio D, Robinson NA, Dashti SR, Eckert RL. The carboxy-terminal hydrophobic domain of TIG3, a class II tumor suppressor protein, is required for appropriate cellular localization and optimal biological activity. Int J Oncol. 2000;17:1195–1203. doi: 10.3892/ijo.17.6.1195. [DOI] [PubMed] [Google Scholar]
- DiSepio D, Ghosn C, Eckert RL, Deucher A, Robinson N, Duvic M, Chandraratna RA, Nagpal S. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc Natl Acad Sci USA. 1998;95:14811–14815. doi: 10.1073/pnas.95.25.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbink HJ, de Waal L, van Haperen R, Verkaik NS, Trapman J, Romijn JC. The human prostate-specific transglutaminase gene (TGM4): genomic organization, tissue-specific expression, and promoter characterization. Genomics. 1998;51:434–444. doi: 10.1006/geno.1998.5393. [DOI] [PubMed] [Google Scholar]
- Duvic M, Helekar B, Schulz C, Cho M, DiSepio D, Hager C, DiMao D, Hazarika P, Jackson B, Breuer-McHam J, Young J, Clayman G, Lippman SM, Chandraratna RA, Robinson NA, Deucher A, Eckert RL, Nagpal S. Expression of a retinoid-inducible tumor suppressor, Tazarotene-inducible gene-3, is decreased in psoriasis and skin cancer. Clin Cancer Res. 2000;6:3249–3259. [PubMed] [Google Scholar]
- Duvic M, Ni X, Talpur R, Herne K, Schulz C, Sui D, Ward S, Joseph A, Hazarika P. Tazarotene-induced gene 3 is suppressed in basal cell carcinomas and reversed in vivo by tazarotene application. J Invest Dermatol. 2003;121:902–909. doi: 10.1046/j.1523-1747.2003.12488.x. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Yaffe MB, Crish JF, Murthy S, Rorke EA, Welter JF. Involucrin–structure and role in envelope assembly. J Invest Dermatol. 1993;100:613–617. doi: 10.1111/1523-1747.ep12472288. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Crish JF, Robinson NA. The epidermal keratinocyte as a model for the study of gene regulation and cell differentiation. Physiol Rev. 1997;77:397–424. doi: 10.1152/physrev.1997.77.2.397. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Sturniolo MT, Broome AM, Ruse M, Rorke EA. Transglutaminase function in epidermis. J Invest Dermatol. 2005;124:481–492. doi: 10.1111/j.0022-202X.2005.23627.x. [DOI] [PubMed] [Google Scholar]
- Efimova T, Broome AM, Eckert RL. Protein kinase Cdelta regulates keratinocyte death and survival by regulating activity and subcellular localization of a p38delta-extracellular signal-regulated kinase 1/2 complex. Mol Cell Biol. 2004;24:8167–8183. doi: 10.1128/MCB.24.18.8167-8183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesus L, Piacentini M. Transglutaminase 2: an enigmatic enzyme with diverse functions. Trends Biochem Sci. 2002;27:534–539. doi: 10.1016/s0968-0004(02)02182-5. [DOI] [PubMed] [Google Scholar]
- Green H. The keratinocyte as differentiated cell type. Harvey Lect. 1980;74:101–139. [PubMed] [Google Scholar]
- Grenard P, Bates MK, Aeschlimann D. Evolution of transglu-taminase genes: identification of a transglutaminase gene cluster on human chromosome 15q15. Structure of the gene encoding transglutaminase X and a novel gene family member, transglutaminase Z. J Biol Chem. 2001;276:33066–33078. doi: 10.1074/jbc.M102553200. [DOI] [PubMed] [Google Scholar]
- Griffin M, Casadio R, Bergamini CM. Transglutaminases: nature’s biological glues. Biochem J. 2002;368:377–396. doi: 10.1042/BJ20021234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi E, Chandraratna RA, Hong WK, Lotan R. Induction of TIG3, a putative class II tumor suppressor gene, by retinoic acid in head and neck and lung carcinoma cells and its association with suppression of the transformed phenotype. Oncogene. 2003;22:4627–4635. doi: 10.1038/sj.onc.1206235. [DOI] [PubMed] [Google Scholar]
- Huang SL, Shyu RY, Yeh MY, Jiang SY. Cloning and characterization of a novel retinoid-inducible gene 1(RIG1) deriving from human gastric cancer cells. Mol Cell Endocrinol. 2000;159:15–24. doi: 10.1016/s0303-7207(99)00207-5. [DOI] [PubMed] [Google Scholar]
- Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SP, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science. 1995;267:525–528. doi: 10.1126/science.7824952. [DOI] [PubMed] [Google Scholar]
- Jahng WJ, Xue L, Rando RR. Lecithin retinol acyltransferase is a founder member of a novel family of enzymes. Biochemistry. 2003;42:12805–12812. doi: 10.1021/bi035370p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans R, Sturniolo MT, Eckert RL. Localization of the TIG3 transglutaminase interaction domain and demonstration that the amino-terminal region is required for TIG3 function as a keratinocyte differentiation regulator. J Invest Dermatol. 2008;128:517–529. doi: 10.1038/sj.jid.5701035. [DOI] [PubMed] [Google Scholar]
- Jarnik M, Simon MN, Steven AC. Cornified cell envelope assembly: a model based on electron microscopic determinations of thickness and projected density. J Cell Sci. 1998;111(Pt 8):1051–1060. doi: 10.1242/jcs.111.8.1051. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Wu MS, Chen LM, Hung MW, Lin HE, Chang GG, Chang TC. Identification and characterization of the retinoic acid response elements in the human RIG1 gene promoter. Biochem Biophys Res Commun. 2005;331:630–639. doi: 10.1016/j.bbrc.2005.03.214. [DOI] [PubMed] [Google Scholar]
- Kim HC, Idler WW, Kim IG, Han JH, Chung SI, Steinert PM. The complete amino acid sequence of the human transglutaminase K enzyme deduced from the nucleic acid sequences of cDNA clones. J Biol Chem. 1991;266:536–539. [PubMed] [Google Scholar]
- Kim IG, McBride OW, Wang M, Kim SY, Idler WW, Steinert PM. Structure and organization of the human transglutaminase 1 gene. J Biol Chem. 1992;267:7710–7717. [PubMed] [Google Scholar]
- Kim IG, Lee SC, Lee JH, Yang JM, Chung SI, Steinert PM. Structure and organization of the human transglutaminase 3 gene: evolutionary relationship to the transglutaminase family. J Invest Dermatol. 1994;103:137–142. doi: 10.1111/1523-1747.ep12392470. [DOI] [PubMed] [Google Scholar]
- Kim SY, Chung SI, Yoneda K, Steinert PM. Expression of transglutaminase 1 in human epidermis. J Invest Dermatol. 1995;104:211–217. doi: 10.1111/1523-1747.ep12612769. [DOI] [PubMed] [Google Scholar]
- Kruszewski FH, Hennings H, Yuspa SH, Tucker RW. Regulation of intracellular free calcium in normal murine keratinocytes. Am J Physiol. 1991;261:C767–C773. doi: 10.1152/ajpcell.1991.261.5.C767. [DOI] [PubMed] [Google Scholar]
- Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- Lorand L, Barnes N, Bruner-Lorand JA, Hawkins M, Michalska M. Inhibition of protein cross-linking in Ca2+-enriched human erythrocytes and activated platelets. Biochemistry. 1987;26:308–313. doi: 10.1021/bi00375a043. [DOI] [PubMed] [Google Scholar]
- Lotz K, Kellner T, Heitmann M, Nazarenko I, Noske A, Malek A, Gontarewicz A, Schafer R, Sers C. Suppression of the TIG3 tumor suppressor gene in human ovarian carcinomas is mediated via mitogen-activated kinase-dependent and -independent mechanisms. Int J Cancer. 2005;116:894–902. doi: 10.1002/ijc.21127. [DOI] [PubMed] [Google Scholar]
- Menon GK, Grayson S, Elias PM. Ionic calcium reservoirs in mammalian epidermis: ultrastructural localization by ion-capture cytochemistry. J Invest Dermatol. 1985;84:508–512. doi: 10.1111/1523-1747.ep12273485. [DOI] [PubMed] [Google Scholar]
- Michel S, Bernerd F, Jetten AM, Floyd EE, Shroot B, Reichert U. Expression of keratinocyte transglutamine mRNA revealed by in situ hybridization. J Invest Dermatol. 1992;98:364–368. doi: 10.1111/1523-1747.ep12499806. [DOI] [PubMed] [Google Scholar]
- Nemes Z, Steinert PM. Bricks and mortar of the epidermal barrier. Exp Mol Med. 1999;31:5–19. doi: 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- Ou CC, Hsu SC, Hsieh YH, Tsou WL, Chuang TC, Liu JY, Kao MC. Downregulation of HER2 by RIG1 involves the PI3K/Akt pathway in ovarian cancer cells. Carcinogenesis. 2008;29:299–306. doi: 10.1093/carcin/bgm263. [DOI] [PubMed] [Google Scholar]
- Petit E, Huber M, Rochat A, Bodemer C, Teillac-Hamel D, Muh JP, Revuz J, Barrandon Y, Lathrop M, de Prost Y, Hohl D, Hovnanian A. Three novel point mutations in the keratinocyte transglutaminase (TGK) gene in lamellar ichthyosis: significance for mutant transcript level, TGK immunodetection and activity. Eur J Hum Genet. 1997;5:218–228. [PubMed] [Google Scholar]
- Phillips MA, Stewart BE, Qin Q, Chakravarty R, Floyd EE, Jetten AM, Rice RH. Primary structure of keratinocyte transglutaminase. Proc Natl Acad Sci USA. 1990;87:9333–9337. doi: 10.1073/pnas.87.23.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MA, Stewart BE, Rice RH. Genomic structure of keratinocyte transglutaminase. Recruitment of new exon for modified function. J Biol Chem. 1992;267:2282–2286. [PubMed] [Google Scholar]
- Phillips MA, Qin Q, Mehrpouyan M, Rice RH. Keratinocyte transglutaminase membrane anchorage: analysis of site-directed mutants. Biochemistry. 1993;32:11057–11063. doi: 10.1021/bi00092a015. [DOI] [PubMed] [Google Scholar]
- Polakowska R, Herting E, Goldsmith LA. Isolation of cDNA for human epidermal type I transglutaminase. J Invest Dermatol. 1991;96:285–288. doi: 10.1111/1523-1747.ep12464554. [DOI] [PubMed] [Google Scholar]
- Rice RH, Rong XH, Chakravarty R. Proteolytic release of keratinocyte transglutaminase. Biochem J. 1990;265:351–357. doi: 10.1042/bj2650351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice RH, Mehrpouyan M, O’Callahan W, Parenteau NL, Rubin AL. Keratinocyte transglutaminase: differentiation marker and member of an extended family. Epithelial Cell Biol. 1992;1:128–137. [PubMed] [Google Scholar]
- Ruse M, Lambert A, Robinson N, Ryan D, Shon KJ, Eckert RL. S100A7, S100A10, and S100A11 are transglutaminase substrates. Biochemistry. 2001;40:3167–3173. doi: 10.1021/bi0019747. [DOI] [PubMed] [Google Scholar]
- Sers C, Emmenegger U, Husmann K, Bucher K, Andres AC, Schafer R. Growth-inhibitory activity and downregulation of the class II tumor-suppressor gene H-rev 107 in tumor cell lines and experimental tumors. J Cell Biol. 1997;136:935–944. doi: 10.1083/jcb.136.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefring GE, Jr, Apostol AB, Velasco PT, Lorand L. Enzymatic basis for the Ca2+-induced cross-linking of membrane proteins in intact human erythrocytes. Biochemistry. 1978;17:2598–2604. doi: 10.1021/bi00606a022. [DOI] [PubMed] [Google Scholar]
- Simon M, Green H. Participation of membrane-associated proteins in the formation of the cross-linked envelope of the keratinocyte. Cell. 1984;36:827–834. doi: 10.1016/0092-8674(84)90032-1. [DOI] [PubMed] [Google Scholar]
- Steven AC, Steinert PM. Protein composition of cornified cell envelopes of epidermal keratinocytes. J Cell Sci. 1994;107:693–700. [PubMed] [Google Scholar]
- Sturniolo MT, Dashti SR, Deucher A, Rorke EA, Broome AM, Chandraratna RA, Keepers T, Eckert RL. A novel tumor suppressor protein promotes keratinocyte terminal differentiation via activation of type I transglutaminase. J Biol Chem. 2003;278:48066–48073. doi: 10.1074/jbc.M307215200. [DOI] [PubMed] [Google Scholar]
- Sturniolo MT, Chandraratna RA, Eckert RL. A novel transglutaminase activator forms a complex with type 1 trans-glutaminase. Oncogene. 2005;24:2963–2972. doi: 10.1038/sj.onc.1208392. [DOI] [PubMed] [Google Scholar]
- Tarcsa E, Marekov LN, Andreoli J, Idler WW, Candi E, Chung SI, Steinert PM. The fate of trichohyalin. Sequential post-translational modifications by peptidyl-arginine deiminase and transglutaminases. J Biol Chem. 1997;272:27893–27901. doi: 10.1074/jbc.272.44.27893. [DOI] [PubMed] [Google Scholar]
- Tarcsa E, Candi E, Kartasova T, Idler WW, Marekov LN, Steinert PM. Structural and transglutaminase substrate properties of the small proline-rich 2 family of cornified cell envelope proteins. J Biol Chem. 1998;273:23297–23303. doi: 10.1074/jbc.273.36.23297. [DOI] [PubMed] [Google Scholar]
- Tsai FM, Shyu RY, Jiang SY. RIG1 inhibits the Ras/mitogen-activated protein kinase pathway by suppressing the activation of Ras. Cell Signal. 2006;18:349–358. doi: 10.1016/j.cellsig.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Tsai FM, Shyu RY, Jiang SY. RIG1 suppresses Ras activation and induces cellular apoptosis at the Golgi apparatus. Cell Signal. 2007;19:989–999. doi: 10.1016/j.cellsig.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Xue L, Rando RR. Roles of cysteine 161 and tyrosine 154 in the lecithin-retinol acyltransferase mechanism. Biochemistry. 2004;43:6120–6126. doi: 10.1021/bi049556f. [DOI] [PubMed] [Google Scholar]
- Yang JM, Ahn KS, Cho MO, Yoneda K, Lee CH, Lee JH, Lee ES, Candi E, Melino G, Ahvazi B, Steinert PM. Novel mutations of the transglutaminase 1 gene in lamellar ichthyosis. J Invest Dermatol. 2001;117:214–218. doi: 10.1046/j.0022-202x.2001.01429.x. [DOI] [PubMed] [Google Scholar]