Abstract
Today we can generate hundreds of gigabases of DNA and RNA sequencing data in a week for less than US$5,000. The astonishing rate of data generation by these low-cost, high-throughput technologies in genomics is being matched by that of other technologies, such as real-time imaging and mass spectrometry-based flow cytometry. Success in the life sciences will depend on our ability to properly interpret the large-scale, high-dimensional data sets that are generated by these technologies, which in turn requires us to adopt advances in informatics. Here we discuss how we can master the different types of computational environments that exist — such as cloud and heterogeneous computing — to successfully tackle our big data problems.
The wave of new technologies in genomics — such as ‘third-generation’ sequencing technologies1, sophisticated imaging systems and mass spectrometry-based flow cytometry2 — are enabling data to be generated at unprecedented scales. As a result, we can monitor the expression of tens of thousands of genes simultaneously3,4, score hundreds of thousands of SNPs in individual samples5, sequence an entire human genome for less than US$5,000 (REF. 6) and relate these data patterns to other biologically relevant information.
In under a year, genomics technologies will enable individual laboratories to generate terabyte or even petabyte scales of data at a reasonable cost. However, the computational infrastructure that is required to maintain and process these large-scale data sets, and to integrate them with other large-scale sets, is typically beyond the reach of small laboratories and is increasingly posing challenges even for large institutes.
Luckily, the computational field is rife with possibilities for addressing these needs. Life scientists have begun to borrow solutions from fields such as high-energy particle physics and climatology, which have already passed through similar inflection points. Companies such as Microsoft, Amazon, Google and Facebook have also become masters of petabyte-scale data sets — as they have been linking pieces of data that are distributed over a massively parallel architecture in response to a user’s requests and presenting them to the user in a matter of seconds. Following these advances made by others, we provide an overview and guidance on the types of computational environments that currently exist and that, in the immediate future, can tackle many of the big data problems now being faced by the life sciences.
Computational solutions range from cloud-based computing to an emerging revolution in high-speed, low-cost heterogeneous computational environments. But are life scientists ready to embrace these possibilities? If you are a life scientist confronted with the task of analysing a Mount Everest of data, and you are wondering how to derive meaning from them using Microsoft Excel 2007’s 1,048,576 row and 16,384 column limit, then this article will take you through the steps needed to allow you to compete with more computationally savvy groups.
In this Review, we define the typical workflows associated with the generation of high-throughput biological data, the challenges in those workflows, and how cloud computing and heterogeneous computational environments can help us to overcome these challenges. We then describe how complex data sets can be distilled to obtain higher-order biological relationships, and we discuss the direction that computation must take to help us to further our understanding at the cellular, tissue, organism, population and community levels.
Challenges posed by large-scale data analysis
Understanding how living systems operate will require the integration of the many layers of biological information that high-throughput technologies are generating.
As an example, the amount of data from large projects such as 1000 Genomes will collectively approach the petabyte scale for the raw information alone. The situation will soon be exacerbated by third-generation sequencing technologies7 that will enable us to scan entire genomes, microbiomes and transcriptomes and to assess epigenetic changes directly8 in just minutes, and for less than US$100. To this should be added data from imaging technologies, other high-dimensional sensing methods and personal medical records. Although processing individual data dimensions is complex (for example, uncovering functional DNA variation in multiple cancer samples using whole-genome sequencing), the true challenge is in integrating the multiple sources of data. Mining such large high-dimensional data sets poses several hurdles for storage and analysis. Among the most pressing challenges are: data transfer, access control and management; standardization of data formats; and accurate modelling of biological systems by integrating data from multiple dimensions.
Data transfer, access control and management
Analysis results can markedly increase the size of the raw data, given that all relationships among DNA, RNA and other variables of interest are stored and mined. Therefore, it is important to efficiently move these big data sets around the internet, to provide for access control if the data are stored centrally (to reduce storage costs) and to properly organize large-scale data in ways that facilitate analyses. Network speeds are too slow to enable terabytes of data to be routinely transferred over the web. Currently, the most efficient mode of transferring large quantities of data is to copy the data to a big storage drive and then ship the drive to the destination. This is inefficient and presents a barrier for data exchange between groups. One solution is to house the data sets centrally and bring the high-performance computing (HPC) to the data. Although this is an attractive solution, it presents access control challenges, as groups generating the data may want to retain control over who can access the data before they are published. Furthermore, controlling access to big data sets requires IT support, which is costly.
Mining the data for discovery crucially requires managing and organizing big data sets — consider, for example, the task of comparing whole-genome sequence data from multiple tumour and matched adjacent normal tissue pairs. Retrieving sequences, over all pairs, that map to many different genomic regions would not be trivial on inappropriately organized data.
Standardizing data formats
Different centres generate data in different formats, and some analysis tools require data to be in particular formats or require different types of data to be linked together. Thus, time is wasted reformatting and re-integrating data multiple times during a single analysis. For example, next-generation sequencing companies do not deliver raw sequencing data in a format common to all platforms, as there is no industry-wide standard beyond simple text files that include the nucleotide sequence and the corresponding quality values. As a result, carrying out sequence analyses across different platforms requires tools to be adapted to specific platforms.
It is therefore crucial to develop interoperable sets of analysis tools that can be run on different computational platforms depending on which is best suited for a given application, and then stitch those tools together to form analysis pipelines.
Modelling the results
A primary goal for biological researchers is to integrate diverse, large-scale data sets to construct models that can predict complex phenotypes such as disease. As mentioned above, constructing predictive models can be computationally demanding. Consider, for example, reconstructing Bayesian networks using large-scale DNA or RNA variation, DNA–protein binding, protein interaction, metabolite and other types of data. As the scales and diversity of the data grow, this type of modelling will become increasingly important for representing complex systems and predicting their behaviour. Computationally, however, the need for this type of modelling poses an intense problem that falls into the category of NP hard problems9 (FIG. 1). Finding the best Bayesian network by searching through all possible networks is a complex process; this is true even in cases in which there are only ten genes (or nodes), given that there would be in the order of 1018 possible networks. As the number of nodes increases, the number of networks to consider grows superexponentially. The computational environments that are required to organize vast amounts of data, build complex models from them and then enable others to interpret their data in the more informative context of existing models are beyond what is available today in the life sciences.
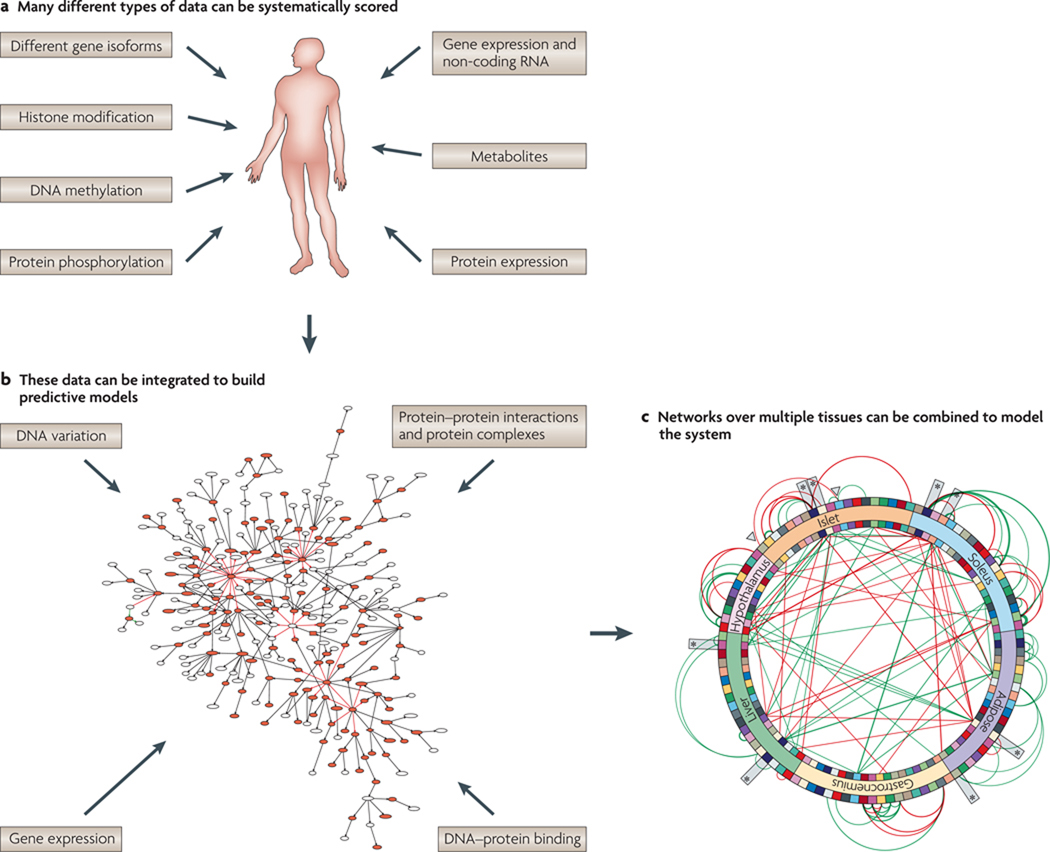
Figure 1. Generating and integrating large-scale, diverse types of data.
Modelling living systems will require generating (a) and integrating (b) multidimensional data sets. In b, large-scale, complex data sets are shown as a network in which the nodes represent variables of biological interest, such as DNA variation, RNA variation, protein levels, protein states, metabolite levels and disease-associated traits, and the edges between these nodes represent causal relationships between the variables. These more granular networks (at the gene level) can be effectively summarized into subnetworks (c) that interact with one another both within and between tissues. In this way, a network-centred view is obtained of how core biological processes interact with one another to define physiological states associated with disease. Part b is adapted, with permission, from REF. 40 © (2009) Macmillan Publishers Ltd. All rights reserved.
Meeting the challenge
Understanding your computational problem
Addressing big data and computational challenges requires efficiently targeting limited resources — money, power, space and people — to solve an application of interest. In turn, this requires understanding and exploiting the nature of the data and the analysis algorithms. Factors that must be taken into account to solve a particular problem most efficiently include: the size and complexity of the data; the ease with which data can be efficiently transported over the internet; whether the algorithm to apply to the data can be efficiently parallelized; and whether the algorithm is simple (for example, an algorithm used to compute the mean and standard deviation of a vector of numbers) or complex (for example, an algorithm applied to reconstructing Bayesian networks through the integration of diverse types of large-scale data) (BOX 1).
Box 1 | Understanding the informatics components of your problem.
Selecting the best computational platform for your problem of interest requires an understanding of the magnitude and complexity of the data, as well as the memory, network bandwidth and computational constraints of the problem. This is because different platforms have different strengths and weaknesses with respect to these constraints. One of the key technical metrics to consider is OPs/byte. Others are described below.
Understanding the nature of the data
Can the data be efficiently copied via the internet to the computational environment in which it will be processed along with other data? The size of your data set, the location of other data sets needed to process your data set and the network speed between the data set locations and computational environment determine whether your problem will be ‘network bound’.
Can the data be efficiently managed on a single disk-storage device for processing, or do they need to be distributed over many disk storage systems? Extremely large data sets that cannot be processed on a single disk, but instead demand a distributed storage solution for processing, are said to be ‘disk bound’.
Can the data be efficiently managed for processing by existing computer memory? Certain applications, such as constructing weighted co-expression networks36, operate on the data most efficiently if they are held in a computer’s random access memory (RAM). If the data set is too large to hold in memory for a particular application, the application is said to be ‘memory bound’.
Disk- and network-bound applications may be more dependent on the broader system than on the type of processor. These types of applications benefit from targeted investment in system components or a distributed approach that assembles large, aggregate memory or disk bandwidth from clusters of low-cost, low-power components37. In some instances, such as during the construction of weighted co-expression networks, expensive special-purpose supercomputing resources may be required.
Understanding the analysis algorithms
Does the processing require algorithms that are computationally intense, such as the class of NP hard algorithms? Reconstructing Bayesian networks is an example of an NP hard problem, that is, one that requires supercomputing resources (resources capable of trillions of floating point operations per second or more) to solve effectively and in a timely manner. Such problems are considered to be ‘computationally bound’.
Computationally bound applications can benefit from a particular processor or the use of a specialized hardware accelerator. This was the case in the early days of DNA and protein sequencing: determining best alignments was a computationally intense operation. Popular algorithms such as the Smith–Waterman alignment algorithm were substantially accelerated using specialized hardware.
One of the most important aspects to consider for computing large data sets is the parallelization of the analysis algorithms. Computationally or data-intensive problems are primarily solved by distributing tasks over many computer processors. Because different algorithms used to solve a problem are amenable to different types of parallelization, different computational platforms (TABLE 1) are needed to achieve the best performance.
Table 1.
Main categories of high-performance computing platforms
| Large-scale computing platform |
computing architectures | Advantages | Disadvantages | example applications |
|---|---|---|---|---|
| Cluster computing | Multiple computers linked together, typically through a fast local area network, that effectively function as a single computer | Cost-effective way to realize supercomputer performance | Requires a dedicated, specialized facility, hardware, system administrators and IT support |
|
| Cloud computing | Computing capability that abstracts the underlying hardware architectures (for example, servers, storage and networking), enabling convenient, on-demand network access to a shared pool of computing resources that can be readily provisioned and released (NIST Technical Report) | The virtualization technology used results in extreme flexibility; good for one-off HPC tasks, for which persistent resources are not necessary | Privacy concerns; less control over processes; bandwidth is limited as large data sets need to be moved to the cloud before processing |
|
| Grid computing | A combination of loosely coupled networked computers from different administrative centres that work together on common computational tasks. Typified by volunteer computing efforts (such as Folding@Home), which ‘scavenge’ spare computational cycles from volunteers’ computers | Ability to enlist large-scale computational resources at low or no cost (large-scale volunteer-based efforts) | Big data transfers are difficult or impossible; minimal control over underlying hardware, including availability |
|
| Heterogeneous computing | Computers that integrate specialized accelerators — for example, GPUs or reconfigurable logic (FPGAs) — alongside GPPs | Cluster-scale computing for a fraction of the cost of a cluster; optimized for computationally intensive fine-grained parallelism; local control of data and processes | Significant expertise and programmer time required to implement applications; not generally available in cluster- and cloud-based services |
|
The above categories are not exclusive. For example, heterogeneous computers are often used as the building blocks of cluster, grid or cloud computing systems; the shared computational clusters available in many organizations could be described as private Platform as a Service (PaaS) clouds. The main differences between the platforms are degree of coupling and tenancy — grid and cloud computers are designed for loosely coupled parallel workloads, with the grid resources allocated exclusively for a single user, whereas the underlying hardware resources in the cloud are typically shared among many users (multi-tenancy). Cluster computers are typically used for tightly coupled workloads and are often allocated to a single user. FPGA, field-programmable gate array; GPP, general purpose processor; GPU, graphics processing unit; GWA, genome-wide association; HPC, high-performance computing; NIST, National Institute of Standards and Technology.
We can classify the types of parallelism into two broad categories: loosely coupled (or coarse-grained) parallelism and tightly coupled (or fine-grained) parallelism. In loosely coupled parallelism, little effort is required to break up a problem into parallel tasks. For example, consider the problem of computing all genetic associations between thousands of gene expression traits and hundreds of thousands of SNP genotypes assayed in a tissue-specific cohort10. Each SNP–trait pair (or a given SNP tested against all traits) can be computed independently of the other pairs, so the computation can be carried out on independent processors or even completely separate computers. Conversely, tightly coupled parallelism requires a substantial programming effort, and possibly specialized hardware, because communication between the different parallel tasks using specialized frameworks must be maintained with minimal delay. The message passing interface (MPI)11 is an example of such a framework. In the context of the example just given involving the genetics of gene expression, instead of testing for SNP–trait associations independently, one could seek to partition the traits into modules of interconnected expression traits that are associated with common sets of SNPs12. This same problem was also solved through a Bayesian approach12 using a Markov chain Monte Carlo (MCMC) method that combines the two types of parallelism: the construction of each Markov chain is a coarsely parallel problem, but the construction of each chain is also an example of a fine-grained parallelism in which the algorithm was coded using MPI.
Computational solutions
Solutions to integrating the new generation of large-scale data sets require approaches akin to those used in physics, climatology and other quantitative disciplines that have mastered the collection of large data sets. Cloud computing and heterogeneous computational environments are relatively recent inventions that address many of the limitations mentioned above relating to data transfer, access control, data management, standardization of data formats and advanced model building (FIG. 2).
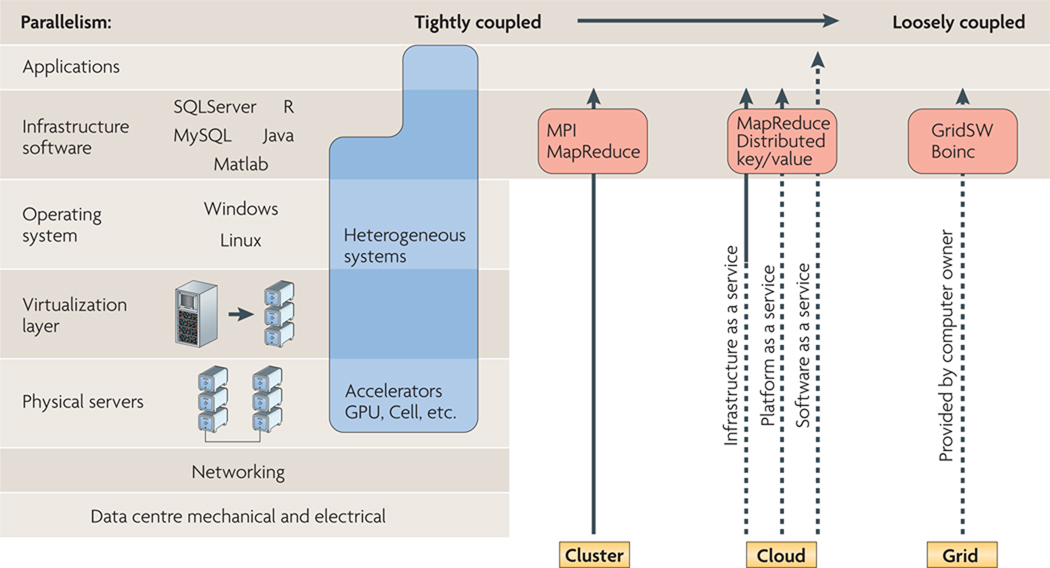
Figure 2. Cluster, cloud, grid and heterogeneous computing hardware and software stacks.
The hardware and software stacks comprise the different layers of a computational environment. At the lowest level of the stack is the physical structure that houses the hardware, with networking infrastructure coming next, and then the physical computers or servers. Sitting on top of the physical hardware is the virtualization layer, and the operating system lies on top of that. Finally, there are the software infrastructure and application layers. The different types of computing can be differentiated by which of these layers are under the user’s direct control (solid line) and which levels are provided by others, for example, the cloud provider and grid volunteer (dashed lines). Cloud and grid services are best suited for applications with loosely coupled, or coarse-grained, parallelism. Heterogeneous systems include specialized hardware accelerators, such as graphics processing units (GPUs). These accelerators are optimized for massive tightly coupled, or fine-grained, parallelism. However, the software that runs on these accelerators differs from its general purpose processor (GPP) counterparts, and often must be specifically written for a particular accelerator. MPI, message passing interface.
Compared to general purpose processors (GPPs), heterogeneous systems can deliver a tenfold increase or greater in peak arithmetic throughput for a few hundred US dollars. Cloud computing can make large-scale computational clusters readily available on a pay-as-you-need basis. But both approaches have trade-offs that result from trying to optimize for peak performance (heterogeneous systems) or low-cost and flexibility (cloud computing).
It is important that the working scientists understand the advantages and disadvantages of these different computational platforms and the problems to which they are best suited (TABLE 1). Computational platforms can be classified by the performance characteristic that they aim to optimize and how they allocate resources to do so. In the following sections we attempt to address exactly this issue, with particular attention to how heterogeneous systems and cloud computing have changed the traditional cost–capability trade-offs that users consider when deciding on the approach that best suits their computing needs. We provide an overview of these computational systems and directions for choosing the ideal computational architecture for an application of interest.
Cloud computing and MapReduce
Cluster-based and grid computing
HPC has been transformed in the past decade by the maturation of ‘cluster-based computing’ (FIG. 2). With a wide range of components available (for example, different networks, computational nodes and storage systems), clusters can be optimized for many classes of computationally intense applications. Consider the annotation of genes predicted in novel bacterial genomes, an important research problem for understanding microbiomes and how they interact with our own genomes to alter phenotypes of interest13–15. Using BLASTP16 to search 6,000 predicted genes against, say, the non-redundant protein (NR) database is a moderately computationally intense problem. Searching one gene against the NR database on a standard desktop computer would take ~30 s, so searching all 6,000 predictions could take 4 days. Distributing the searches over 1,000 central processing units (CPUs) would complete the search in less than 10 min. Although not all applications experience improvements on this scale, cluster-based computing can markedly accelerate these types of operations. However, there is a significant cost associated with building and maintaining a cluster. Even after the hardware components are acquired and assembled to build an HPC cluster, substantial costs are associated with operating the cluster, including those of space, power, cooling, fault recovery, backup and IT support. In this sense a cluster contrasts with grid computing, an earlier form of cluster-like computing. Here, a combination of loosely coupled networked computers that are administrated independently work together on common computational tasks at low or no cost, as individual computer owners volunteer their systems for such efforts (for example, the Folding@Home project).
Cloud computing
More recently, supercomputing has been made more accessible and affordable through the development of virtualization technology. Virtualization software allows systems to behave like a true physical computer, but with the flexible specification of details such as number of processors, memory and disk size, and operating system. Multiple virtual machines can often be run from a single physical server, providing significant cost savings in server hardware, administration and maintenance.
The use of these on-demand virtual computers17,18 is known as cloud computing. The combination of virtual machines and large numbers of affordable CPUs has made it possible for internet-based companies such as Amazon, Google and Microsoft to invest in ‘mega-scale’ computational clusters and advanced, large-scale data-storage systems. These companies offer on-demand computing to tens of thousands of users simultaneously, with flexible computer architectures that can manipulate and process petabyte scales of data (BOX 2).
Box 2 | Should I run my analysis in the cloud?
With the rise of remote, or cloud, computational resources, the ‘where’ has become as important a question for scientific computing as the ‘how’. At the terabyte, or even gigabyte, scale it is often more efficient to bring the computation to the data.
As a rule of thumb, 100,000 central processing unit (CPU) cycles are required to amortize the cost of transferring a single byte of data to a remote computational resource via the internet (equivalent to roughly 1 second per megabyte on current high-end laptops)38. For example, constructing a Bayesian network using state-of-the-art algorithms can require in the order of 1018 computational cycles on current high-end general purpose processors (GPPs). Given such computational demands on problems that may involve no more than hundreds of megabytes of data3,10,39, it is cost effective to pipe such data through the internet to high-end computational environments to carry out such an operation.
However, if summarizing the distribution of allele frequencies in a population of 50,000 individuals genotyped at 1,000,000 SNPs (about one trillion bytes, or a terabyte, of data), only 1012 computational cycles are required, orders of magnitude less than the 1017 computational cycles that one would need to run to break even in piping a terabyte of data over the internet.
Even so, remote systems can offer compelling ease of use and significant cost efficiencies, and in many cases large shared data sets are already being hosted in the cloud. Again, understanding how best to spend one’s resources is key. The online presentation associated with this paper (‘Computational solutions to large-scale data management’) provides a decision tree that can be used to help users decide on the most appropriate platform for their problem.
Advantages of cloud computing
Cloud computing platforms offer a convenient solution for addressing computational challenges in modern genetics research, beyond what could be achieved with traditional on-site clusters. For example, paying for cloud computing is typically limited to what you use — that is, the user requests a computer system type on demand to fit their needs and only pays for the time in which they used an instance of that system. Administrative functions such as backup and recovery are included in this cost. This pay-as-you-go model provides enormous flexibility: a computational job that would normally take 24 hours to complete on a single virtual computer can be sped up to 1 hour on 24 virtual computers for the same nominal cost. Such flexibility is difficult to achieve in the life sciences outside the large data centres in the genome sequencing centres or larger research institutes, but it can be achieved by anyone leveraging the cloud computing services offered by large information centres such as Amazon or Microsoft. In fact, Microsoft Research and the US National Science Foundation (NSF) recently initiated a programme to offer individual researchers and research groups selected through NSF’s merit review process free access to advanced cloud computing resources (see ‘Microsoft and the National Science Foundation enable research in the cloud’ on the Microsoft News Center website).
In addition to flexibility, cloud computing addresses one of the challenges relating to transferring and sharing data, because data sets and analysis results held in the cloud can be shared with others. For example, Amazon Web Services provides access to many useful data sets, such as the Ensembl and 1000 Genomes data. Also, to minimize cost and maximize flexibility, cloud vendors offer almost all aspects of the computer system, not just the CPU, as a service. For example, persistent data are often maintained in networked storage services, such as Amazon S3.
Disadvantages of cloud computing
The downsides of cloud computing are a reduced control over the distribution of the computation and the underlying hardware, and the time and cost that are required to transfer large volumes of data to and from the cloud. Although cloud computing offers flexibility and easy access to big computational resources, it does not solve the data transfer problem. Network bandwidth issues make the transfer of large data sets into and out of the cloud or between clouds impractical, making it difficult for groups using different cloud services to collaborate easily.
Furthermore, there are privacy concerns relating to the hosting of data sets on publicly accessible servers, as well as issues related to storage of data from human studies (for example, those posting human data must ensure compliance with the Health Insurance Portability and Accountability Act (HIPAA)).
MapReduce
Several years ago, a distributed computing paradigm known as MapReduce emerged to simplify the development of massively parallel computing applications and provide better scalability and fault tolerance for computing procedures involving many (>100) simultaneous processes19. In the context of distributed computing, MapReduce refers to the splitting of a problem into many homogeneous sub-problems in a ‘map’ step, followed by a ‘reduce’ step that combines the output of the smaller problems into the whole desired output. Whereas the Google implementation of distributed MapReduce remains proprietary, an open-source implementation of the concept is widely available through the Hadoop project19. In addition to meeting the required computational demand, this project addresses several of the data access and management challenges, as it provides for useful abstractions such as distributed file systems, distributed query language and distributed databases. An example of a problem that can be solved using MapReduce is that of aligning raw sequence reads that have been generated from a whole-genome sequencing run against those of a reference genome. The homogeneous sub-problems in this case (the map step) consist of aligning individual reads against the reference genome. Once all reads are aligned, the reduce step consists of aggregating all of the aligned reads into a single alignment file. In a later section, we cover in detail how you could implement this problem using the Amazon Web Services’ Elastic MapReduce resource.
Combining MapReduce and cloud computing
The combination of distributed MapReduce and cloud computing can be an effective answer for providing petabyte-scale computing to a wider set of practitioners. Several recent papers have demonstrated the feasibility of this concept by implementing MapReduce workflows on cloud-based resources for searching sequence databases20 and aligning raw sequencing reads to reference genomes21,22. The work by Langmead21 advances this trend one step further by converting a traditional two-stage workflow — sequence alignment followed by consensus calling — into a single application of a MapReduce workflow. The resultant pipeline scales well with additional computational resources: a whole-genome SNP analysis that started with 38× sequencing coverage of the human genome was completed in less than 3 hours using a 320-CPU cluster.
The MapReduce model is often a straightforward fit for data-parallel approaches in which a single task, such as read alignment, is split into smaller identical sub-tasks that operate on an independent subset of the data. Many large-scale analyses in biology fit into this ‘embarrassingly parallel’ decomposition (for example, sequence searches, image recognition, read alignments and protein ID by mass spectrometry). The use of distributed file systems (implicit in distributed MapReduce) becomes a necessity when operating on data sets of a terabyte scale or larger.
One of the bigger advantages of combining MapReduce and cloud computing is the reuse of a growing community of developers and software tools, and with just a few mouse clicks you can set up a MapReduce cluster with many nodes. For example, the Amazon EC2 cloud-computing environment provides a specific service for streamlining the set-up and running of Hadoop-based workflows (see the Amazon Machine Images website). The ease with which these workflows can be established has no doubt fuelled the movement of companies such as Eli Lilly to carry out their bioinformatics analyses on Amazon’s EC2 cloud-computing environment23.
As a measure of how ready this platform is for widespread use, Hadoop is now being taught to undergraduate students across the United States24. A new generation of computer users is therefore being trained to view the internet, rather than a laptop computer, as its computing device.
However, not all large-scale computations are best treated with a simple MapReduce approach. For example, algorithms with complex data access patterns, such as those for reconstructing probabilistic gene networks, require special treatment. Distributed MapReduce should be viewed as an emerging successful model for petabyte-scale computing, but not necessarily the only model to consider when mapping a particular problem to larger-scale computing.
Future developments and applications
Several emerging trends in cloud computing will influence petabyte-scale computing for biology. The analysis of large biological data sets retrieved from standard gateways, such as Ensembl, GenBank, Protein Data Bank (PDB), Gene Expression Omnibus (GEO), UniGene and the database of Genotypes and Phenotypes (dbGAP), can incur high network traffic and, consequently, generate redundant copies of the data that are distributed around the world. By placing these types of data sets into cloud-based storage and attaching them to cloud-based analysis clusters, biological analyses can be brought to the data without creating redundant copies and with a greatly reduced transfer start-up time25. The use of cloud resources in this context is also being seen as an ideal solution for data storage from biomedical consortia efforts that require the consolidation of large data sets from multiple distributed sites around the globe26,27.
Standardization and easy access to petabyte-scale computing will soon enable an ecosystem of reusable scientific tools and workflows. One would simply transfer or point to their large data set, select their favourite workflow of choice and receive results back in the form of files in cloud-based storage. The foundations for this model of reusable petabyte-scale workflows can be seen in the success of virtual appliance marketplaces such as those hosted by Amazon, 3Tera, VMware and Microsoft.
Heterogeneous computational environments
A complementary system to cloud-based computing is the use of heterogeneous multiple core (one-CPU) computers — integrated specialized accelerators that increase peak arithmetic throughput by 10-fold to 100-fold and can turn individual computers, the work-horses of both desktop and cluster computers, into mini-supercomputers28.
The accelerators consist of graphics processing units (GPUs) that operate alongside the multi-core GPPs that are commonly found in desktop and laptop computers. Similar to cloud computing, heterogeneous systems are helping to expand access to HPC capabilities over a broad range of applications. Modern GPUs were driven by the videogame industry to deliver ever more realistic, real-time gaming environments to consumers. Although the working scientist is more likely to encounter GPUs, some vendors sell field-programmable gate array (FPGA)-based accelerators for genomic applications (for example, the CLC Bioinformatics Cube). Given that every modern computer includes a GPU, many of which are usable for general purpose computing, GPUs can be purchased for between US$100 and US$1,500. GPUs from NVIDIA and AMD/ATI offer ‘cluster-scale’ performance (more than one trillion floating point operations (FLOPS) per second) in an add-on card that costs only several hundred dollars.
General purpose heterogeneous computing has become feasible only in the past few years, but it has already yielded notable successes for biological data processing. The Folding@Home project uses a distributed client for protein folding simulation, with clients available for GPUs. Although GPUs constitute only 5% of the processors that are actively engaged in this project, they contribute 60% of the total FLOPS29,30. The GPU port of NAMD, a widely used program for molecular dynamics simulation, running on a 4-GPU cluster outperforms a cluster with 16 quad-core GPPs (48 cores). A GPU version of the sequence alignment program MUMmer was developed and achieved a roughly three- to fourfold speed increase over the serial-CPU version31. Several important algorithms have been developed for the GPU for annotating genomes, identifying SNPs and identifying RNA isoforms. They include CUDASW++, a GPU version of the Smith–Waterman sequence database search algorithm32, and Infernal, a novel RNA alignment tool33. Finally, in our own work in Bayesian network learning, the GPU implementation is 5 to 7.5 times faster than the heavily optimized GPP implementation34.
In addition to these types of applications, programmers are writing GPU-accelerated plug-ins for commonly used functions in high-level programming frameworks such as R and Matlab. Furthermore, GPU-optimized libraries of software functions are also being developed, including cuBLAS for basic linear algebra operations, cuFFT for carrying out fast Fourier transforms and Thrust for large vector operations.
Advantages of heterogeneous computing
With speed increases of fivefold to tenfold, a laptop can be used in place of a dedicated workstation, a workstation in place of a small cluster, or a small cluster in place of a larger cluster. The improved performance results in significant cost savings to the researcher. Data can be stored and analysed locally without requiring any specialized infrastructure, such as heavy-duty cooling, high-amperage power or professional system administration. In this respect, heterogeneous systems complement cloud computing by radically changing the cost–capability trade-offs.
Because GPUs were primarily designed for real-time graphics and gaming, they are optimal for solving problems involving tightly coupled (or fine-grained) parallelism (TABLE 1). However, for such an application to benefit from a GPU, it must require sufficient computation to amortize the cost of transferring data between the GPP and the accelerator (the local analogue of computing on a local machine versus transferring data to the cloud to carry out computations on the data). For example, in constructing Bayesian networks with fewer than 25 nodes, the overhead is too great, in relation to the amount of work done by the GPU, to achieve an advantage over a GPP. By contrast, for networks with more than 25 nodes, the GPUs provide a substantial advantage. This highlights an important point: ideally, applications such as Bayesian network reconstructions would be implemented to run on several different computer architectures, as the characteristics for any given problem define the architecture that is best suited to solve that particular instance. Although not all applications can take advantage of heterogeneous systems, when they can they will benefit from an efficient alternative to assembling similar capabilities from clusters of general purpose computers.
Disadvantages of heterogeneous computing
One of the primary challenges in leveraging heterogeneous computational environments for scientific computing is that most applications of interest to geneticists and others in the life sciences have not been ported to these environments. Significant informatics expertise is required to develop or modify applications to run effectively on GPUs or FPGAs. Heterogeneous systems improve performance and efficiency by exposing to programmers certain architectural features, such as vector arithmetic units, that are unavailable in GPPs. Not only is the code that executes on these accelerators different from its GPP counterpart, but often entirely different algorithms are needed to take advantage of the unique capabilities of the specialized accelerators. As a result, developing applications for these architectures is more challenging than for traditional GPPs.
The implementation of algorithms to be run in a general purpose GPU environment is carried out using specialized programming languages, such as CUDA programming (a proprietary programming model for NVIDIA GPUs)35. To implement an algorithm using CUDA, the programmer must write sequential code for a single ‘thread’ that processes a small subset of the total data set. Then, when the associated program is run on a data set of interest, large numbers of these individual processes are created in a grid to process the entire data set. The GPU operates in a separate memory space from the GPP, and often uses its own, much faster graphics memory. However, as noted above, the disadvantage here is that, before launching the program, the programmer must explicitly copy to the GPU the necessary data, and then copy back the results after completion.
A tutorial for computing in the clouds
The number of cloud service providers, big data centres and third-party vendors aiming to facilitate your entry into cloud computing is growing quickly. The options for transferring data to a cloud and begin computing on it can be overwhelming (TABLE 2). However, service providers have now found simpler means of allowing users to access the cloud. An example of a simpler approach to cloud computing is the Amazon Web Services Management Console (FIG. 3). The management console — which can be accessed from any web browser — provides a simple and intuitive interface for several uses: moving data and applications into and out of the Amazon S3 storage system; creating instances in Amazon EC2; and running data-processing programs on those instances, including the analysis of big data sets using MapReduce-based algorithms.
Table 2.
Examples of cloud and heterogeneous computational environments
| Environment | URL |
|---|---|
| Cloud computing | |
| Amazon Elastic Compute Cloud | http://aws.amazon.com/ec2 |
| Bionimbus | http://www.bionimbus.org |
| NSF CluE | http://www.nsf.gov/cise/clue/index.jsp |
| Rackspace | http://www.rackspacecloud.com |
| Science Clouds | http://www.scienceclouds.org |
| Heterogeneous computing | |
| NVIDIA GPUs | http://www.nvidia.com |
| AMD/ATI GPUs | http://www.amd.com |
| Heterogeneous cloud computing | |
| SGI Cyclone Cloud | http://www.sgi.com/products/hpc_cloud/cyclone |
| Penguin Computing On Demand | http://www.penguincomputing.com/POD/Summary |
GPU, graphics processing unit; NSF, US National Science Foundation.
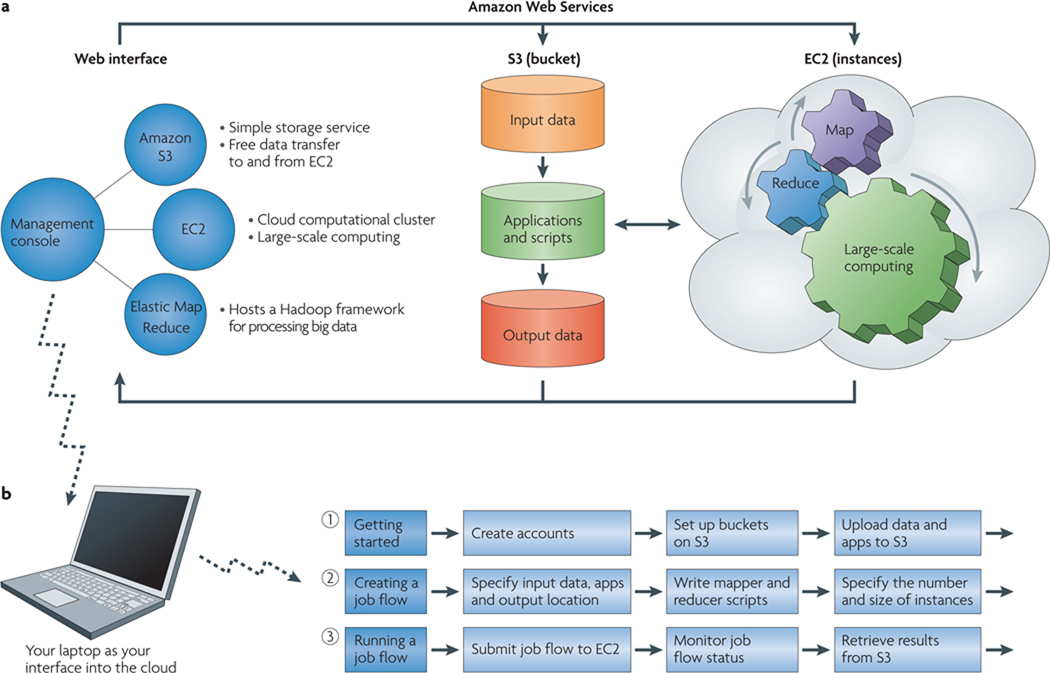
Figure 3. Amazon Web Services.
Amazon Web Services provides a simple and intuitive web-based interface into the Amazon S3 storage services and Amazon EC2 cloud resources. a | The management console available in Amazon Web Services provides a convenient interface into Amazon’s cloud-based services, including direct access to Amazon S3 and Amazon EC2 for data storage and large-scale computing, respectively. b | Steps for using the management console to compute big data using Amazon’s Elastic MapReduce resource (see main text for details).
The best way to understand how to begin processing data in the cloud is to run through an example: aligning raw sequencing reads to a reference genome. This application is on the rise owing to the drop in whole-genome sequencing costs and the development of third-generation sequencing technologies, such as the Pacific Biosciences single-molecule, real-time sequencing (SMRT sequencing) approach1,8. One of the first processing steps to be carried out on raw sequencing reads is to align the reads to a reference genome to derive a consensus sequence that can then be used to identify novel mutations, structural variations or other interesting genomic features.
As discussed above, this application fits well into the MapReduce framework21. The input data are the raw reads from the sequencing run and a reference genome to which to map the reads. Because the reads are mapped independently to the reference genome, the input reads can be partitioned into sets that are then distributed over multiple processors or cores (the map part of MapReduce) to carry out the alignments more rapidly than could be done on a single processor. The alignment files generated in this way can then be aggregated into a single alignment file after all reads are mapped (the reduce part of MapReduce). The lower panel of FIG. 3 details the steps needed to carry out this type of process using the Amazon Elastic MapReduce (EMR) resource. These steps are done in a user-guided manner using the management console to create a job flow consisting of three simple steps: uploading your input data and applications to Amazon S3; configuring the job flow and submitting it; and retrieving results from S3.
Getting started: uploading input data and applications
In our example, the first step consists of assembling the input files and applications to carry out the alignment procedure and uploading these files into your own personal ‘bucket’ on Amazon S3. Using the management console, you simply select an option to create buckets for the input files, for the applications and scripts used to process the data and for the output results (FIG. 3). In the alignment problem for SMRT reads, the input data are the raw reads from the SMRT sequencer and a reference genome of interest (in FASTA format). For MapReduce, the raw reads need to be split into, say, hundreds of raw reads files so that they can be distributed by the Elastic MapReduce resource over many different cores on Amazon EC2. The long-read aligner application (ReadMatcher) and documentation for this tool can be downloaded from the Pacific Biosciences Developers Network website. With all files assembled, the management console can be used to upload the files onto Amazon S3.
Defining the job flow
The next step is defining the job flow via the management console, which again is a user-guided series of steps that are initiated by clicking a ‘create workflow’ button in the management console. In configuring the workflow the user specifies which buckets contain the input files, the applications and the output location, and then specifies the size and number of instances on Amazon EC2 to allocate for the job flow, which will define the amount of memory and number of cores to make available for computing the alignments. In the map portion of MapReduce, a wrapper script calls the ReadMatcher application with each of the segmented raw read files. Because each segmented file contains different information, multiple instances of ReadMatcher can operate on each file independently on each Amazon EC2 instance. In the reduce component of MapReduce, another script takes all of the independent alignment results and combines — or reduces — them into a single alignment file. The mapper and reducer scripts are the only programming steps that need to be taken for this example. The scripting for this example is straightforward and involves substantially less development time than would traditional multi-process or multi-threading programming.
Running the job flow
After the job flow is configured it can be launched, again by submitting the job flow via the management console. Several tools are provided in the management console that can be used to monitor the progress of the run. The final step is retrieving the alignment results from Amazon S3. The results can be downloaded to your local system or can remain on S3 for shared use or for additional processing. For example, the alignment results in this example can be fed into another application, EviCons (available at the Pacific Biosciences Development Network), to derive a consensus sequence from the alignments, which can then be used to identify SNPs and other types of variation of interest.
Perspectives
The problems with data storage and analysis will continue to grow at a superexponential pace, with the complexity of the data only increasing as we are able to isolate and sequence individual cells, monitor the dynamics of single molecules in real time and lower the cost of the technologies that generate all of these data, such that hundreds of millions of individuals can be profiled. Sequencing DNA, RNA, the epigenome, the metabolome and the proteome from numerous cells in millions of individuals, and sequencing environmentally collected samples routinely from thousands of locations a day, will take us into the exabyte scales of data in the next 5–10 years. Integrating these data will demand unprecedented high-performance computational environments. Big genome centres, such as the Beijing Genomics Institute (BGI), are already constructing their own cloud-based computational environments with exabyte-scale data-storage capabilities and hundreds of thousands of cores.
Choosing the optimal computer architecture for storing, organizing and analysing big data sets requires an understanding of the problems one wishes to solve and the advantages of each of the architectures or mixture of architectures for efficiently solving such problems. The optimal solution may not always be obvious or may require a mixture of advanced computational environments. We anticipate the creation of more versatile cloud-based services that make use of computer architectures that are not limited to map-reducible problems, as well as low-cost, high-performance heterogeneous computational solutions that can serve as an adequate local solution for many laboratories.
Ultimately, our ability to consider very large-scale, diverse types of data collected on whole populations for the construction of predictive disease models will demand an open, data-sharing environment — not only from the perspective of industry but from academic communities as well, in which there are strong incentives to restrict data distribution to maintain competitive advantages. This will require the development of tools and software platforms that enable the integration of large-scale, diverse data into complex models that can then be operated on and refined by experimental researchers in an iterative fashion. This is perhaps the most crucial milestone we must achieve in the biomedical and life sciences if large-scale data and the results derived from them are to routinely affect biological research at all levels.
Glossary
- Petabyte
Refers to 1012 bytes. Many large computer systems now have many petabytes of storage.
- Cloud-based computing
The abstraction of the underlying hardware architectures (for example, servers, storage and networking) that enable convenient, on-demand network access to a shared pool of computing resources that can be readily provisioned and released.
- Heterogeneous computational environments
Computers that integrate specialized accelerators, for example, graphics processing units (GPUs) or field-programmable gate arrays (FPGAs), alongside general purpose processors (GPPs).
- High-performance computing
A catch-all term for hardware and software systems that are used to solve ‘advanced’ computational problems.
- Bayesian network
A network that captures causal relationships between variables or nodes of interest (for example, transcription levels of a gene, protein states, and so on). Bayesian networks enable the incorporation of prior information in establishing relationships between nodes.
- NP hard
For the purposes of this paper, NP hard problems are some of the most difficult computational problems; as such, they are typically not solved exactly, but with heuristics and high-performance computing.
- Algorithm
A well-defined method or list of instructions for solving a problem.
- Parallelization
Parallelizing an algorithm enables different tasks that are carried out by its implementation to be distributed across multiple processors, so that multiple tasks can be carried out simultaneously.
- Markov chain Monte Carlo
A general method for integrating over probability distributions so that inferences can be made around model parameters or predictions can be made from a model of interest. The sampling from the probability distributions required for this process draws samples from a specially constructed Markov chain: a discrete time random process in which the distribution of a random variable at a given point in time given the random variables at all previous time points is only dependent on the distribution of the random variable directly preceding it.
- General purpose processor
A microprocessor designed for many purposes. It is typified by the ×86 processors made by Intel and AMD and used in most desktop, laptop and server computers.
- OPs/byte
A technical metric that describes how many computational operations (OPs) are performed per byte of data accessed, and where those bytes originate.
- Random access memory
Computer memory that can be accessed in any order. It typically refers to the computer system’s main memory and is implemented with large-capacity, volatile DRAM modules.
- Cluster
Multiple computers linked together, typically through a fast local area network, that effectively function as a single computer.
- Cluster-based computing
An inexpensive and scalable approach to large-scale computing that lowers costs by networking hundreds to thousands of conventional desktop central processing units together to form a supercomputer.
- Computational node
The unit of replication in a computer cluster. Typically it consists of a complete computer comprising one or more processors, dynamic random access memory (DRAM) and one or more hard disks.
- Central processing unit
(CPU). A term often used interchangeably with the term ‘processor’, the CPU is the component in the computer system that executes the instructions in the program.
- Virtualization
Refers to software that abstracts the details of the underlying physical computational architecture and allows a virtual machine to be instantiated.
- Operating system
Software that manages the different applications that can access a computer’s hardware, as well as the ways in which a user can manipulate the hardware.
- Health Insurance and Portability and Accountability Act
(HIPAA.) United States legislation that regulates, among many things, the secure handling of health information.
- Distributed file system, distributed query language and distributed database
A file system, query language or database that allows access to files, queries and databases, respectively, from many different hosts that are networked together and that enable sharing via the network. In this way, many different processes (or users) running on many different computers can share data, share database and storage resources and execute queries in a large grid of computers.
- Core
Typically used in the context of multi-core processors, which integrate multiple cores into a single processor.
- Graphics processing unit
(GPU.) A specialized processor that is designed to accelerate real-time graphics. Previously narrowly tailored for that application, these chips have evolved so that they can now be used for many forms of general purpose computing. GPUs can offer tenfold higher throughput than traditional general purpose processors (GPPs).
- Field-programmable gated array
(FPGA). Digital logic that can be reconfigured for different tasks. It is typically used for prototyping custom digital integrated circuits during the design process. Modern FPGAs include many embedded memory blocks and digital signal-processing units, making them suitable for some general purpose computing tasks.
- Floating point operations
(FLOPS). The count of floating point arithmetic operations (an approximation of operations on real numbers) in an application.
- Single-molecule, real-time sequencing
(SMRT sequencing). Pacific Biosciences’ proprietary sequencing platform in which DNA polymerization is monitored in real time using zero-mode waveguide technology. SMRT sequencing produces much longer reads than do current second-generation technologies (averaging 1,000 bp or more versus 150–400 bp). It also produces kinetic information that can be used to detect base modifications such as methyl-cytosine.
- Bucket
Fundamental storage unit provided to Amazon S3 users to store files. Buckets are containers for your files that are similar conceptually to a root folder on your personal hard drive, but in this case the file storage is hosted on Amazon S3.
- Exabyte
Refers to 1018 bytes. For context, CISCO estimates that the monthly global internet traffic in the spring of 2010 was 21 exabytes.
Footnotes
Competing interests statement
E.E.S., J.S. and L.L. declare competing financial interests: see Web version for details.
FURTHER INFORMATION
1000 Genomes Project: http://www.1000genomes.org
3Tera Application Store: http://appstore.3tera.com
Amazon Machine Images: http://developer.amazonwebservices.com/connect/kbcategory.jspa?categoryID=171
Amazon Web Services Management Console (MC): http://aws.amazon.com/console
CLC Bioinformatics Cube: http://www.clccube.com
Collaboration between the National Science Foundation and Microsoft Research for access to cloud computing: http://www.microsoft.com/presspass/press/2010/feb10/02-04nsfpr.mspx
Condor Project: http://www.cs.wisc.edu/condor
Database of Genotypes and Phenotypes (dbGAP): http://www.ncbi.nlm.nih.gov/gap
Ensembl: http://www.ensembl.org/index.html
GenBank: http://www.ncbi.nlm.nih.gov/genbank
Gene Expression Omnibus (GEO): http://www.ncbi.nlm.nih.gov/geo
Nature Reviews Genetics audio slide show on ‘Computational solutions to large-scale data management’: http://www.nature.com/nrg/multimedia/compsolutions/index.html
NIST Technical Report: http://csrc.nist.gov/groups/SNS/cloud-computing
NVIDIA Bio WorkBench: http://www.nvidia.com/object/tesla_bio_workbench.html
Pacific Biosciences Developers Network: http://www.pacbiodevnet.com
Protein Data Bank (PDB): http://www.pdb.org
Public data sets available through Amazon Web Services: http://aws.amazon.com/publicdatasets
UniGene: http://www.ncbi.nlm.nih.gov/unigene
VMware Virtual Applicances: http://www.vmware.com/appliances
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 2.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 5.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 7.Munroe DJ, Harris TJ. Third-generation sequencing fireworks at Marco Island. Nature Biotech. 2010;28:426–428. doi: 10.1038/nbt0510-426. [DOI] [PubMed] [Google Scholar]
- 8. Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nature Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. Shows how SMRT sequencing will add an important time dimension to DNA and RNA sequencing data. Maximizing the information that can be derived from the data will demand substantially increased data-storage requirements and computational resources.
- 9.Garey MR, Johnson DS. Computers and Intractability: A Guide to the Theory of NP-Completeness. New York: W. H. Freeman; 1979. [Google Scholar]
- 10.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snir M. MPI-The Complete Reference. 2nd edn. Cambridge, Massachusetts: MIT Press; 1998. [Google Scholar]
- 12.Zhang W, Zhu J, Schadt EE, Liu JS. A Bayesian partition method for detecting pleiotropic and epistatic eQTL modules. PLoS Comput. Biol. 2010;6:e1000642. doi: 10.1371/journal.pcbi.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuczynski J, et al. Direct sequencing of the human microbiome readily reveals community differences. Genome Biol. 2010;11:210. doi: 10.1186/gb-2010-11-5-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armbrust M, et al. Above the Clouds: A Berkeley View of Cloud Computing. Berkeley: University of California; 2009. [Google Scholar]
- 18.Buyya R, Yeo CS, Venugopal S, Broberg J, Brandic I. Cloud computing and emerging IT platforms: vision, hype, and reality for delivering computing as the 5th utility. Future Generation Comput. Syst. 2009;25:599–616. [Google Scholar]
- 19. Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters; 6th Symp. on Operating Systems Design and Implementation; 2004. [online], http://www.usenix.org/events/osdi04/tech/full_papers/dean/dean_html. Introduces the MapReduce concept, which was developed at Google. MapReduce is one of the leading large-scale parallel computing technologies, both in terms of the size of data it can handle and the size of the computational infrastructure that is available to process such data.
- 20.Matsunaga A, Tsugawa M, Fortes J. 4th IEEE International Conference on eScience. Indianapolis, Indiana: IEEE; 2008. pp. 222–229. [Google Scholar]
- 21. Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. An early example in genomics of using standard cloud-based services to detect SNPs — in this case, by aligning whole-genome sequence data to a reference genome.
- 22.Schatz MC. CloudBurst: highly sensitive read mapping with MapReduce. Bioinformatics. 2009;25:1363–1369. doi: 10.1093/bioinformatics/btp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sansom C. Up in a cloud? Nature Biotech. 2010;28:13–15. doi: 10.1038/nbt0110-13. [DOI] [PubMed] [Google Scholar]
- 24.Vance A. Training to climb an Everest of digital data. New York Times. 2009 Oct 11;B1 [Google Scholar]
- 25. Stein LD. Towards a cyberinfrastructure for the biological sciences: progress, visions and challenges. Nature Rev. Genet. 2008;9:678–688. doi: 10.1038/nrg2414. A comprehensive review of the informatics infrastructure that will be required to achieve success in biological research, both now and in the future.
- 26.Constable H, Guralnick R, Wieczorek J, Spencer C, Peterson AT. VertNet: a new model for biodiversity data sharing. PLoS Biol. 2010;8:e1000309. doi: 10.1371/journal.pbio.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal A, et al. Cloud computing: a new business paradigm for biomedical information sharing. J. Biomed. Inform. 2009;43:342–353. doi: 10.1016/j.jbi.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Owens JD, et al. A survey of general-purpose computation on graphics hardware. Comput. Graph. Forum. 2007;26:80–113. [Google Scholar]
- 29.Friedrichs MS, et al. Accelerating molecular dynamic simulation on graphics processing units. J. Comput. Chem. 2009;30:864–872. doi: 10.1002/jcc.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luttmann E, et al. Accelerating molecular dynamic simulation on the cell processor and Playstation 3. J. Comput. Chem. 2009;30:268–274. doi: 10.1002/jcc.21054. [DOI] [PubMed] [Google Scholar]
- 31. Schatz MC, Trapnell C, Delcher AL, Varshney A. High-throughput sequence alignment using Graphics Processing Units. BMC Bioinformatics. 2007;8:474. doi: 10.1186/1471-2105-8-474. One of the first genomics applications to use GPUs to substantially speed up the process of high-throughput sequence alignments.
- 32.Liu Y, Maskell DL, Schmidt B. CUDASW + + : optimizing Smith–Waterman sequence database searches for CUDA-enabled graphics processing units. BMC Res. Notes. 2009;2:73. doi: 10.1186/1756-0500-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. One of the first GPU-based RNA sequence aligners.
- 34. Linderman MD, et al. High-throughput Bayesian network learning using heterogeneous multicore computers; Proc. of the 24th ACM Int. Conf. on Supercomputing; Japan; 2–4 Jun 2010; Tsukuba, Ibaraki. New York: ACM; 2010. pp. 95–104. http://doi.acm.org/10.1145/1810085.1810101. Describes a high-throughput GPU-based application for Bayesian network learning. The network learner was built with a novel software tool, the Merge compiler, that helps programmers to integrate multiple implementations of the same algorithm, targeting different processors, into a single application that optimally chooses at run-time which implementation to use based on the problem and hardware available.
- 35.Nickolls J, Buck I, Garland M, Skadron K. Scalable parallel programming with CUDA. Queue. 2008;6:40–53. [Google Scholar]
- 36.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 37. Barroso LA, Holzle U. The Datacenter as a Computer: An Introduction to the Design of Warehouse-Scale Machines. Morgan & Claypool Publishers; 2009. pp. 1–108. Highlights the important future role of large-scale data centres in hosting big data sets and facilitating computing on those sets.
- 38.Bell G, Gray J. Petascale computational systems: balanced cyberinfrastructure in a data-centric world. Microsoft Research. 2005 [online], http://research.microsoft.com/en-us/um/people/gray/papers/Petascale%20computational%20systems.pdf. [Google Scholar]
- 39. Zhu J, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nature Genet. 2008;40:854–861. doi: 10.1038/ng.167. An example of an integrative genomics network-reconstruction method that is among the most computationally demanding methods in biological research.
- 40.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nature Rev. Drug Discov. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]