Abstract
Background:
Polymorphisms in the drug-metabolizing enzymes are found to be associated with the inter-individual variation in response to a particular drug. Glutathione S-transferases (GSTs) are involved in the metabolism of several anticancer drugs, including alkylating agents, anthracyclines, and cyclophosphamides.
Aim:
The present study is aimed to examine the association of GST and CYP1A1*2A polymorphisms in the susceptibility to acute lymphoblastic leukemia (ALL) and the prognostic significance.
Materials and Methods:
A total of 92 immunophenotyped patients and 150 cord blood controls were genotyped by PCR for GSTM1 and GSTT1, RQ-PCR allelic discrimination assay for GSTP1 and PCR-RFLP for CYP1A1*2A polymorphism.
Results:
We have previously reported the significant association of GSTM1 (null) and combined GSTP1 {(Ile/Val)/ (Val/Val)} /GSTM1 (null) genotype with the susceptibility to ALL. No significant association was observed with GSTT1 (P=0.75) and CYP1A1*2A (P=0.61 for +/- and P=0.86 for -/- respectively) in the susceptibility to ALL. Survival analysis was performed in 50 of the 92 patients who were followed for three years. Kaplan-Meier survival analysis for three years showed significant lower event-free survival in patients harboring GSTP1 (Ile/Val) and GSTP1 (Val/Val) (P=0.038 and 0.0001, respectively) genotype. Cox regression analysis revealed GSTP1 as an independent prognostic marker with 6-fold higher risk with Val/Val genotype (P=0.003).
Conclusions:
Our results show that GSTP1 (Ile/Val) polymorphism has a role in the susceptibility to ALL and also influence treatment outcome.
Keywords: CYP1A1*2A, GSTM1, GSTP1, GSTT1, polymorphisms
INTRODUCTION
Pediatric acute lymphoblastic leukemia (ALL) is the most common malignant disease occurring during childhood and can be cured in more than 70% of cases using intensive multi-agent chemotherapeutic regimens.[1] Intensive treatment also has significant long-term consequences, including second malignancies and cognitive impairment. Thus, there is a need to identify factors associated with both the risk of relapse and chemotherapeutic toxicity.[2]
The glutathione S transferase (GST) family (phase II drug-metabolizing enzymes) is involved in the metabolism of a wide range of chemicals including environmental carcinogens, reactive oxygen species, and chemotherapeutic agents.[3] Four different gene families of GSTs (cytosolic enzymes) are known, namely α, μ, π, and θ, which have different but often overlapping substrate specificities.[4] Polymorphism in GST genes lead to either decreased activity of the enzyme or complete loss of enzyme activity. The most common variant of the GSTM1 and GSTT1 genes is homozygous deletion (null genotype), which has been associated with the loss of the enzyme activity.[5] GSTM1 activity is absent in about 40% to 60% of the Caucasian population and 20% to 30% of Caucasians show absence of GSTT1 activity.[6] GSTP1 (Ile105val) single nucleotide polymorphism affects substrate specific catalytic activity of the enzyme and its thermal stability.[6–8] Cytochrome P-450 (CYP) 1A1 is a key enzyme in phase I bioactivation of xenobiotics and its polymorphism is associated with elevated enzymatic activity.[9,10]
Polymorphisms in GSTs and CYPs are associated with increased risk for several types of cancers as well as influence the treatment outcome.[4–6,9,11–19] We have previously reported the association of GSTM1 and GSTP1 in the susceptibility to ALL.[20] The present study is aimed at examining the association of GSTM1, GSTT1, GSTP1, and CYP1A1*2A polymorphisms in the susceptibility to ALL as well as their prognostic significance. This is the first study to our knowledge to report the prognostic significance of GST polymorphisms in South Indian ALL patients.
MATERIALS AND METHODS
Study population
Our study included 92 immunophenotyped ALL patients (less than 25 years of age) treated during the period 2004–2007 at Cancer Institute (Women India Association), No. 38, Sardar Patel Road, Guindy, Chennai. Patients were treated with modified MCP 841 and BFM 86 protocols. At diagnosis, 8 ml of peripheral blood and 4 ml of bone marrow aspirate were collected in EDTA after obtaining informed consent from parents or patients as applicable. Controls comprised 150 cord blood samples collected in EDTA after obtaining informed consent from mothers delivering in Andhra Mahila Sabha hospital, Adyar, Chennai. Ethical committee clearances were obtained from both the institutions. The reason for selecting cord blood samples as controls instead of age-matched controls was explained elsewhere.[20]
Methods
DNA was extracted from lymphocytes using QIAmp DNA blood kit (QIAGEN Inc., Valencia, CA, USA) according to the manufacturer's instructions and quantitated using the Nanodrop spectrophotometer (ND 1000, Nanodrop Technologies.Inc, USA). The integrity of DNA was checked by PCR amplification of ABL gene. Genotyping for GSTM1 and GSTP1 was performed as described previously.[20] Genotyping of GSTT1 was performed by the PCR method described by Krajinovic et al.,[2] with modification. The reaction mixture consisted of 50 ng of genomic DNA, 10 pmoles of forward and reverse primers (Forward 5′-GCCCTGGCTAGTTGCTGAAG-3′ and Reverse 5′ GCATCTGATTTGGGGACCACA-3′), 25 mM Mgcl2, 200 mM dNTPs, 0.5 units of Amplitaq gold polymerase (Life Technologies, Foster city, CA, USA) in a 25μL reaction volume. The cycling conditions included an initial denaturation at 95°C for 10 minutes, followed by 35 cycles of denaturation, annealing and extension at 95°C, 64°C, and 72°C for 15 seconds, 30 seconds and 45 seconds each, and a final extension at 72°C for 5 minutes. The PCR products were run in a 2.5% agarose gel containing ethidium bromide. The presence of one or both the alleles was identified by a 112-bp PCR product fragment, whereas its absence indicates the null genotype. Positive and negative controls were included in each run.
Genotyping of CYP1A1*2A was performed as described by Krajinovic et al.[2] Briefly, PCR reaction mixture consisted of 50 ng of genomic DNA, 200 mM dNTPs, 25 mM Mgcl2, 1 unit of Taq polymerase, 10 pmoles of both forward and reverse primers (Forward 5′-GGCTGAGCAATCTGACCCTA-3′ and Reverse 5′-TAGGAGTCTTGTCTCATGCCT-3′). The cycling conditions included an initial denaturation at 95°C for 10 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 63°C for 1 minute, and extension at 72°C for 1 minute, and final extension at 72°C for 5 minutes. PCR product of 899-bp was digested with Msp1 and run in a 2% agarose gel. Polymorphism introduces Msp1-restricting site resulting in the presence of two fragments of 693 bp and 206 bp.
Statistics
The association of GST and CYP1A1*2A polymorphisms with the risk to develop ALL has been estimated by chi-square test. Logistic regression analysis was used to obtain the odds ratios with 95% confidence intervals. Combined analysis was performed by analyzing two genes at a time to check the gene–gene interactions. Event is defined as relapse at any site, induction failure or the death in remission. Survival duration corresponds to the time from the start of treatment to the occurrence of an event or the end of 3 years of treatment. The survival probabilities of different genotypes on event-free survival were estimated by Kaplan–Meier survival analysis. Survival curves were generated and log rank P values were used to test the significance of different polymorphisms. Univariate cox regression analysis was performed to test the prognostic significance of clinical factors and genetic variants.
RESULTS
The frequencies of GSTT1 and CYP1A1*2A genotypes among the cases and controls are given in Table 1. The allelic frequencies of CYP1A1*2A polymorphisms in cases and controls have been analyzed. The distributions of alleles were found to be in Hardy–Weinberg equilibrium.
Table 1.
Distribution of GSTT1 and CYP1A1*2A (rs4646903 3801T>C) polymorphisms among cases and controls
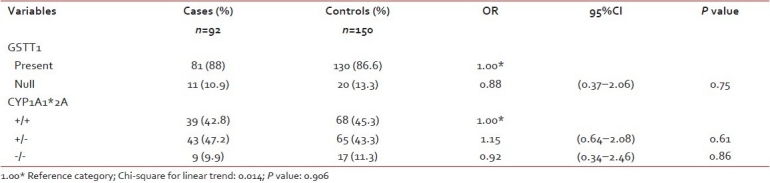
Our results show GSTT1 (present/null) polymorphism is not found to be associated with the susceptibility to ALL (P=0.75). CYP1A1*2A polymorphism also did not show any significant association with the risk to develop ALL.
Three years follow-up data was available for 66 patients. Fifty patients were treated with modified MCP-841 protocol and 16 patients with BFM-86 protocol.[21,22] As the number of patients treated under BFM-86 protocol is very small for analysis, survival analysis was performed in 50 patients treated under modified MCP-841 protocol. Cox regression analysis was performed to assess the prognostic significance of clinical characteristics like age, gender, immunophenotype, and WBC in this group of patients. No significant association was observed with any of the above mentioned variables (data not shown).
The frequencies of genotypes among patients with event and without event are presented in Table 2. Kaplan–Meier survival analysis was performed for the genotypes and log-rank P values were derived. GSTP1 (Ile/Val) and GSTP1 (Val/Val) genotypes were significantly associated with the outcome of the patient (log rank P=0.038 and 0.0001, respectively). Survival curves of ALL patients with GSTP1(Ile105val) polymorphism were shown in Figure 1. Cox regression analysis showed GSTP1 polymorphism as an independent prognostic factor associated with 6-fold increased risk with Val/Val genotype and 2-fold increased risk with Ile/Val genotype [Table 2]. Of the 5 patients who had induction failure, 3 patients had GSTP1 (Ile/Val) and 2 had GSTP1 (Val/Val) genotype. GSTM1 (present/null), GSTT1 (present/null) and CYP1A1*2A polymorphism did not reveal any prognostic significance.
Table 2.
Distribution of GSTM1, GSTT1, GSTP1 (rs1695) and CYP1A1 polymorphisms among the patients with and without event and its association with clinical outcome by univariate analysis
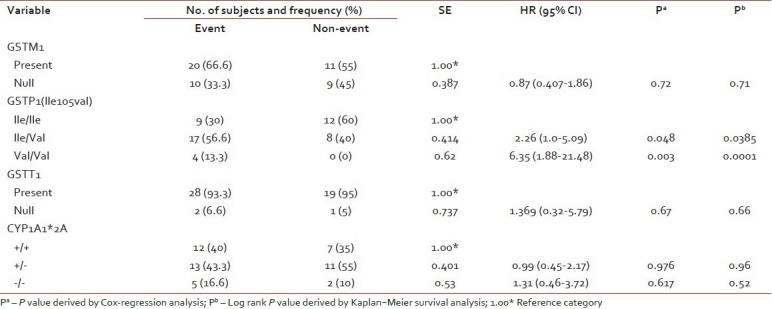
Figure 1.
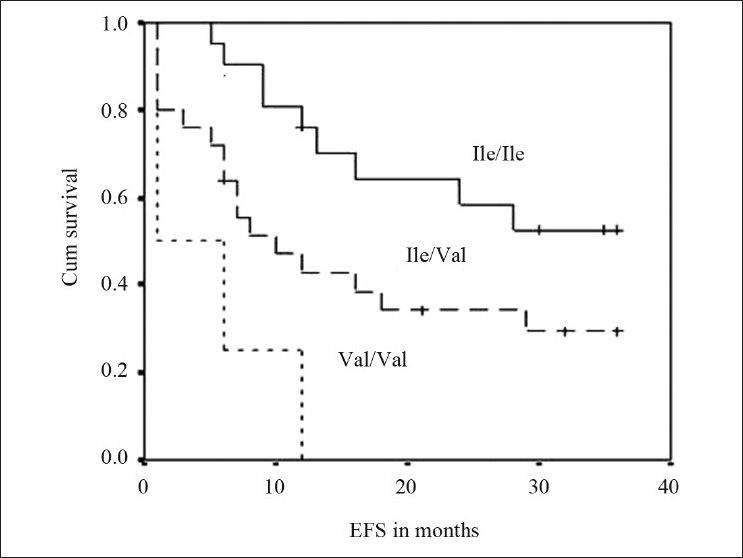
Event free survival curves of ALL patients with GSTP1(Ile105val) polymorphism
DISCUSSION
The significant association of GSTM1 polymorphism and the combination of GSTM1 null and GSTP1 variant genotype with the risk to develop ALL has been reported previously. In the present study, we have shown that GSTT1 is not associated with the susceptibility to develop ALL. This is in concordance to the reports by Krajinovic et al.,[2] Davies et al.,[23] Alves et al.,[11] Pakakasama et al.,[19] and Joseph et al.,[18] whereas bolufer et al.,[13] Haranatha et al.,[17] have reported significant increase in the risk to develop ALL with this polymorphism. Rollinson et al.,[24] has showed significant increase in risk to develop adult ALL associated with GSTT1 polymorphism. Combined analysis of GSTT1 (null) with other polymorphisms also did not show any significant association. Our results show that CYP1A1*2A is not associated with the susceptibility to develop ALL. This is in agreement with bolufer et al.,[13] and Pakakasama et al.,[19] whereas Krajinovic et al.,[2] Joseph et al.,[18] Gallegos et al.,[15] have reported significant association of this polymorphism to the risk to develop ALL. Balta et al.,[12] in their study in a Turkish population, have reported that homozygous CYP1A1*2A genotype was insignificantly lower in ALL patients compared to controls.
Our results reveal GSTP1 polymorphism as an independent prognostic factor in ALL. GSTP1 (Val allele) either in heterozygous or homozygous condition was associated with significant poor outcome. This is in contrast to Stanulla et al.,[1,25] as reported in their case control study, a non-significant decrease in risk of relapse and a significant decrease in risk of central nervous system relapse associated with GSTP1(Val/Val) genotype in ALL patients. Our study differs from their study in terms that this is not a case-control study. Krajinovic et al.,[18] has reported no significant association of GSTP1 polymorphism with clinical outcome in ALL patients. Improved outcomes in patients with ALL depend on interactions among drugs, ALL blast sensitivity, and host factors. The GSTP1 (Ile105Val) single nucleotide polymorphism is associated with reduced enzymatic activity for certain substrates and altered thermo stability.[6–8] GSTP1 is the most abundant GST in both tumour tissues and cell lines.[26] As there is no information on whether any chemotherapeutic agent acts as a substrate for GSTP1, its role in the detoxification of drugs is not clear. Blasts with high glutathione levels were found to have significant resistance to vincristine and ifosfamide.[27] The possible explanation for the association of GSTP1 (Ile/Val)/(Val/Val) genotype to the reduced EFS among ALL patients may be that the lower activity of GSTP1 has indirect effect on the cells by increasing the levels of GSH in the cells, decreasing the cells sensitivity towards certain chemotherapeutic drugs. All the patients who had induction failure in the present study showed GSTP1 variant genotype either in heterozygous or homozygous condition, which supports this hypothesis. Glutathione has been shown to affect cell proliferation.[28] GSTP1 Val allele was found to be an independent prognostic factor for lower survival in other cancers like breast and oesophagus.[29,30] Moureau et al.,[29] have hypothesized that low GSTP1 expression would reduce the global activity of GSTs, and consequently reduce glutathione (GSH) consumption in GST catalyzed reactions, thereby leading to higher levels of GSH, which would block apoptosis and promote proliferation of tumor cells.
Our study shows that GSTM1 did not show any significant effect on the outcome of the patient. Rocha et al.,[31] have reported a greater risk of hematological relapse associated with GSTM1(present) genotype in high-risk group, whereas Krajinovic et al.,[18] and Davies et al.,[23] have reported no significant association of GSTM1 polymorphisms on the outcome. GSTT1 polymorphism also was not found to have any significant association with the outcome of the patients. This is in concordance with Krajinovic et al.,[18] and Davies et al.,[23] but Stanulla et al.,[1] reported a significant decrease in the risk of relapse associated with the GSTT1 null genotype.
CYP1A1 plays an important role in the metabolic activation of polycyclic aromatic hydrocarbons, carcinogenic components of air pollution and CYP1A1*2A polymorphism is associated with elevated enzymatic activity.[10] Krajinovic et al.,[16] have reported that children with CYP1A1*2A genotype had shorter survival probabilities. In our study, CYP1A1*2A polymorphism was not found to be associated with the outcome of the patient.
In conclusion, our results show GSTP1 polymorphism as an independent prognostic factor associated with an inferior outcome in ALL patients. Studies with a large sample and other treatment protocols are needed to arrive at a definite conclusion.
Footnotes
Source of Support: DST (DST No. SR/SO/HS-26/2004)
Conflict of Interest: None declared.
REFERENCES
- 1.Stanulla M, Schrappe M, Brechlin AM, Zimmermann M, Welte K. Polymorphisms within glutathione S-transferase genes (GSTM1, GSTT1, GSTP1) and risk of relapse in childhood B-cell precursor acute lymphoblastic leukemia: A case-control study. Blood. 2000;95:1222–8. [PubMed] [Google Scholar]
- 2.Krajinovic M, Labuda D, Richer C, Karimi S, Sinnett D. Susceptibility to childhood acute lymphoblastic leukemia: Influence of CYP1A1, CYP2D6, GSTM1 and GSTT1 genetic polymorphisms. Blood. 1999;93:1496–501. [PubMed] [Google Scholar]
- 3.Kellen E, Hemelt M, Broberg K, Golka K, Kristensen VN, Hung RJ, et al. Pooled analysis and meta-analysis of the glutathione S-transferase P1 Ile 105Val polymorphism and bladder cancer: A HuGE-GSEC review. Am J Epidemiol. 2007;165:1221–30. doi: 10.1093/aje/kwm003. [DOI] [PubMed] [Google Scholar]
- 4.Ryberg D, Skaug V, Hewer A, Phillips DH, Harries LW, Wolf CR, et al. Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis. 1997;18:1285–9. doi: 10.1093/carcin/18.7.1285. [DOI] [PubMed] [Google Scholar]
- 5.White DL, Li D, Nurgalieva Z, El-Serag HB. Genetic variants of glutathione S-transferase as possible risk factors for hepatocellular carcinoma: A HuGE systematic review and meta-analysis. Am J Epidemiol. 2008;167:377–89. doi: 10.1093/aje/kwm315. [DOI] [PubMed] [Google Scholar]
- 6.Ntais C, Polycarpou A, Ioannidis JP. Association of GSTM1, GSTT1 and GSTP1 gene polymorphisms with the risk of prostate cancer: A meta-analysis. Cancer Epidemiol Biomarkers Prev. 2005;14:176–81. [PubMed] [Google Scholar]
- 7.Sundberg K, Johansson AS, Stenberg G, Widersten M, Seidel A, Mannervik B, et al. Differences in the catalytic efficiencies of allelic variants of glutathione transferase P1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocarbons. Carcinogenesis. 1998;19:433–6. doi: 10.1093/carcin/19.3.433. [DOI] [PubMed] [Google Scholar]
- 8.Hu X, Xia H, Srivastava SK, Pal A, Awasthi YC, Zimniak P, et al. Catalytic efficiencies of allelic variants of human glutathione S-transferase Pl-1 toward carcinogenic anti-diol epoxides of benzo[c]phenanthrene and benzo[g]chrysene. Cancer Res. 1998;58:5340–3. [PubMed] [Google Scholar]
- 9.Masson LF, Sharp L, Cotton SC, Little J. Cytochrome P-450 1A1 gene polymorphisms and risk of breast cancer: A HuGE review. Am J Epidemiol. 2005;161:901–15. doi: 10.1093/aje/kwi121. [DOI] [PubMed] [Google Scholar]
- 10.Georgiadis P, Topinka J, Vlachodimitropoulos D, Stoikidou M, Gioka M, Stephanou G, et al. Interactions between CYP1A1 polymorphisms and exposure to environmental tobacco smoke in the modulation of lymphocyte bulky DNA adducts and chromosomal aberrations. Carcinogenesis. 2005;26:93–101. doi: 10.1093/carcin/bgh294. [DOI] [PubMed] [Google Scholar]
- 11.Alves S, Amorim A, Ferreira F, Norton L, Prata MJ. The GSTM1 and GSTT1 genetic polymorphisms and susceptibility to acute lymphoblastic leukemia in children from north Portugal. Leukemia. 2002;16:1565–7. doi: 10.1038/sj.leu.2402543. [DOI] [PubMed] [Google Scholar]
- 12.Balta G, Yuksek N, Ozyurek E, Ertem U, Hicsonmez G, Altay C, et al. Characterization of MTHFR, GSTM1, GSTP1 and CYP1A1 genotypes in childhood acute leukemia. Am J Hematol. 2003;73:154–60. doi: 10.1002/ajh.10339. [DOI] [PubMed] [Google Scholar]
- 13.Bolufer P, Collado M, Barragan E, Cervera J, Calasanz MJ, Colomer D, et al. The potential effect of gender in combination with common genetic polymorphisms of drug-metabolizing enzymes on the risk of developing acute leukaemia. Haematologica. 2007;92:308–14. doi: 10.3324/haematol.10752. [DOI] [PubMed] [Google Scholar]
- 14.Chen CL, Liu Q, Pui CH, Rivera GK, Sandlund JT, Ribeiro R, et al. Higher frequency of glutathione S-transferase deletions in black children with acute lymphoblastic leukemia. Blood. 1997;89:1701–7. [PubMed] [Google Scholar]
- 15.Gallegos-Arreola MP, González-García JR, Figuera LE, Puebla-Pérez AM, Delgado-Lamas JL, Zúñiga-González GM. Distribution of CYP1A1*2A polymorphism in adult patients with acute lymphoblastic leukemia in a Mexican population. Blood Cells Mol Dis. 2008;41:91–4. doi: 10.1016/j.bcmd.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Krajinovic M, Labuda D, Mathonnet G, Labuda M, Moghrabi A, Champagne J, et al. Polymorphisms in genes encoding drugs and xenobiotic metabolizing enzymes, DNA repair enzymes, and response to treatment of childhood acute lymphoblastic leukemia. Clin Cancer Res. 2002;8:802–10. [PubMed] [Google Scholar]
- 17.Haranatha RP, Kaiser J. Polymorphisms in the GST (M1 and T1) gene and their possible association with susceptibility to childhood acute lymphocytic leukemia in Indian population. Afr J Biotechnol. 2006;5:1454–6. [Google Scholar]
- 18.Joseph T, Kusumakumary P, Chacko P, Abraham A, Radhakrishna Pillai M. Genetic polymorphism of CYP1A1, CYP2D6, GSTM1 and GSTT1 and susceptibility to acute lymphoblastic leukaemia in Indian children. Pediatr Blood Cancer. 2004;43:560–7. doi: 10.1002/pbc.20074. [DOI] [PubMed] [Google Scholar]
- 19.Pakakasama S, Mukda E, Sasanakul W, Kadegasem P, Udomsubpayakul U, Thithapandha A, et al. Polymorphisms of drug-metabolizing enzymes and risk of childhood acute lymphoblastic leukemia. Am J Hematol. 2005;79:202–5. doi: 10.1002/ajh.20404. [DOI] [PubMed] [Google Scholar]
- 20.Suneetha KJ, Nancy KN, Rajalekshmy KR, Sagar TG, Rajkumar T. Role of GSTM1 (Present/Null) and GSTP1 (Ile105Val) polymorphisms in susceptibility to acute lymphoblastic leukemia among the South Indian population. Asian Pac J Cancer Prev. 2008;9:733–6. [PubMed] [Google Scholar]
- 21.Shanta V, Maitreyan V, Sagar TG, Gajalakshmi CK, Rajalekshmy KR. Prognostic variables and survival in pediatric acute lymphoblastic leukemia:Cancer institute experience. Pediatr Hematol Oncol. 1996;13:205–16. doi: 10.3109/08880019609030819. [DOI] [PubMed] [Google Scholar]
- 22.Möricke A, Zimmermann M, Reiter A, Henze G, Schrauder A, Gadner H. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000. Leukemia. 2010;24:265–84. doi: 10.1038/leu.2009.257. [DOI] [PubMed] [Google Scholar]
- 23.Davies SM, Bhatia S, Ross JA, Kiffmeyer WR, Gaynon PS, Radloff GA, et al. Glutathione S transferase genotypes, genetic susceptibility and outcome of therapy in childhood acute lymphoblastic leukemia. Blood. 2002;100:67–71. doi: 10.1182/blood.v100.1.67. [DOI] [PubMed] [Google Scholar]
- 24.Rollinson S, Roddam P, Kane E, Roman E, Cartwright R, Jack A, et al. Polymorphic variation within the glutathione-S-transferase genes and risk of adult acute leukemia. Carcinogenesis. 2002;21:43–7. doi: 10.1093/carcin/21.1.43. [DOI] [PubMed] [Google Scholar]
- 25.Stanulla M, Schäffeler E, Arens S, Rathmann A, Schrauder A, Welte K, et al. GSTP1 and MDR1 genotypes and central nervous system relapse in childhood acute lymphoblastic leukemia. Int J Hematol. 2005;81:39–44. doi: 10.1532/ijh97.e0418. [DOI] [PubMed] [Google Scholar]
- 26.Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994;54:4313–20. [PubMed] [Google Scholar]
- 27.Kearns PR, Pieters R, Rottier MM, Pearson AD, Hall AG. Raised blast glutathione levels are associated with an increased risk of relapse in childhood acute lymphocytic leukemia. Blood. 2001;97:393–8. doi: 10.1182/blood.v97.2.393. [DOI] [PubMed] [Google Scholar]
- 28.Noelle RJ, Lawrence DA. Determination of glutathione in lymphocytes and possible association of redox state and proliferative capacity of lymphocytes. Biochem J. 1981;198:571–9. doi: 10.1042/bj1980571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moureau-Zabotto L, Ricci S, Lefranc JP, Coulet F, Genestie C, Antoine M, et al. Prognostic impact of multidrug resistance gene expression on the management of breast cancer in the context of adjuvant therapy based on a series of 171 patients. Br J Cancer. 2006;94:473–80. doi: 10.1038/sj.bjc.6602958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JM, Wu MT, Lee YC, Yang SY, Chen JS, Hsu HH, et al. Association of GSTP1 polymorphism and survival for esophageal cancer. Clin Cancer Res. 2005;11:4749–53. doi: 10.1158/1078-0432.CCR-04-2333. [DOI] [PubMed] [Google Scholar]
- 31.Rocha JC, Cheng C, Liu W, Kishi S, Das S, Cook EH, et al. Pharmacogenetics of outcome in children with acute lymphoblastic leukemia. Blood. 2005;105:4752–8. doi: 10.1182/blood-2004-11-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]


