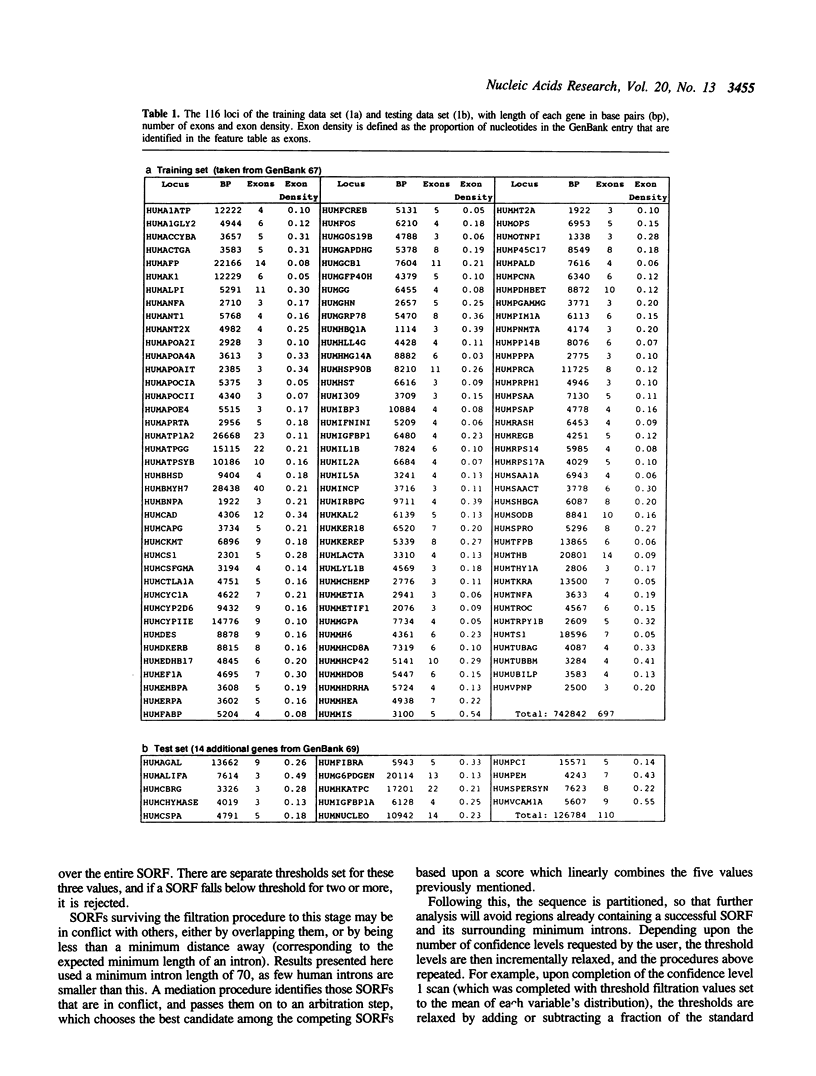
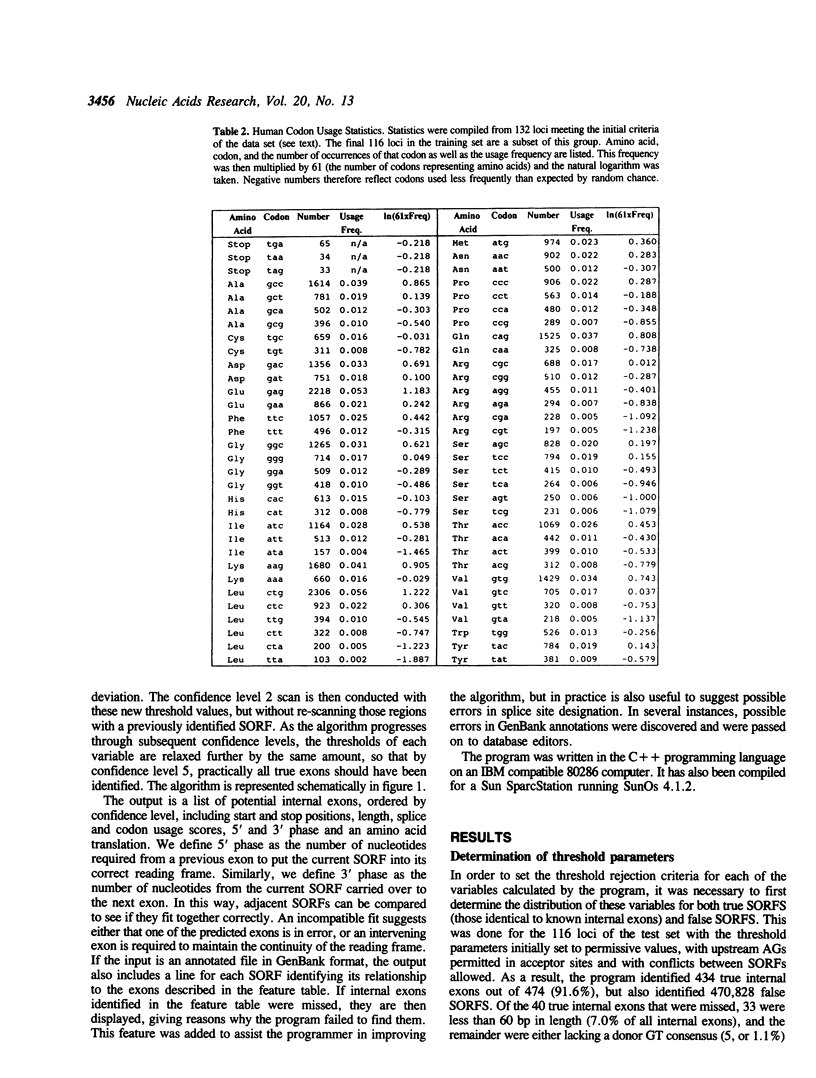
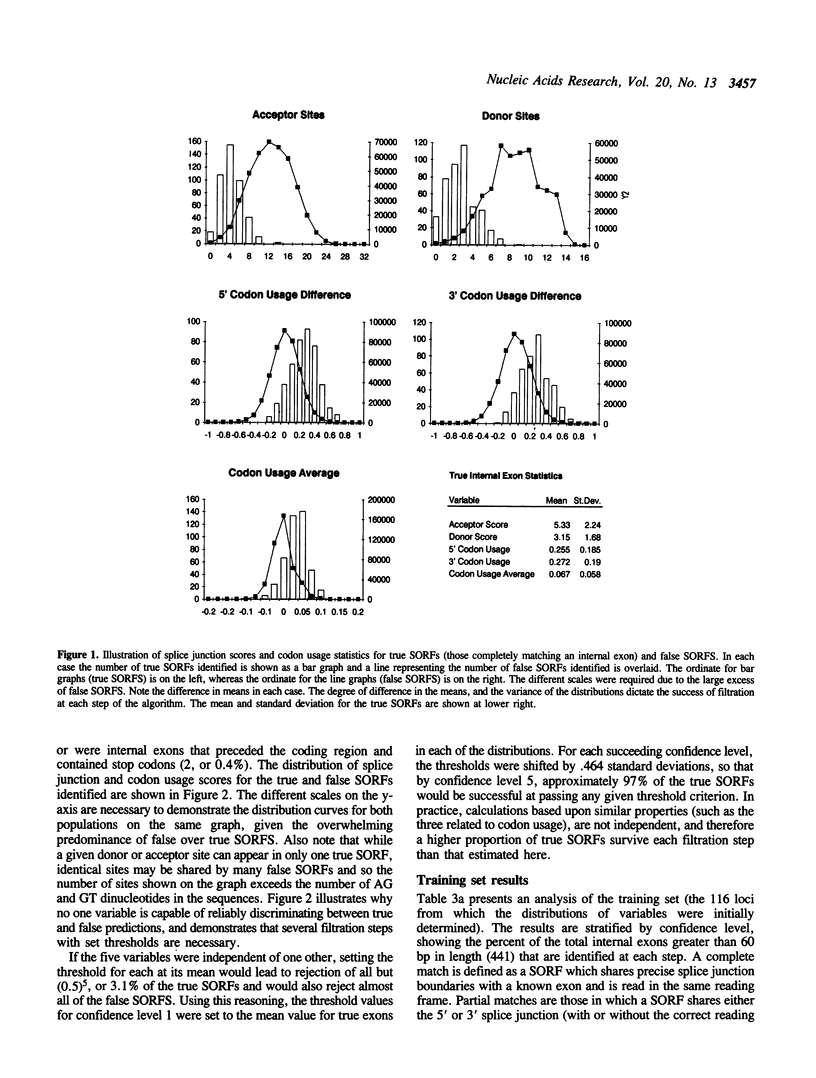
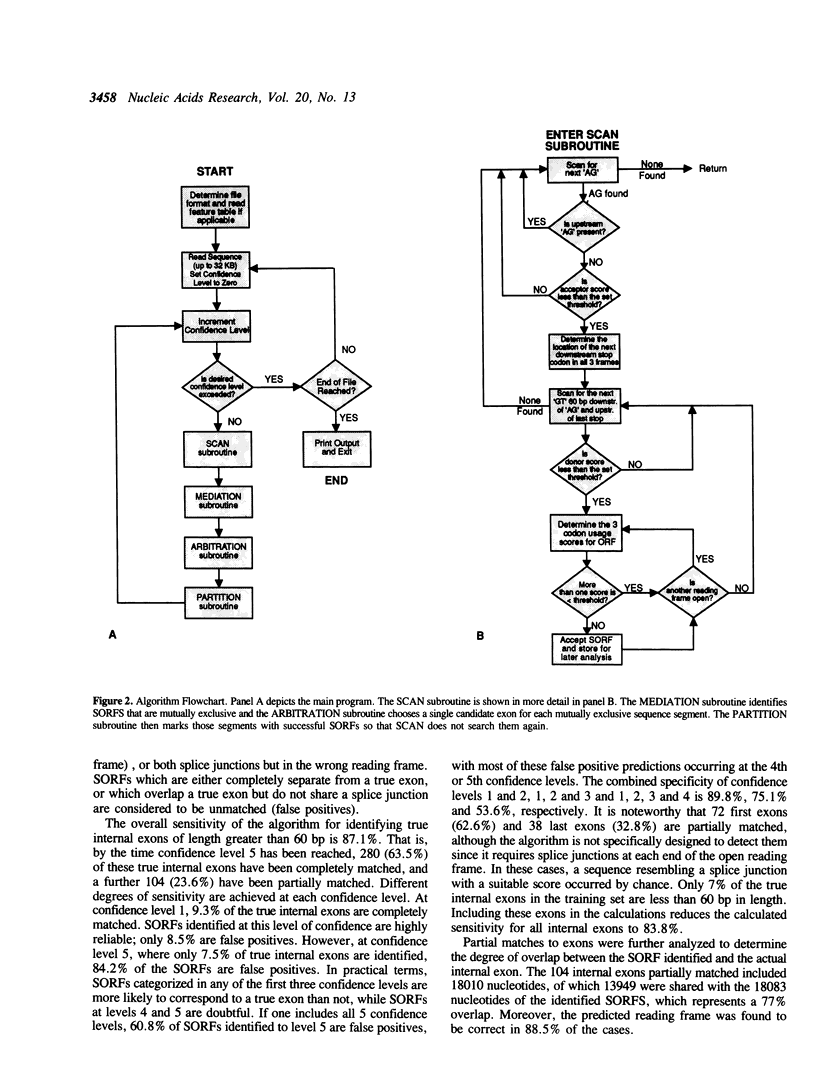
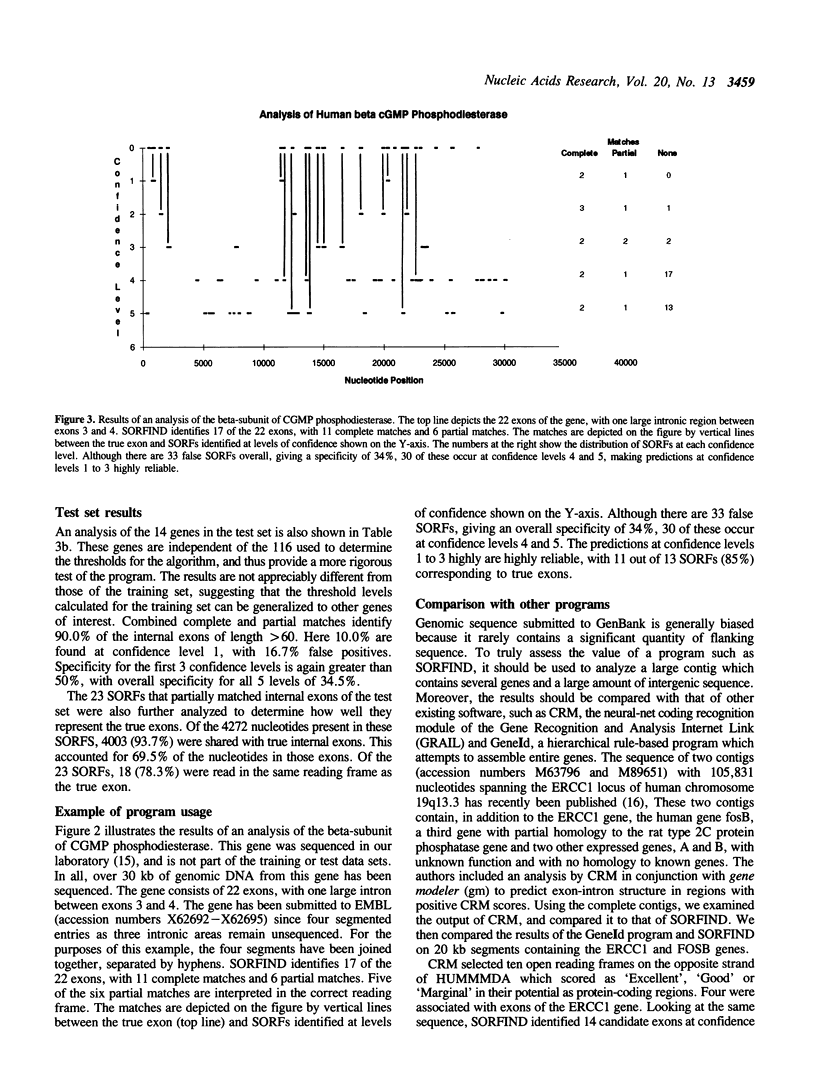
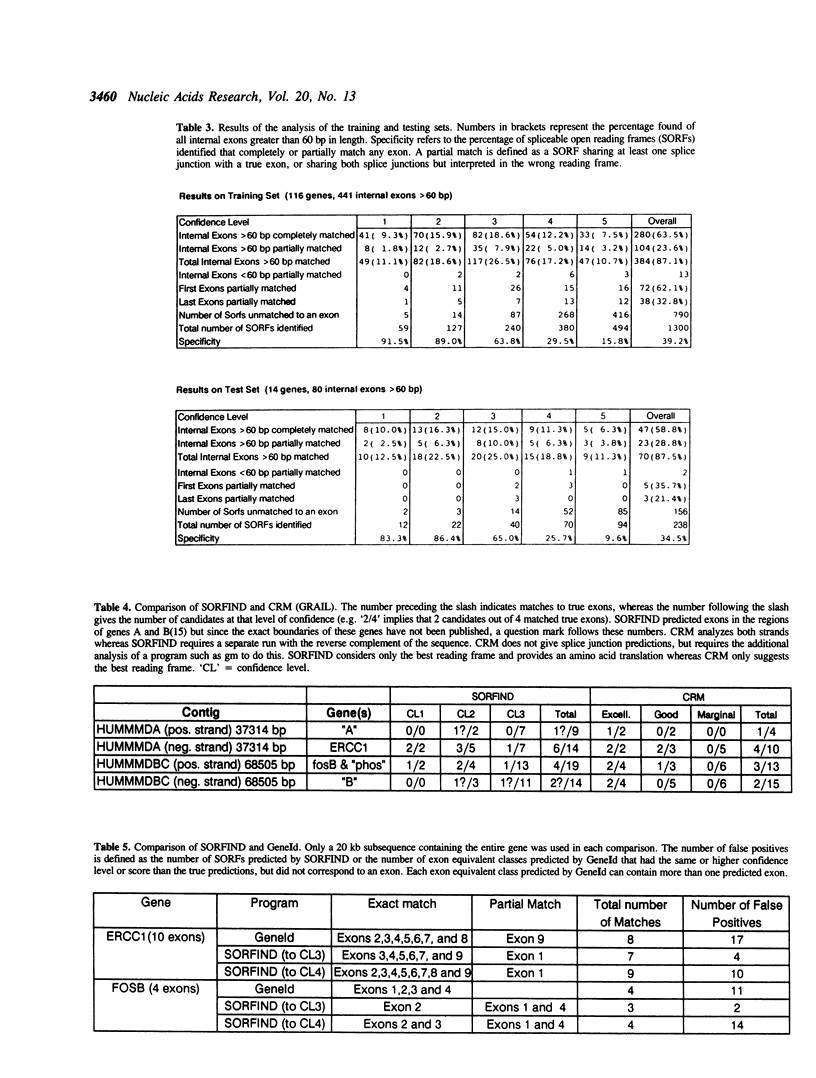
Abstract
We have developed a computer program which predicts internal exons from naive genomic sequence data and which will run on any IBM-compatible 80286 (or higher) computer. The algorithm searches a sequence for 'spliceable open reading frames' (SORFs), which are open reading frames bracketed by suitable splice-recognition sequences, and then analyzes the region for codon usage. Potential exons are stratified according to the reliability of their prediction, from confidence levels 1 to 5. The program is designed to predict internal exons of length greater than 60 nucleotides. In an analysis of 116 genes of a training set, 384 out of 441 such exons (87.1%) are identified, with 280 (63.5%) of predictions matching the true exon exactly (at both 5' and 3' splice junctions and in the correct reading frame), and with 104 (23.6%) exons matching partially. In a similar analysis of 14 genes in a test set unrelated to the genes used to generate the parameters of the program, 70 out of 80 internal exons greater than 60 bp in length are identified (87.5%), with 47 completely and 23 partially matched. SORFs that partially match true internal exons share at least one splice junction with the exon, or share both splice junctions but are interpreted in an incorrect reading frame. Specificity (the percentage of SORFs that correspond to true exons) varies from 91% at confidence level 1 to 16% at confidence level 5, with an overall specificity of 35-40%. The output displays nucleotide position, confidence level, reading frame phase at the 5' and 3' ends, acceptor and donor sequences and scoring statistics and also gives an amino acid translation of the potential exon. SORFIND compares favourably with other programs currently used to predict protein-coding regions.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988 Apr 20;200(4):709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987 Feb 20;193(4):723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991 Jul 5;220(1):49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Claverie J. M., Sauvaget I., Bougueleret L. K-tuple frequency analysis: from intron/exon discrimination to T-cell epitope mapping. Methods Enzymol. 1990;183:237–252. doi: 10.1016/0076-6879(90)83017-4. [DOI] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields C. A., Soderlund C. A. gm: a practical tool for automating DNA sequence analysis. Comput Appl Biosci. 1990 Jul;6(3):263–270. doi: 10.1093/bioinformatics/6.3.263. [DOI] [PubMed] [Google Scholar]
- Gelfand M. S. Computer prediction of the exon-intron structure of mammalian pre-mRNAs. Nucleic Acids Res. 1990 Oct 11;18(19):5865–5869. doi: 10.1093/nar/18.19.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka A. K., Owens J. Complexity charts can be used to map functional domains in DNA. Genet Anal Tech Appl. 1990 Apr;7(2):35–38. doi: 10.1016/0735-0651(90)90010-d. [DOI] [PubMed] [Google Scholar]
- Martin-Gallardo A., McCombie W. R., Gocayne J. D., FitzGerald M. G., Wallace S., Lee B. M., Lamerdin J., Trapp S., Kelley J. M., Liu L. I. Automated DNA sequencing and analysis of 106 kilobases from human chromosome 19q13.3. Nat Genet. 1992 Apr;1(1):34–39. doi: 10.1038/ng0492-34. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penotti F. E. Human pre-mRNA splicing signals. J Theor Biol. 1991 Jun 7;150(3):385–420. doi: 10.1016/s0022-5193(05)80436-9. [DOI] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Staden R. Finding protein coding regions in genomic sequences. Methods Enzymol. 1990;183:163–180. doi: 10.1016/0076-6879(90)83012-x. [DOI] [PubMed] [Google Scholar]
- Stormo G. D. Consensus patterns in DNA. Methods Enzymol. 1990;183:211–221. doi: 10.1016/0076-6879(90)83015-2. [DOI] [PubMed] [Google Scholar]
- Uberbacher E. C., Mural R. J. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B., Riess O., Hutchinson G., Collins C., Lin B. Y., Kowbel D., Andrew S., Schappert K., Hayden M. R. Genomic organization and complete sequence of the human gene encoding the beta-subunit of the cGMP phosphodiesterase and its localisation to 4p 16.3. Nucleic Acids Res. 1991 Nov 25;19(22):6263–6268. doi: 10.1093/nar/19.22.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]



