Abstract
Aim and Background:
Staphylococcus aureus is a major human pathogen that also causes important infections in cattle and sheep. The present study aimed to test genetic diversity among strains of S. aureus isolated from cattle (n=34) and humans (n=22) by DNA typing.
Materials and Methods:
Fluorescent amplified fragment length polymorphism (FAFLP) is the genotyping tool used in the study. The presence of the mecA and Panton-Valentine leukocidin (PVL) genes among these strain groups was also checked.
Results:
A dendrogram deduced from FAFLP showed that all the strains clustered into 10 groups (A–J) with a relative genetic divergence of less than 8%. Sixty-seven percent of the isolates from bovine sources clustered together in two clades (A and H), while another major cluster with 13 isolates (59%) (Cluster G) had all strains from a human host. The remaining strains from both the hosts clustered independently into smaller clusters with the exception of two strains of human origin, which clustered along with a bovine cluster. Thirteen strains belonging to cluster G were highly clonal. About 77% of strains obtained from human infections were methicillin-resistant S. aureus (MRSA), whereas only 29% of strains from bovine origin were MRSA. Only three strains from human origin showed PVL positive, while no strain from cattle had PVL genes. The complete absence of PVL genes in all the bovine strains in the study appears to be significant.
Conclusions:
FAFLP can be successfully applied to assess the genetic relationship of S. aureus isolates from different hosts. The study also provided the valuable epidemiological data on S. aureus from bovine sources in India, which is lacking.
Keywords: Bovine mastitis, Human infections, Staphylococcus aureus
INTRODUCTION
Staphylococcus aureus is a serious human pathogen responsible for life-threatening infections like septicemia, nosocomial pneumonia, endocarditis, and toxic shock syndrome (TSS).[1] In humans, this organism has become a major cause of nosocomial and community-acquired infections with increased hospital morbidity and mortality.[2,3] In the last decade, there has been tremendous increase in the prevalence of methicillin-resistant S. aureus (MRSA) strains that has become an additional problem in infection control practices in modern medicine[3–5] It is also one of the most significant pathogens causing mastitis in dairy cattle worldwide thus causing immense economic damage.[6–8]
S. aureus strains vary considerably in virulence and epidemiological potential. There are considerable genetic variations in natural populations of S. aureus.[3,7] Effective practical strategies to control human and bovine infections might require efforts that direct action against clones that are highly virulent and commonly cause disease. It is therefore important to type the S. aureus strains associated with human and animals. Many sources of S. aureus exist, including housing materials and fodder, equipment and air, bovine skin, nonbovine animals, and humans.[9] Studies have predicted human-to-bovine transmission by recovering S. aureus clones from cattle that are closely related to clones obtained from humans.[9] However, a few investigators have suggested that such transmission is unlikely under natural conditions.[7] Yet a recent study claimed that MRSA from an animal reservoir has entered the human population and it is responsible for causing >20% of all MRSA infections.[10]
In veterinary field, many techniques have been applied for characterization of bovine S. aureus strains such as single-gene typing systems like detection of coagulase gene polymorphism,[11] ribotyping, arbitrarily primed PCR[12] and PFGE.[13] In clinical microbiology, many other methods like FAFLP, MLST, and binary typing[8,14] are being used for enhanced resolution. Cuteri et al,[13] and Trovo Fabiano et al[15] reported the use of FAFLP for a limited number of S. aureus strains of animal origin. Previously, it was shown that there is close genetic relatedness among S. aureus isolates originated from human and animal sources obtained from same geographical locations.[15] There is a paucity of epidemiological data on S. aureus obtained from bovine sources in India and very little information available regarding S. aureus clones circulating in this country.[16] The present investigation attempted FAFLP genotyping to examine the diversity of S. aureus isolated from various clinical specimens obtained from humans and from bovine mastitis cases occurring in India. We also compared MRSA and methicillin-sensitive S. aureus (MSSA) strains from both cattle and human origins and verified their genetic differences. Finally, an effort was also made to identify genes that are implicated for enhanced virulence in S. aureus such as mecA for invasive and resistant properties and cytotoxin genes Panton valentine leukocidin (PVL) responsible for leukocyte destruction and tissue necrosis.
MATERIALS AND METHODS
Bacterial isolates
S. aureus isolates from human infections were obtained from a Medical College Hospital, Aligarh, India. Twenty-two isolates of S. aureus representing all kinds of human infections were randomly selected from a collection of 50 cultures obtained from both clinical specimens and nares of patients [Table 1]. An additional 34 cultures of S. aureus that were obtained during the year 2005 from a referral animal husbandry lab of an Agricultural University located in Hyderabad, India, were included in the study. These cultures were from infected udder of cows suffering from mastitis collected from cattle reared at different places of India. A total of 56 selected isolates were again characterized by growing them on sheep blood agar and mannitol salt agar (Oxoid) and they were tested for catalase, coagulase, and DNAse and phenotypically identified with Api Staph (Bio Merieux, France). All the strains were subjected to 16S-23S rRNA amplification for species identification prior to FAFLP analysis.[17,18]
Table 1.
Details of strain origin and results of antibiotic resistance, PCR and FAFLP typing
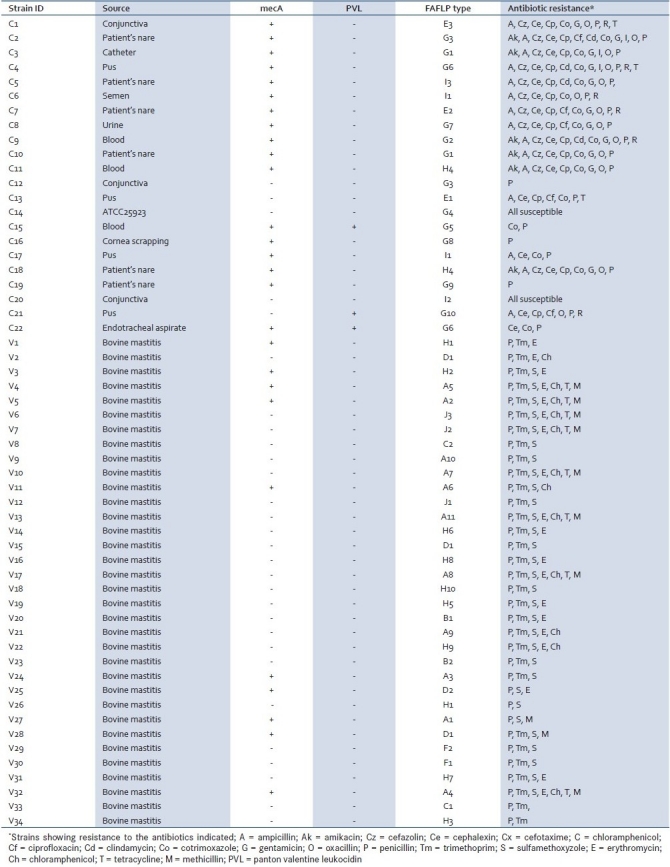
Susceptibility testing
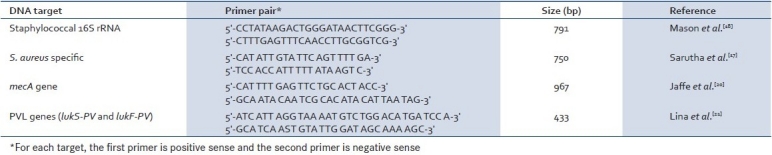
Antimicrobial susceptibility testing was performed for human clinical isolates with 14 antibiotics by the disk diffusion method on Mueller-Hinton agar that included important broad-spectrum antimicrobials. The following antimicrobials obtained from Hi Media (Mumbai) were used: ampicillin (10mcg), amikacin (30mcg), cefazolin (30mcg), clindamycin (2mcg), cotrimoxazole (1.25/23.75mcg), gentamicin (10mcg), penicillin (10units), tetracycline (30mcg), rifampicin (5mcg), methicillin (5mcg), and vancomycin (30mcg). Antimicrobial susceptibility testing for veterinary isolates included penicillin (10units), trimethoprim (5mcg), sulfamethoxyzole (300mcg), erythromycin (15mcg), chloramphenicol (30mcg), tetracycline (30mcg), and methicillin (5mcg). Methicillin resistance was screened by oxacillin (1μmg) disk susceptibility testing according to the guidelines of the Clinical and Laboratory Standards Institute.[19] S. aureus ATCC 25923 was included as a control. All strains were checked for the presence of mecA which codes for a unique penicillin-binding protein (PBP2a or PBP2’) and is directly associated with oxacillin resistance using PCR.[18,20] Detection of methicillin resistance was demonstrated by the amplification of the 967-bp fragment of the mecA gene. The frequency of PVL-producing S. aureus strains in different hosts was checked by screening all the strains for PVL genes (lukS-PV and lukF-PV) that were responsible for human infections and bovine mastitis.[21] The details of primers used in the study are given in Table 2.
Table 2.
Primer sets used in this study
FAFLP DNA fingerprinting
Fifty-six S. aureus strains obtained from diverse origins were genotyped by FAFLP. The strains were grown at 37°C in brain heart infusion (BHI) broth and harvested by centrifugation during the logarithmic phase. The resulting cell pellet was then subjected to DNA extraction, performed according to the protocol described earlier with slight modification.[22] In brief, the isolates were grown in BHI broth, harvested during the logarithmic phase and the cell pellet was washed with tris EDTA (TE). Twenty microliters of lysostaphin (1mg/ml), 20 μl of lysozyme (25mg/ml), and 5μl of RNAase (10mg/ml) were added and tubes were kept for incubation at 37°C for 1 h. Remaining downstream steps were as described by the authors.[22] The extracted DNA was dissolved in 0.1ml of TE and stored at 4°C until required. The quality and concentration of DNA were verified by 1% agarose gel electrophoresis.
Fifty-six isolates along with the standard reference strain (S. aureus ATCC 25923) were characterized by FAFLP as described previously.[23,24] Enzyme combination of EcoRI-MseI was used and fragments analyzed were within the size range of 50 to 500 bp. Primer combinations used were EcoRI+C and MseI+0. The selective reverse primer for the EcoRI adapter site was labeled with a fluorophore (FAM [6-carboxyfluorescein]). The FAFLP experiment and analysis (AFLP Microbial Fingerprinting kit; Applied Biosystems, Inc., Foster City, CA) were performed according to the manufacturer's instructions. Molecular relationships between the isolates were determined by the Genescan and Genotyper software (Applied Biosystems, Inc., CA) analysis of FAFLP profiles generated for all the isolates.
FAFLP fragments were detected and compiled by the ABI Data Collection software (Applied Biosystems). Gel images were generated and all lanes were extracted to make individual electropherograms. Fragment analysis was performed with Genescan 3.7. Peak height thresholds were set at 50; any peak heights lesser than these values were not included in the analysis. Electropherograms of all fragment profiles were visually inspected for polymorphisms, with the presence and absence of fragments were noted.[23] FAFLP was also performed for replicates of DNA derived before and after five subcultures of the standard ATCC strain and examined. They shared a minimum of 96% inter-gel similarity for approximately 75 fragments. Using criteria formulated in earlier studies for S. aureus,[25] individual isolates of S. aureus that shared 96% similarity are considered as identical clones. Genescan data of all the isolates were imported to genotyper and the presence or absence of amplicons within the categories was scored by a user-defined Genotyper macro. Allele scores (the presence or absence of amplicons) were converted into binary format (1 or 0). This binary format was used to construct a dendrogram by the FreeTree software program. Standard sequenced S. aureus referral strains could not be included in the FAFLP analysis; however, the predictive insilico AFLP method was used on genome sequences of six S. aureus referral strains namely S. aureus MW2, S. aureus Mu50, S. aureus N315, S. aureus subsp. aureus COL, S. aureus subsp. aureus MRSA252 and S. aureus subsp. aureus MSSA476 that are in the public domain for comparative extrapolation.
RESULTS
Standard tests used for biochemical characterization of all the isolates obtained from both human and bovine-origin confirmed the isolates as S. aureus. The disc diffusion susceptibility testing results of all the isolates are given in the form of resistograms in Table 1, that show the different patterns of resistance profiles. The antibiotics that are not shown in the antibiotype profile represent the susceptibility of the isolates to such antibiotics. High levels of resistance were recorded for penicillin, ampicillin, cefazolin, cephalexin, gentamicin, cefotaxime, and co-trimaxazole that were in the range of about 92% to 98% resistance among clinical strains of human origin. Amikacin, clindamycin, ciprofloxacin, and chloramphenicol showed maximum activity against all the strains. Strains that were resistant to oxacillin were found to be more resistant to all other antibiotics tested in the present study, as compared to oxacillin-susceptible strains. Twenty-seven strains of S. aureus were mecA positive (48%) among the total of 56 strains. The mecA possessing strains were more in the human group (17 of 22; 77%) than the bovine group, where only 10 strains were mecA positive out of 34 strains (29%). Three strains belonging to the human group showed amplification of PVL genes, while no strain from the bovine group was positive for PVL genes.
FAFLP for all strains in this study was reproducible over a time. Different DNA preparations from the same strain were subjected to the same FAFLP reaction conditions at different time intervals and analyzed accordingly. Profiles were stable for different DNAs extracted successively from a strain. Under all these experimental conditions, the characteristic amplified fragment profile was reproducibly detected, and none of the fragment sizes varied in any instance by more than 2 bp in these experiments. Thirty-four bovine strains were compared with 22 human S. aureus strains. FAFLP generated a total of 65 to 124 differently sized fragments experimentally ranging in size from 50 to 500 bp for all the strains. Electropherograms of all the strains obtained from Genescan were able to detect polymorphic and monomorphic markers. Typical group-specific amplitypes are shown in Figure 1. About 70% of the fragments were smaller than 350 bp. Many amplitypes showed more number of smaller fragments within a range of 50 bp to 350 bp. All these smaller fragment amplifications were reproduced twice to rule out the noise in FAFLP.
Figure 1.
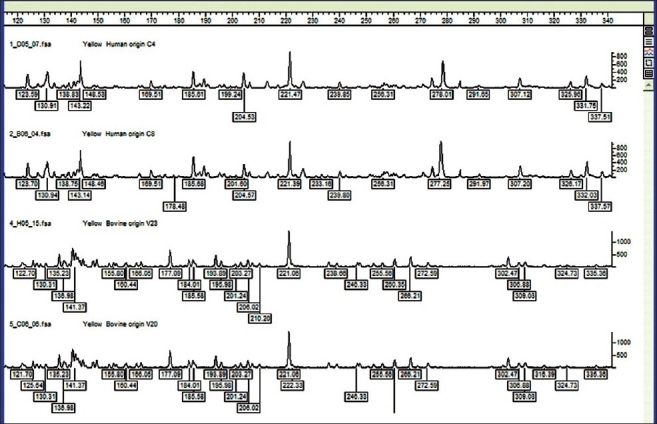
Typical group specific FAFLP profiles or amplitypes for MseI+0 and EcoRI+C selectivity showing the number of amplicons between the range of 120 and 340 base pairs. The genotyper plots of representative strains belonging to two host groups. FAFLP patterns in order (top to bottom) are C-4 (human origin), C-8 (human origin A), V-23 (bovine origin), and V-20 (bovine origin)
FAFLP profiles were sized using genotyper, and a matrix of binary data representing the presence or absence of amplified fragments was generated for all the strains, and a dendrogram was constructed using the FreeTree software. This dendrogram depicted the genetic relatedness among the 56 strains by showing three main clusters and seven small clusters [Figure 2]. FAFLP defined ten clone complexes designated A–J that were generated from the strains derived from the two groups, bovine and human. Two large clusters (A and H) of strains from bovine sources, comprising 11 and 10 strains, respectively, were branched together, whereas cluster G was another large cluster that contained only the strains from human infections with 13 strains. The remaining strains from the two groups clustered separately into many small clusters. Interestingly, two human strains (C11 and C18) clustered along with the bovine cluster H.
Figure 2.
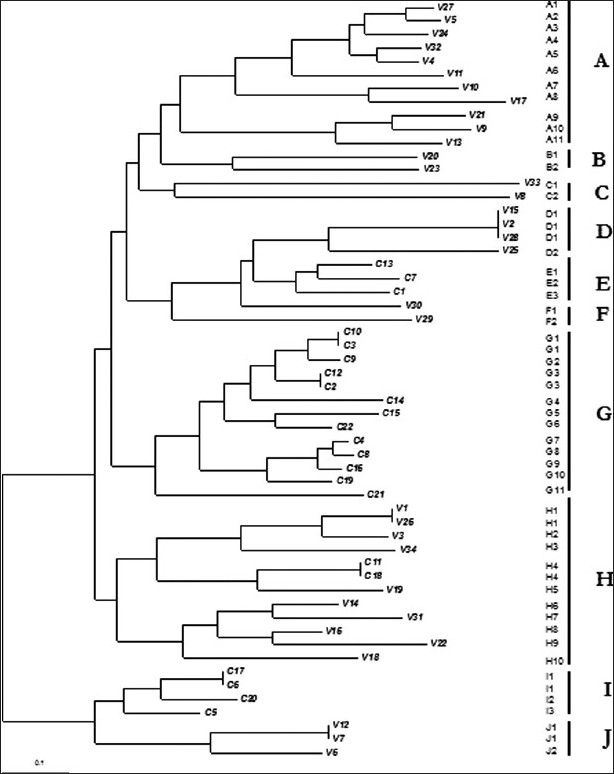
Dendrogram deduced from the binary data of genotyper analysis derived from FAFLP profiles depicting genetic relatedness among the strains from the two host backgrounds. The scale at the bottom represents the genetic distance between the isolates in percentages (1% divergence). FAFLP experiment had selectivity primers EcoRI+C and MseI+0
Cluster analysis identified many distinct genetic lineages in the two groups of S. aureus strains. Cluster A comprised bovine strains seemed to be very different from the rest of the clusters by falling away from rest of the clusters. Other strains separately formed clusters that corresponded to the two different sources from which these strains were obtained. Large clusters A and H comprised strains showing more intragenetic variations than the cluster G. We observed genetic differences among the four big clusters (A, D, G, H) at the level of approximately 8%. Major branches in the dendrogram segregated the large clusters of bovine and human strains separately. However, cluster E comprised human isolates (C1, C7, and C13) grouped closely with bovine isolates and cluster F (V29 and V30) grouped vice-versa. The intra-specific genetic distance among the S. aureus strains was wider among the clones obtained from the cattle (>4%). Thus, relatively high genetic heterogeneity was witnessed among the organisms recovered from dairy herds. The results showed that many different clones of S. aureus are widely distributed in infected mammary glands of cattle. Two clusters, D and J, had three and two strains respectively that were 100% identical. D and E clusters, comprising human and bovine isolates respectively, clustered closely, branching out from a common bigger clade in the dendrogram. Clinical strains from human infections were highly clonal which formed distinct cluster G in the dendrogram with a genetic distance of less than 4%. Within this, there were two mini clusters of two strains each, that were 100% identical. E and I were two other clusters from the human group that constituted three and five strains, respectively. Thirteen strains from the cluster G and strains from other two clusters (E and I), all are of human origin, were very closely related and appeared to be clonal in origin. S. aureus strains from human infections showed very less genetic variations.
Distribution of fragments among the ten clusters varied significantly. Predictive insilico methods used on genome sequences of six S. aureus strains that are in the public domain generated fragments ranging from 94 to 115 in number in the range between 50 and 500 bp upon selective PCR with single selectivity of EcoRI +C.[26] Strains belonging to human and bovine infections were subjected to FAFLP, and the results were extrapolated to the computer-predicted AFLP data of the S. aureus sequenced strains. Approximately around 85 fragments amplified were common among the different human and bovine strains analyzed. Most of these genomic regions were mapped to house-keeping genes. Strains from human origin showed differential amplification of 11 genomic regions in comparison to bovine strains. Most of the bovine strains lacked amplification of these 11 genomic regions; this might be due to the mutation (insertion or deletion) in the DNA region bearing EcoRI and MseI restriction sites. These corresponding polymorphisms are mapped to genes such as ebh, guaB, alsS, cap5H, and genes coding for enzymes such as GTP pyrophosokinase, proline dehydrogenase, and a hypothetical protein (coding DNA sequences[CDS] -MW0626 or SAV0156) predicted to be low-affinity inorganic phosphate transporter and a protein that is similar to low temperature requirement B protein (SAV0504 or SA0462).
DISCUSSION
Infections due to S. aureus are of major importance to veterinary and human medicine. Infections caused by MRSA in humans are mainly nosocomial and are increasingly being reported in many countries.[1] Earlier studies have shown that considerable variation in MRSA proportions exist not only between countries but also between hospitals within a country.[5] In the present study, about 54% of strains obtained from human infections were MRSA, whereas 29% of strains from bovine origin were MRSA. In contrast, markedly lower percentage of MRSA (<20%) among the bovine population has been observed till now.[10,13] High levels of methicillin resistance are understandable in strains recovered from hospital. Increasing levels of methicillin resistance in bovine strains can be explained by the increased use of beta-lactam antibiotics in treatment of mastitis. A relative comparison of the mecA presence in two host backgrounds showed less prevalence of mecA in bovine strains and this shows that there may be many methicillin-sensitive S. aureus (MSSA) lineages still exist in this setting. FAFLP detected markedly lower genomic variations between MRSA and MSSA strains obtained from the two groups. Strains containing mecA were distributed in many clusters defined by FAFLP.
The importance of PVL as a potential virulence factor led us to investigate the frequency of PVL-producing S. aureus strains obtained from diverse origins. Recent reports show an association of SCCmec type IV element with lysogenic phage-encoded PVL genes in community-acquired MRSA (CA-MRSA) strains.[27] We hypothesized that MRSA from bovine hosts could also contain PVL genes since cattle has a close contact with the cattle rearing community. However, when tested no strain from bovine background had PVL amplification. The absence of PVL genes in both MRSA and non-MRSA isolated bovine sources appears to be very significant, though their absence in non-MRSA bovine S. aureus has been recently reported.[28] The genes encoding for PVL toxin are shown to be responsible for severe necrotic lesions of the skin and soft tissues and are predominantly present in CA-MRSA.[27] Only three strains from human origin showed the presence of PVL genes. This is not significant and might be due to the fact that most of the strains studied were recovered from hospital and not from the community.
An emerging concept in clinical microbiology is that extensive variation exists in gene content among strains of many pathogenic bacterial species. Accordingly, one of the studies showed that genetic variation in S. aureus is very extensive, with approximately 22% of the genome comprising dispensable genetic material.[2] This might also be true with the strains of a species isolated in different hosts. In our study, the strains from human and bovine could be divided into distinct groups based on their origin of the isolates in concordance other studies.[29,30] FAFLP characterization of strains causing mastitis infection in a dairy showed a greater genetic difference than the strains obtained from clinical sources. Bovine strains studied in this study were responsible for severe mastitis. This study provides evidence of existence of variant virulent clones in India that were able to cause severe disease, having devastating impact on lactating cattle. Relatively a high level of genetic differences was observed among the S. aureus bovine strains in our study. In contrast, another study showed the genetic diversity among the S. aureus strains from bovine hosts is relatively low compared to that of strain-derived human hosts.[31] Our bovine strain collection was obtained from different locations in Andhra Pradesh and nearby states. High genetic variations in bovine strains amply prove that there is extensive dissemination of different clones in this region, and these clones are responsible for the cases of severe mastitis. In contrast, the number of indistinguishable strains from humans observed in the present study leads to the notion that there are very few prevalent clones in a particular geographic location or single hospital setup represented in our sample.
Strains from human origin showed lower genetic variation in the present study. With the exception of a few isolates belonging to cluster E that arise out of a common bigger clade, no human clones showed high similarity to the clones obtained from veterinary infections. Interestingly a similar observation made by van Leeuwen et al,[32] revealing that mastitis-associated S. aureus isolated from diverse farm animals formed a unique distinct genetic cluster when compared with the invasive infective S. aureus strains from pets and humans which formed closely related clonal complexes.[32] Interestingly, a recent study observed that the majority of bovine mastitis strain lineages that are unique and different from human lineages but the same study also contrarily claimed that these two lineages are closely related except with respect to a handful of genes that are distinct and responsible for host specificity.[30] In our study, strains from the human host appear to be specifically associated only with human infections and not with bovine infections. The results are consistent with the concept of host specialization among S. aureus clones and imply that successful transfer between humans and cattle is very limited.[7] Nonetheless, two clones from human C11 and C18 grouped with bovine cluster H in our investigation.
FAFLP results of human and bovine strains were extrapolated to the computer-predicted AFLP data of 6 S. aureus sequences that localized the differentially amplified fragments to the corresponding genes in the genomic regions. Eighty-five genomic regions amplified were common among the different human and bovine isolates. These were analyzed and most of them were mapped to housekeeping and essential genes such as iron metabolism, enzyme, toxin production, etc. Many of earlier studies were in concordance with our analysis, as there were no significant differences among bovine and human isolates noted in them.[30,31]Nevertheless,strains from the human host showed differential amplification of 11 genomic regions when compared with that of bovine strains in our study. When these DNA fragments were mapped to corresponding genes, they were coding for Ebh, Gua B, AlsS, C5H polysaccharide, and several other hypothetical proteins. Two of these molecules namely Ebh (ECM-binding protein homologue) and CP5 (capsular polysaccharide) seem to have definite role in human infections.[33–35] The recombinant Ebh protein was found to specifically bind human fibronectin.[33] Ebh is also produced during human infection, as serum samples taken from patients with confirmed S. aureus infections were found to contain anti-Ebh antibodies.[33] Ebh is cell envelope associated and it is also presumed to be involved in cellular adhesion. The cap5H-K genes were shown to be responsible for CP5 serotype specificity in S. aureus that encodes for serotype five capsular polysaccharide.[34] The cap5H gene coded CP5 is shown to facilitate the colonization in the host and is implicated as one of the virulence factors.[35] The lack of amplification of fragments that are mapped to ebh] and cap5H genes in most of the bovine strains indicate the incidence of mutation in restriction sites or there might be indels within these genes. Bovine S. aureus clones seem to possess modified sequences of ebh and cap5H genes and these two genes might not play vital role in the disease process of cattle. There may be other genes that are important in pathogenesis of S. aureus in mastitis. Our observation suggests that those host-specific genomic modifications might be occurring according to the requirement of the pathogens. Earlier studies have also shown association between distinct genotypes and severity of disease, suggesting strain and host-specific virulence.[2,8]
The present study described type of bovine S. aureus clones existing in India as well revealed genotypic variations among the S. aureus strains isolated from different origins in India. FAFLP appears to be an efficient tool for strain characterization and for resolution of clonal relationships of bacteria within and between host species. The present study demonstrated that the strains from human and veterinary pathology are different, and the results also provided a valuable insight into molecular specificities of different lineages of this important pathogen. Our analyses showed that there are notable differences in the genomes of human clones and bovine mastitis-associated clones of S. aureus. Identification of mutations in certain genes examined in this study may provide a lead for the identification of specific factors associated with host specificity in this major human and animal pathogen.
Footnotes
Source of Support: Authors would like to thank UGC, Govt. of India for Special Assistance Programme funds (UGC-SAP) provided to Department of Biotechnology, Pondicherry University
Conflict of Interest: None declared.
REFERENCES
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;8:520–32. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. Evolutionary genomics of Staphylococcus aureus: Insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci U S A. 2001;98:8821–6. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musser JM, Kapur V. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: Association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J Clin Microbiol. 1992;30:2058–63. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayliffe GA. The progressive intercontinental spread of methicillin resistant Staphylococcus aureus. Clin Infect Dis. 1997;24:S74–9. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 5.Tiemersma EW, Bronzwaer LA, Lyytikainen O, Degener JE, Schrijnemakers P, Bruinsma N, et al. Methicillin-resistant Staphylococcus aureus in Europe, 1999-2002. Emerg Infect Dis. 2004;10:1627–34. doi: 10.3201/eid1009.040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sischo WM, Heider LE, Miller GY, Moore DA. Prevalence of contagious pathogens of bovine mastitis and use of mastitis control practices. J Am Vet Med Assoc. 1993;202:595–600. [PubMed] [Google Scholar]
- 7.Kapur V, Sischo WM, Greer RS, Whittam TS, Musser JM. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J Clin Microbiol. 1995;33:376–80. doi: 10.1128/jcm.33.2.376-380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zodaks R, Leeuwen WV, Barkema H, Sampimon O, Verbrugh H, Schukken YH, et al. Application of Pulse-field Gel Electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular epidemiologic analysis of bovine and human Staphylococcus aureus isolates. J Clin Microbiol. 2000;38:1931–9. doi: 10.1128/jcm.38.5.1931-1939.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberson JR, Fox LK, Hancock DD, Gay JM, Besser TE. Ecology of Staphylococcus aureus isolated from various sites on dairy farms. J Dairy Sci. 1994;77:3354–64. doi: 10.3168/jds.S0022-0302(94)77277-5. [DOI] [PubMed] [Google Scholar]
- 10.van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, et al. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007;13:1834–9. doi: 10.3201/eid1312.070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su C, Herbelin C, Frieze N, Skardova O, Sordillo LM. Coagulase gene polymorphism of Staphylococcus aureus isolates from dairy cattle in different geographical areas. Epidemiol Infect. 1999;122:329–36. doi: 10.1017/s0950268899002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald JR, Meaney WJ, Hartigan PJ, Smyth CJ, Kapur V. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol Infect. 1997;119:261–9. doi: 10.1017/s0950268897007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuteri V, Mezzasoma P, Valente C. Application of biomolecular methods to Staphylococcus aureus strains from dairy cows. Vet Res Commun. 2003;27:335–8. doi: 10.1023/b:verc.0000014174.17108.f5. [DOI] [PubMed] [Google Scholar]
- 14.Grady R, Desai M, O’neill G, Cookson B, Stanley J. Genotyping of epidemic methicillin-resistant Staphylococcus aureus phage type 15 isolates by Fluorescent Amplified-Fragment Length Polymorphism analysis. J Clin Microbiol. 1999;37:3198–203. doi: 10.1128/jcm.37.10.3198-3203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trovo Fabiano TL, Lemos MV, Givisiez PE. Fluorescent amplified fragment length polymorphism genotyping of human and animal Staphylococcus aureus isolates from dairy farms with manual milking. Vet Microbiol. 2005;109:57–63. doi: 10.1016/j.vetmic.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Kalorey DR, Shanmugam Y, Kurkure NV, Chousalkar KK, Barbuddhe SB. PCR-based detection of genes encoding virulence determinants in Staphylococcus aureus from bovine subclinical mastitis cases. J Vet Sci. 2007;8:151–4. doi: 10.4142/jvs.2007.8.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saruta K, Matsunaga T, Kono M, Hoshina S, Ikawa S, Sakai O, et al. Rapid identification and typing of Staphylococcus aureus by nested PCR of amplified ribosomal DNA spacer region. FEMS Microbiol Lett. 1997;146:271–8. doi: 10.1111/j.1574-6968.1997.tb10204.x. [DOI] [PubMed] [Google Scholar]
- 18.Mason WJ, Blevins JS, Beenken K, Noroyono W, Ojha NA, Smeltzer MS. Multiplex PCR Protocol for the Diagnosis of Staphylococcal Infection. J Clin Microbiol. 2001;39:3332–8. doi: 10.1128/JCM.39.9.3332-3338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 16th informational supplement. Wayne, PA: CLSI; 2006. pp. M100–S16. [Google Scholar]
- 20.Jaffe RL, Lane JD, Albury SV, Niemeyer DM. Rapid extraction from and direct identification in clinical samples of methicillin-resistant Staphylococci using the PCR. J Clin Microbiol. 2000;38:3407–12. doi: 10.1128/jcm.38.9.3407-3412.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 22.Goto T, Nagamune H, Miyazaki A, Kawamura Y, Ohnishi O, Hattori K, et al. Rapid identification of Streptococcus intermmedius by PCR with the ily gene as a species marker gene. J Med Microbiol. 2002;51:178–86. doi: 10.1099/0022-1317-51-2-178. [DOI] [PubMed] [Google Scholar]
- 23.Kenchappa P, Sangwan VS, Ahmed N, Rao KR, Pathengay A, Mathai A, et al. High-resolution genotyping of Pseudomonas aeruginosa strains linked to acute post cataract surgery endophthalmitis outbreaks in India. Ann Clin Microbiol Antimicrobial. 2005;4:19. doi: 10.1186/1476-0711-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenchappa P, Duggirala A, Ahmed N, Pathangey A, Das T, Hasnain SE, et al. Fluorescent amplified fragment length polymorphism genotyping demonstrates the role of biofilm-producing methicillin-resistant periocular Staphylococcus epidermidis strains in postoperative endophthalmitis. BMC Ophthalmol. 2006;6:1. doi: 10.1186/1471-2415-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloos JH, Janssen P, van Boven CP, Dijkshoorn L. AFLP™ typing of Staphylococcus epidermidis in multiple sequential blood cultures. Res Microbiol. 1998;149:221–8. doi: 10.1016/s0923-2508(98)80082-x. [DOI] [PubMed] [Google Scholar]
- 26.Bikandi J, San Millán R, Rementeri A, Garaizar J. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease restriction. Bioinformatics. 2004;20:798–99. doi: 10.1093/bioinformatics/btg491. [DOI] [PubMed] [Google Scholar]
- 27.Rankin S, Roberts S, O'shea K, Maloney D, Lorenzo M, Benson CE. Panton valentine leukocidin (PVL) toxin positive MRSA strains isolated from companion animals. Vet Microbiol. 2005;108:145–8. doi: 10.1016/j.vetmic.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Yamada T, Tochimaru N, Nakasuji S, Hata E, Kobayashi H, Eguchi M, et al. Leukotoxin family genes in Staphylococcus aureus isolated from domestic animals and prevalence of lukM-lukF-PV genes by bacteriophages in bovine isolates. Vet Microbiol. 2005;110:97–103. doi: 10.1016/j.vetmic.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Reinoso EB, El-Sayed A, Lämmler C, Bogni C, Zschöck M. Genotyping of Staphylococcus aureus isolated from humans, bovine subclinical mastitis and food samples in Argentina. Microbiol Res. 2008;163:314–22. doi: 10.1016/j.micres.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 30.Sung JM, Lloyd DH, Lindsay JA. Staphylococcus aureus host specificity: Comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology. 2008;154:1949–59. doi: 10.1099/mic.0.2007/015289-0. [DOI] [PubMed] [Google Scholar]
- 31.Reinoso E, Bettera S, Frigerio C, DiRenzo M, Calzolari A, Bogni C. RAPD-PCR analysis of Staphylococcus aureus strains isolated from bovine and human hosts. Microbiol Res. 2004;159:245–55. doi: 10.1016/j.micres.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 32.van Leeuwen WB, Melles DC, Alaidan A, Al-Ahdal M, Boelens HA, Snijders SV, et al. Host- and tissue-specific pathogenic traits of Staphylococcus aureus. J Bacteriol. 2005;187:4584–91. doi: 10.1128/JB.187.13.4584-4591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke SR, Harris LG, Richards RG, Foster SJ. Analysis of Ebh, a 1.1-Megadalton Cell Wall-Associated Fibronectin-Binding Protein of Staphylococcus aureus. Infect Immun. 2002;70:6680–7. doi: 10.1128/IAI.70.12.6680-6687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wann ER, Dassy B, Fournier JM, Foster TJ. Genetic analysis of the cap5 locus of Staphylococcus aureus. FEMS Microbiol Lett. 1999;170:97–103. doi: 10.1111/j.1574-6968.1999.tb13360.x. [DOI] [PubMed] [Google Scholar]
- 35.Bhasin N, Albus A, Michon F, Livolsi PJ, Park JS, Lee JC. Identification of a gene essential for O-acetylation of the Staphylococcus aureus type 5 capsular polysaccharide. Mol Microbiol. 1998;27:9–21. doi: 10.1046/j.1365-2958.1998.00646.x. [DOI] [PubMed] [Google Scholar]


