Abstract
Genetic information in eukaryotes is managed by strategic hierarchical organization of chromatin structure. Primary chromatin structure describes an unfolded nucleosomal array, often referred to as ‘beads on a string’. Chromatin is compacted by the non-linear rearrangement of nucleosomes to form stable secondary chromatin structures. Chromatin conformational transitions between primary and secondary structures are mediated by both nucleosome-stacking interactions and the intervening linker DNA. Chromatin model system studies find that the topography of secondary structures is sensitive to the spacing of nucleosome within an array. Understanding the relationship between nucleosome spacing and higher order chromatin structure will likely yield important insights to the dynamic nature of secondary chromatin structure as it occurs in vivo. Genome-wide nucleosome mapping studies find the distance between nucleosomes varies, and regions of uniformly spaced nucleosomes are often interrupted by regions of nonuniform spacing. This type of organization is found at a subset of actively transcribed genes in which a nucleosome depleted region near the transcription start site is directly adjacent to uniformly spaced nucleosomes in the coding region. Here, we evaluate secondary chromatin structure and discuss the structural and functional implications of variable nucleosome distributions in different organisms and at gene regulatory junctions.
Keywords: nucleosome, chromatin, transcription, 30 nm fiber
Introduction
Eukaryotic nuclear processes, such as DNA replication, repair, recombination and gene expression, function within the constraints of a highly compacted chromatin fiber consisting of histones, non-histone proteins, and DNA. Although the precise path that the DNA follows for a highly folded chromatin fiber is controversial, intrinsic properties of the DNA will either set limits on the degree of chromatin compaction or will provide opportunities for dynamic conformational transitions required in vivo. Chromatin remodeling and modifying mechanisms function together to alter fiber structure and liberate regions of the genome for appropriate metabolic outcomes (Cairns 2009; Clapier and Cairns 2009; Kouzarides 2007). These mechanisms act to either disrupt or restore intrinsic histone-histone and histone-DNA interactions for chromatin structure fluidity. Here, we evaluate our current understanding of higher order chromatin structure and discuss how primary chromatin structure (i.e. the linear organization of nucleosomes) might impact subsequent levels of folded secondary chromatin structure in vivo.
Hierarchical chromatin organization
Chromatin primary structure
The first level of organization of a eukaryotic genome consists of a multifaceted and highly dynamic nucleoprotein complex known as the nucleosome. The nucleosome consists of an octamer of core histone proteins (two copies each of H2A, H2B, H3 and H4) wrapped ~1.65 times by 147 base pairs (bp) of DNA (Luger et al. 1997). A linker histone (e.g. H1 and H5) bound to a single nucleosome is known as a chromatosome. Linker histones associate with DNA located at the entry/exit sites of the nucleosome and influence the orientation of linker DNA with respect to the nucleosome (Hamiche et al. 1996; Simpson 1978; Syed et al. 2010). Typically, linker DNA describes the non-nucleosomal DNA connecting two or more nucleosomes in an array. Linker DNA length ranges between ~20–90 bp and varies among different species, tissues, and even fluctuates within a single cellular genome (van Holde 1988). In low salt, arrays of nucleosomes connected by linker DNA have the appearance of ‘beads on a string’ by electron microscopy, and form an extended primary structure that is 10 nm in diameter (10 nm fiber) (Olins and Olins 1974; van Holde 1988). Although electron microscopy (EM) has visualized 10 nm fibers using both endogenous and reconstituted chromatin, this conformation does not represent the most favored conformation under physiological conditions (Hansen 2002; Horowitz-Scherer and Woodcock 2006; Thoma et al. 1979). In the presence of physiological salt, linear chromatin condenses into a helical rearrangement of nucleosomes, referred to as chromatin folding, or formation of chromatin secondary structure (Woodcock and Dimitrov 2001).
Chromatin secondary structure
Chromatin secondary structure is driven by salt and intrinsic nucleosome-nucleosome and nucleosome-DNA interactions (Hansen 2002), and is stabilized by linker histones (Carruthers et al. 1998). EM and solution-state techniques have characterized both endogenous chromatin and reconstituted model systems to advance our understanding of chromatin secondary structure (Hansen 2002; Horowitz-Scherer and Woodcock 2006). Analytical ultracentrifugation experiments have identified two distinct secondary conformational states based on their hydrodynamic properties. These include a moderately folded intermediate characterized by close approach of adjacent nucleosomes, and a maximally folded conformation (Hansen 2002). In physiological salt (100–150 mM NaCl or 2–5 mM Mg2+), chromatin compacts into its maximally folded structure, comparable to the “30 nm fiber” (Hansen et al. 1989). The 30 nm fiber is of particular interest because of its physiological relevance as a local regulator of DNA metabolic pathways (Horowitz-Scherer and Woodcock 2006).
Chromatin tertiary structure
Chromatin tertiary structures are formed from interactions between discreet secondary chromatin structures, also referred to as fiber-fiber interactions. Fibrous chromatin loops and other suprastructures found in both metaphase chromosomes and specialized regions of interphase chromosomes, such as gene enhancers and insulators (Fraser and Grosveld 1998; Razin 1999; Woodcock and Dimitrov 2001; Woodcock and Ghosh 2010) are examples of chromatin tertiary structures. Although the structural properties of chromatin tertiary structure are largely unknown, reconstituted nucleosomal arrays fold and self-associate in the presence of salt to form large oligomers. In addition, chromatin fiber oligomerization is both cooperative and reversible (Schwarz et al. 1996) and, analogous to chromatin secondary structure, requires core histone amino-terminal ‘tail’ domains (Hansen 2002).
The role DNA plays in chromatin structure
What is DNA contributing to the intrinsic properties of the chromatin fiber at the primary and secondary structure levels?
A single nucleosome contains 14 non-covalent histone-DNA contacts (Luger et al. 1997). Several chromatin remodeling and modifying mechanisms target these regions of the nucleosome to reposition nucleosomes, disassemble nucleosomes and exchange variant histones in vivo (Cairns 2009; Clapier and Cairns 2009; Kouzarides 2007; Park and Luger 2008). Furthermore, DNA is required for histone octamer stability. In the absence of DNA, the histone octamer dissociates into two dimers of H2A-H2B and a tetramer of H3-H4 in physiological salt concentrations (Chung et al. 1978; Luger et al. 1999).
Sequence-dependent curvature of DNA can either favor or disfavor histone-DNA interactions (Lowary and Widom 1998; Travers et al. 2010; Zhang et al. 2009). For example, a sequence pattern containing ~ 10 bp phased arrangement of alternating AA/TT and GC dinucleotides creates intrinsic DNA curvature that promotes nucleosome assembly (Satchwell et al. 1986). DNA sequences that strongly favor histone-DNA interactions are nucleosome positioning sequences, such as the 5S rDNA sequence (Gottesfeld and Bloomer 1980; Simpson and Stafford 1983; Simpson et al. 1985) or the 601 sequence (Lowary and Widom 1998). These sequences are frequently used in constructing chromatin model systems. Nucleosome positioning in vivo may result from both exclusion of nucleosomes at rigid polydA/dT sequence tracks as well as favored positioning elsewhere in the genome (Kaplan et al. 2009; Kunkel and Martinson 1981; Segal and Widom 2009; Yuan et al. 2005). A positioned nucleosome may provide an anchor for the ‘statistical’ positioning of flanking nucleosomes (Kornberg 1981; Zhang et al. 2009); (Kaplan et al. 2009; Tillo et al. 2010). However, the observed nucleosome distributions in vivo likely result from a culmination of biological mechanisms including sequence-dependent positioning (Travers et al. 2010), chromatin remodeling and modification (Cairns 2009; Clapier and Cairns 2009; Kouzarides 2007), nucleosome disassembly (Hansen et al. 2010; Park and Luger 2008), chromatin architectural proteins (Horowitz-Scherer and Woodcock 2006; McBryant et al. 2006) including linker histone (Blank and Becker 1995; Rodriguez-Campos et al. 1989; Siriaco et al. 2009), and possibly, DNA replication (Siriaco et al. 2009), and transcription (Schones et al. 2008; Weiner et al. 2010) mechanisms.
The linear organization of nucleosomes and intervening linker DNA (i.e., chromatin primary structure) influence chromatin secondary structure (Arya et al. 2010; Grigoryev et al. 2009). The trajectory of linker DNA at the entry/exit site of the nucleosome determine the spatial orientation of successive nucleosomes in a folded array, and linker length dictates the tolerable distance between adjoining nucleosomes.
Nucleosome repeat lengths (NRL) of endogenous chromatin samples differ by multiples of ~10 bp (a DNA helical repeat) (Valouev et al. 2008; Wang et al. 2008a; Widom 1992). The nucleosome repeat length is a measure of nucleosomal DNA (147 bp) plus linker DNA (NRL = 147 bp + linker DNA) produced from limited nuclease digestion of isolated chromatin. The ~10 bp linker periodicity suggests that the orientation of consecutive nucleosomes is sensitive to DNA rotational phasing for higher order structure formation (Scipioni et al. 2010; Wu et al. 2007; Yao et al. 1993).
Chromatin model systems have been used to explore the compaction properties of endogenous or reconstituted 30 nm fibers harboring various NRLs and have compared fiber topography, diameter, and nucleosome packing density. Although the route DNA follows in a chromatin fiber remains speculative, as will be discussed in the next section, data suggest that linker DNA orientation and NRL influence fiber architecture.
Models for the 30 nm fiber and the path of linker DNA
The folded 30 nm chromatin fiber is a superhelical structure present in both interphase and metaphase chromosomes. X-ray diffraction and electron microscopy (EM) of isolated nuclei revealed 30 nm chromatin fibers present in select cell types, such as chicken erythrocytes (Langmore and Schutt 1980) (Langmore and Paulson 1983), HeLa metaphase chromosomes (Paulson and Langmore 1983) and the Balbiani ring genes in Chironomus tentans (Andersson et al. 1982). Subsequent EM studies in combination with biophysical analysis using endogenous or reconstituted chromatin have firmly established that 30 nm chromatin fibers are both stable and ubiquitous secondary structures (Horowitz-Scherer and Woodcock 2006). However, due to the structural complexity of the 30 nm fiber, details relating to its organization remain controversial.
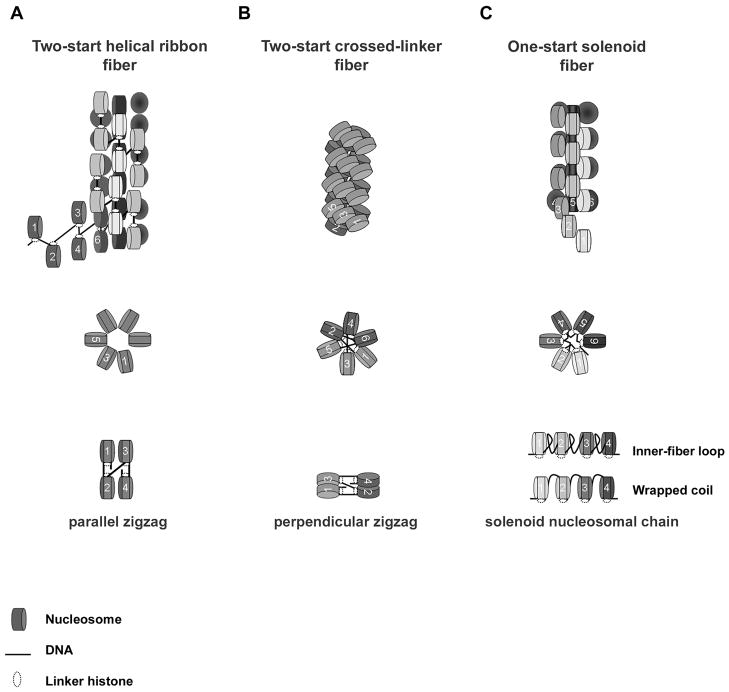
Many models for the 30 nm fiber have been proposed. The most enduring models include the two-start helical ribbon model (Woodcock et al. 1984; Worcel et al. 1981), the two-start crossed-linker model (Williams et al. 1986), and the one-start solenoid model (Finch and Klug 1976; Thoma et al. 1979; Van Holde et al. 1974; Widom and Klug 1985) (Fig 1). Here, we compare the path of DNA for both of the two-start models and the one-start solenoid.
Fig. 1.
Schematic representation of three different models for secondary chromatin structure and linker histone. (a) Two-start helical ribbon. (b) Two-start crossed-linker. (c) One-start solenoid fiber. Fiber long axes (top), cross sectional views (middle) and basic nucleosome arrangements (bottom) are shown for each model.
The two-start helix consists of repeating units of nucleosomes folded into a zigzag arrangement (Fig. 1a, b) (Horowitz et al. 1997). The zigzag arrangement of the two-start helix stacks alternate, nonsequential nucleosomes across from one another (i + 2) and twists, ultimately creating two stacks of winding nucleosomes in a superhelix (Williams et al. 1986). The two-start helical ribbon and the two-start crossed-linker differ by the orientation of the zigzag with respect to the long axis of the fiber. The two-start helical ribbon builds upon a parallel-zigzag arrangement (Fig. 1a) (Woodcock et al. 1984; Worcel et al. 1981) and the two-start crossed-linker builds upon a perpendicular-zigzag arrangement (Fig. 1b) (Williams et al. 1986). Although data discriminating between the two-start helical ribbon and the two-start crossed-linker fibers is limited, compelling experimental evidence suggests that nucleosomal arrays can adopt a zigzag pattern. The crystal structure of a tetra-nucleosome with an NRL of 167 bp at 9 Å resolution orientates nucleosomes in a zigzag conformation with a nucleosome-stacking assignment and orientation consistent with a two-start type of fiber (Schalch et al. 2005). Furthermore, pair-wise disulfide cross-linking of nucleosomes from compacted reconstituted arrays containing H4-V21C and H2A-E64C amino acid substitutions is also in agreement with a zigzag orientation (Dorigo et al. 2004).
The solenoid fiber model, however, does not adopt a zigzag orientation but rather positions consecutive nucleosomes in a hand-to-hand orientation (Fig.1c). Solenoid nucleosomal chains coil around an inner cavity with six to eight nucleosomes per turn and ~ 11 nm pitch, ultimately forming a one-start solenoid superhelix (Finch and Klug 1976; McGhee et al. 1983; Thoma et al. 1979). Nucleosome-stacking interactions are predicted to occur between nearest neighbor nucleosomes (i + 1). Nucleosomes residing in adjacent helical gyres are not necessarily in contact and the superhelix is proposed to stretch analogous to a Hookean spring (Kruithof et al. 2009). However, a variant of the solenoid structure incorporates an added degree of compaction whereby nucleosomes of neighboring helical gyres interdigitate between consecutive nucleosomes (Daban and Bermudez 1998; Robinson et al. 2006). Whatever the case may be, linker DNA of the solenoid either follows the superhelical path of the nucleosomal chain (Felsenfeld and McGhee 1986), as a ‘wrapped coil’, or loops/kinks into the inter-fiber space (Butler 1984), as an ‘inter-fiber loop’ (Fig. 1c).
Fiber topography and nucleosome repeat length
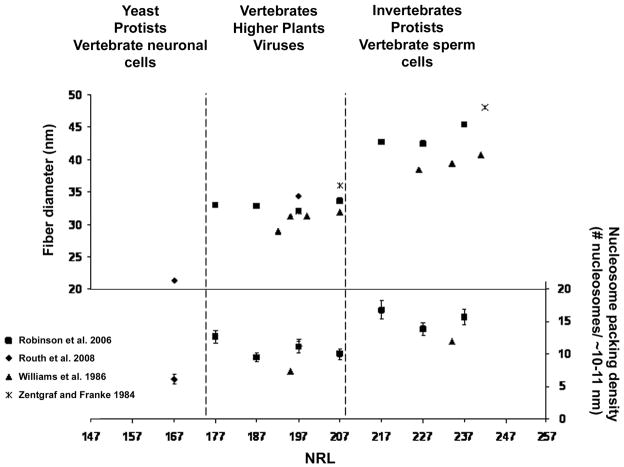
A key distinction among the two-start helical ribbon, the two-start crossed-linker, and the one-start solenoid fibers is the path of the DNA and its relationship to fiber diameter and nucleosome packing density (defined as the number of nucleosomes per 10–11 nm, where 11 nm is the diameter of a nucleosome) (Wu et al. 2007). For the two-start helical ribbon, the linker DNA is parallel to the long axis of the fiber (Fig.1a). Therefore, variation of NRL is predicted to affect fiber length and nucleosome packing density, but not fiber diameter. For the two-start crossed linker fiber, the linker DNA is perpendicular to the long axis of the fiber (Fig. 2b). Thus the diameter of the two-start crossed-linker fiber changes in proportion to changes in NRL. In experiments combining X-ray diffraction and EM of endogenous chromatin samples with varying NRLs, Williams et al. (1986) concluded that fiber diameter correlated nearly linearly with linker length (Fig. 2). In addition to increasing diameter, an increase in NRL also resulted in more compacted fibers. The calculated nucleosome packing densities of compacted fibers from Necturus erythrocytes and Thyone sperm cells (with NRLs of 195 bp and 234 bp, respectively) was reported as 7.4 and 11.9 nucleosomes/10 nm, respectively (Williams et al. 1986).
Fig. 2.
Fiber geometry as a function of nucleosome repeat length (NRL). Average fiber diameter (left) and nucleosome packing density (right) obtained from compacted nucleosomal arrays using EM or X-ray scattering techniques are plotted against their respective NRL. Approximation of NRLs from various eukaryotes and cell-types obtained from endogenous chromatin samples (van Holde, 1989) are indicated by dashed lines.
The diameter of the one-start solenoid fiber was once predicted to be independent of linker length, as intervening DNA loops out between adjacent nucleosomes (Butler 1984) (Fig.1c). Therefore, only the size of the loop would change incrementally with respect to linker length. However, linker DNA that is either too short or too long may not be compatible with nucleosome-stacking interactions (Butler 1984). In three separate papers, Rhodes and colleagues tested the relationship between the length of linker DNA and fiber compaction using a reconstituted chromatin model system based on the strong 601 nucleosome positioning sequence. Linker lengths were increased by ~10 bp increments (NRL of 177–237 bp; linker lengths of 30–90 bp). All nucleosomal arrays tested formed compact ~30 nm fibers in the presence of Mg2+ (1.0 to 1.6 mM MgCl2) and linker histone (H5). Average fiber diameters were determined statistically by EM measurements (Fig. 2) (Robinson et al. 2006). Unexpectedly two classes of fibers were generated based on physical criteria, including length, diameter and nucleosome packing density. The first class of fibers consisted of arrays with NRLs of 177–207 bp (30–60 bp linkers). They had an ~ 33 nm diameter and 11 nucleosomes/11 nm nucleosome packing density. The second class of arrays with NRLs of 217–237 bp (70–90 bp linkers) had an ~ 43 nm diameter and 15 nucleosomes/11 nm nucleosome packing density. Due to the non-linear relationship between NRL and fiber diameter, the authors conclude that fiber dimensions were not consistent with the two-start crossed-linker fiber model. Instead, fiber diameter and nucleosome packing density more closely fit dimensions of the solenoid fiber. A follow up study by the same group compared compacted nucleosomal arrays consisting of an even shorter NRL (167 bp; 20 bp linker) to a 197 bp-NRL (50 bp linker) (Routh et al. 2008). Mg2+-dependent compaction of the 167 bp-NRL array resulted in a third class of fiber with distinct fiber topography. The 167 NRL fibers were longer, thinner (~21 nm) and less compact (6.1 nucleosomes/11 nm nucleosome packing density) compared to the 197-fibers that were shorter, wider (34 nm) and more compact 11.2 nucleosomes/11 nm. In addition, the 167 bp NRL-fiber adopted an ordered zigzag arrangement, consistent with a two-start helix, and relied more on nucleosome-stacking interactions and less on linker histone. Compliance measurements of similar 167 and 197 bp-NRL arrays by single-molecule force spectroscopy supported the EM analysis and concluded that the 167 bp-NRL and the 197 bp-NRL fibers are consistent with a two-start helical ribbon and a solenoid, respectively (Kruithof et al. 2009). Collectively, these data suggest that arrays containing shorter NRL (< 30 bp linker length) prefer a two-start fiber consisting of a zigzag arrangement, while longer NRL (≥ 30 bps linker length) favor one of two classes of one-start solenoid fibers. However, it should be noted that previous analysis of 30 nm fibers with linker DNA >30 bp do not support the one-start solenoid fiber, but rather argue for derivations of the two-start fiber (Dorigo et al. 2004, Woodcock, 1984 #193; Horowitz et al. 1997; Williams et al. 1986; Wu et al. 2007). Thus an unresolved question still remains: whether the 30 nm fiber is organized as a one-start solenoid or two-start fiber? One explanation for the observed experimental inconsistencies has been attributed to H1 content (Routh et al. 2008). However, compelling evidence suggests that linker histones are not required for 30 nm fiber formation or compaction. Single-molecule force spectroscopy analysis of nucleosomal arrays (197 bp NRL) found that linker histones (H1 or H5) were not required for proper folding of chromatin into 30 nm fibers (Kruithof et al. 2009) and linker histone (H5) is not required for maximum condensation of reconstituted nucleosomal arrays examined in solution (Carruthers et al. 1998). Also, dinucleosomes compact to the same degree when in the presence or in the absence of linker histones H1 and H5 (Yao et al. 1991). Thus, linker histones do not have a pivotal role in fiber formation or chromatin compaction but likely play a key role in fiber stability, possibly favoring a specific conformation.
Heteromorphic chromatin fibers
It is possible that multiple DNA conformations simultaneously exist in a single chromatin fiber. Structural variation within a single fiber is described as ‘heteromorphic’. The formation of heteromorphic chromatin fibers is supported experimentally for reconstituted 207 bp-NRL (60 bp linker) nucleosomal arrays using EM-assisted nucleosome interaction capture (EMANIC) with Monte Carlo simulation modeling (Grigoryev et al. 2009) and mesoscale modeling of 173 and 209 bp- NRLs arrays (Schlick and Perisic 2009). In addition to linker length, model systems find both linker histones and cations (both monovalent and divalent) influence the path of DNA within a fiber. Grigoryev et al. (2009) found that a straight-linker, zigzag orientation in a fraction of nucleosomes transitions to a bent-linker solenoid-type orientation in the presence of Mg2+ and linker histone. The transition of a straight linker to a bent linker is consistent with a conversion from a zigzag arrangement to a solenoid-type arrangement. Thus, in addition to linker length, external influences such as linker histone or salt may generate local and/or global fiber polymorphism within a genome.
Distribution of nucleosomes and higher order chromatin structure
As a key component in determining higher order structure, would we expect linker DNA length to be similar among different species?
The NRL of isolated nuclei from a wide variety of organisms including viral, protist, plant, yeast and animal cells has been reported to range from ~150 bp to ~260 bps (van Holde 1988). Recently, genome-wide tiling and/or sequencing of nucleosomal DNA by ChiP-ChiP and ChiP-Seq techniques have estimated the NRLs of S. pombe (~154 bp-NRL) (Lantermann et al. 2010), S. cerevisiae (~165 bp-NRL) (Yuan et al. 2005), C. elegans (~175 bp-NRL) (Valouev et al. 2008), and human (~200 bp-NRL) (Schones et al. 2008). In addition to species variation, variation of NRL is also found among tissues, cell-types, and even within a single cellular genome (van Holde 1988). Global changes to cellular NRL correlate with developmental programs (Brown 1978; Hammoud et al. 2009; Jaeger and Kuenzle 1982; Sperling and Weiss 1980; Weintraub 1978), DNA replication (Siriaco et al. 2009) and RNA polymerase II (RNAPII) activity (Schones et al. 2008; Weiner et al. 2010). Notably, a reduction in cellular NRL correlates linearly with a depletion of linker histone (reviewed in Woodcock et al. 2006). Cells with short NRL (~165 bp) (e.g. yeast (Downs et al. 2003; Freidkin and Katcoff 2001) and neurons (Pearson et al. 1984)) contain <0.5 H1 molecules/nucleosome, while long NRL (> 200 bp) correlate with > 1.0 H1 molecules/nucleosome (Bates and Thomas 1981; Pearson et al. 1984). Further experimental investigation will be necessary to understand how NRL and linker histone content, including the expression and activity of specific variants, impact dynamic changes in chromatin structure.
To better understand the relationship between cellular NRL and secondary chromatin structure, NRLs for yeast, protists, animals, plants, viruses, and specific cell-types were superimposed on fiber dimension plots generated from model systems using EM and X-ray scattering techniques (Fig. 2.) Roughly, three classifications of fibers are indicated based on fiber diameter ranges, including the least compacted fibers with diameters between ~ 20–25 nm (NRLs between ~167–177 bp), intermediate compacted fibers with diameters between ~29–36 nm (NRLs between ~177–207 bps), and the highest compacted fibers with diameters between ~39–48 nm (NRLs between 217–242 bps). NRLs for most vertebrates, higher plants, and viruses correlate with intermediate compacted fibers, while yeast, protists, and certain metazoan cell-types correlate with alternative levels of compaction. Thus, in some settings, fiber diameter is fairly constant, with clear exceptions for certain eukaryotes, such as yeast (~21 nm diameter), and certain cell-types, such as sperm cells (≥40 nm diameter).
What does the cellular NRL of bulk chromatin represent?
Previous analysis of bulk chromatin relied on the enzymatic properties of micrococcal endonuclease (MNase), which may bias DNA sequence, or the accessibility or solubility of particular genomic regions. Thus, the measured NRL of bulk chromatin may exclude nucleosomes from highly compact or insoluble regions, such heterochromatin, or may not be an accurate measure with respect to a particular genomic locus (Henikoff et al. 2009; van Holde 1988; Weiner et al. 2010). Disparities between the NRLs of bulk chromatin and specific genomic regions have been reported. For example, the NRL for Xenopus erythrocytes (187 bp) differed from 5S rDNA genes (178 bps) (Humphries 1979; Gottesfeld, 1980) and the NRL of rat liver cells (198 bp) differed from rat satellite chromatin (185 bp) (Omori 1980). Alternatively, NRLs obtained from bulk chromatin may reflect either the average or the mean of a diverse population of uniformly and non-uniformly spaced nucleosomes. Genome-wide nucleosome mapping of S. Pombe finds that the NRL of whole cells (~154 bp) is nearly equivalent to the coding regions of active genes (also ~154) (Lantermann et al. 2010). Additionally, RNAPII activity correlates with both increased MNase digestion and an increase in bulk chromatin NRL (Weiner et al. 2010). Therefore, the observed NRL for bulk chromatin in yeast may reflect nucleosome spacing mediated by RNAPII at coding regions (estimated at ~66% of the yeast genome).
Chromatin organization at genes
The primary chromatin structure at gene promoters has been intensely investigated. Promoters have been classified based on their linear organization of nucleosomes (Cairns 2009; Tirosh and Barkai 2008). ‘Open’ promoters contain a nucleosome depleted region (NDR) and correlate with constitutive or highly transcribed genes. ‘Covered’ promoters harbor nucleosomes at promoter regions and correlate with highly regulated genes. ‘Mixed’ promoters contain elements of both open and covered promoters. For active genes with open promoters, at least three aspects of chromatin organization are shared among yeast and metazoa. 1) Genes with open promoters contain a NDR at the -1 nucleosome position (~100–200 bps upstream from the transcription start site and ~150 bp in length). 2) A positioned nucleosome is generally located immediately 3′ (downstream) from the transcription start site (+1 nucleosome). 3) A group of ~6–10 consecutive nucleosomes in the coding region are uniformly spaced and ‘statistically’ positioned from the +1 nucleosome. Concomitantly, this tripartite pattern of nucleosome organization correlates with both transcription activity and/or RNAP II occupancy (Lantermann et al. 2010; Schones et al. 2008; Weiner et al. 2010). Importantly, evidence in yeast (Lantermann et al. 2010; Weiner et al. 2010) and human (Schones et al. 2008) suggest that uniform spacing of nucleosomes in gene coding regions is directional and mediated by RNAPII. Thus, it is possible that RNAPII is actively maintaining nucleosome phasing within, and limited to, the transcribed region.
How might the distribution of nucleosomes at genes influence secondary chromatin structure?
Over the past 6 years, whole-genome nucleosome mapping studies have revealed the seemingly intentional positioning of nucleosomes in various regions of the genome. Although particular attention has been paid to the mechanisms governing nucleosome position, nucleosome mapping studies in combination with statistical analysis has the potential to determine nucleosome spacing for specific regions of the genome. In general, certain regions may favor or disfavor higher order chromatin structure formation based on their nucleosome spacing properties. Uniformly spaced nucleosomes, for example, may better support nucleosome-stacking interactions between adjacent nucleosomes and promote chromatin fiber stability (Wu et al. 2007). Model nucleosomal arrays assembled with substoichiometric levels of histone octamer to DNA repeat (< 0.9) produced arrays with nucleosome-free sites and exhibited reduced levels of salt-mediated folding (Hansen and Lohr 1993). Nucleosome-free sites within a reconstituted nucleosomal array may provide a model for the NDRs in gene promoters. From a structural perspective, NDRs at open promoters may control transcription at two levels. At the primary level, the removal of the -1 nucleosome may ‘free-up’ the underlying DNA sequence for transcription factor binding. At the secondary structure level, NDRs reduce the number of nucleosome-stacking interactions near the TSS and may destabilize local higher order chromatin structure. As an alternative route, disruption of nucleosome-stacking interactions may occur via nucleosome remodeling or histone post-translational modification. These alternative mechanisms are likely at play within highly regulated genes with ‘covered’ promoters. Under these circumstances, the – 1 nucleosome is retained but modified. Genome-wide mapping of 36 different histone modifications, including acetylation and methylation in CD4(+) T cells using ChiP-Seq techniques identified a group of 17 modifications that cluster and co-localize to 25% of active gene promoters (Wang et al. 2008b). Importantly, pair-wise correlation analysis revealed that 14 out of the 17 modifications likely occurred on a single nucleosome. Significantly, this same group of modifications did not correlate with gene coding regions. Gene coding regions correlated with an alternative pattern of histone modifications (Wang et al. 2008b). Thus, active gene promoters differ significantly from their coding region with respect to both histone modifications and primary chromatin structure.
Concluding remarks
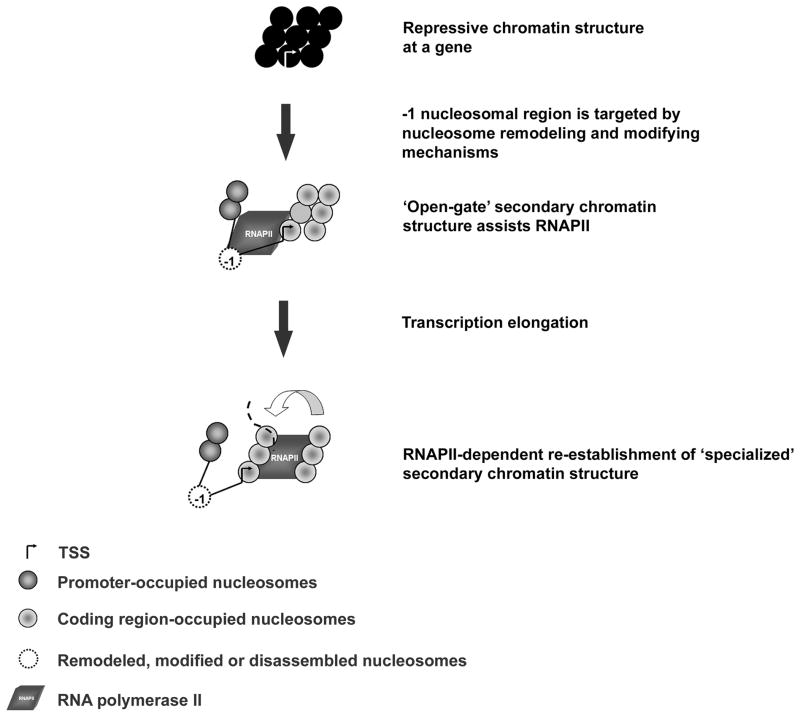
Nuclear signal transduction pathways rely on the coordinated exchanges between trans-acting factors and DNA recognition elements. Associations of this type are tightly regulated, in part, by dynamic changes in chromatin structure. Chromatin structure can assume multiple degrees of compaction potentiated by the linear organization of nucleosomes. Model system studies find that fiber topography is sensitive to the spacing of nucleosomes. An example of dramatic alteration of nucleosome spacing in vivo occurs at gene regulatory junctions between promoters and coding regions. We propose that gene activation requires both dynamic and local changes to secondary chromatin structure. Chromatin at a targeted locus is transformed from a repressive secondary chromatin structure to a bipartite secondary chromatin structure, referred to here as an ‘open gate’ conformation (Fig. 3). An ‘open gate’ secondary chromatin structure consists of a destabilized -1 nucleosomal region ~100–200 bp upstream of a ‘specialized’ folded secondary chromatin structure in the coding region consisting of uniformly spaced nucleosomes. Thus, we propose that a heterotypic secondary chromatin structure at a subset of gene loci mediate the temporal and/or spatial regulation of RNAPII transcription.
Fig. 3.
Schematic representation of dynamic secondary chromatin structure at gene regulatory junctions. Repressive chromatin structure establishes the inactive state. Gene activation by various nuclear signal transduction pathways target the -1 nucleosome region. Gene secondary chromatin structure transitions to a bipartite ‘open-gate’ conformation consisting of a disrupted -1 nucleosomal region in the gene promoter followed by a ‘specialized’ secondary chromatin structure in the coding region. The ‘open-gate’ conformation allows RNAPII access to the TSS for transcription initiation. The ‘specialized’ secondary chromatin structure facilitates transcription via unknown mechanisms. RNAPII-dependent processes actively re-establish nucleosome phasing in the coding region and maintain the ‘open-gate’ secondary chromatin structure for multiple rounds of mRNA synthesis.
Acknowledgments
I am grateful to Dr. Steven McBryant for helpful discussions and for critically reading the article. This work was supported by National Institutes of Health grant GM45916 to J.C.H. and an American Heart Society postdoctoral fellowship to H.S.
References
- Andersson K, Mahr R, Bjorkroth B, Daneholt B. Rapid reformation of the thick chromosome fiber upon completion of RNA synthesis at the Balbiani ring genes in Chironomus tentans. Chromosoma. 1982;87:33–48. doi: 10.1007/BF00333508. [DOI] [PubMed] [Google Scholar]
- Arya G, Maitra A, Grigoryev SA. A structural perspective on the where, how, why, and what of nucleosome positioning. J Biomol Struct Dyn. 2010;27:803–820. doi: 10.1080/07391102.2010.10508585. [DOI] [PubMed] [Google Scholar]
- Bates DL, Thomas JO. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank TA, Becker PB. Electrostatic mechanism of nucleosome spacing. J Mol Biol. 1995;252:305–313. doi: 10.1006/jmbi.1995.0498. [DOI] [PubMed] [Google Scholar]
- Brown IR. Postnatal appearance of short DNA repeat length in neurons of the cerebral cortex. Biochem Biophys Res Commun. 1978;84:285–292. doi: 10.1016/0006-291x(78)90168-7. [DOI] [PubMed] [Google Scholar]
- Butler PJ. A defined structure of the 30 nm chromatin fibre which accommodates different nucleosomal repeat lengths. Embo J. 1984;3:2599–2604. doi: 10.1002/j.1460-2075.1984.tb02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- Chung SY, Hill WE, Doty P. Characterization of the histone core complex. Proc Natl Acad Sci U S A. 1978;75:1680–1684. doi: 10.1073/pnas.75.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Daban JR, Bermudez A. Interdigitated solenoid model for compact chromatin fibers. Biochemistry. 1998;37:4299–4304. doi: 10.1021/bi973117h. [DOI] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, McGhee JD. Structure of the 30 nm chromatin fiber. Cell. 1986;44:375–377. doi: 10.1016/0092-8674(86)90456-3. [DOI] [PubMed] [Google Scholar]
- Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Grosveld F. Locus control regions, chromatin activation and transcription. Curr Opin Cell Biol. 1998;10:361–365. doi: 10.1016/s0955-0674(98)80012-4. [DOI] [PubMed] [Google Scholar]
- Freidkin I, Katcoff DJ. Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin. Nucleic Acids Res. 2001;29:4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld JM, Bloomer LS. Nonrandom alignment of nucleosomes on 5S RNA genes of X. laevis. Cell. 1980;21:751–760. doi: 10.1016/0092-8674(80)90438-9. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci U S A. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- Hansen JC, Ausio J, Stanik VH, van Holde KE. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- Hansen JC, Lohr D. Assembly and structural properties of subsaturated chromatin arrays. J Biol Chem. 1993;268:5840–5848. [PubMed] [Google Scholar]
- Hansen JC, Nyborg JK, Luger K, Stargell LA. Histone chaperones, histone acetylation, and the fluidity of the chromogenome. J Cell Physiol. 2010 doi: 10.1002/jcp.22150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz RA, Koster AJ, Walz J, Woodcock CL. Automated electron microscope tomography of frozen-hydrated chromatin: the irregular three-dimensional zigzag architecture persists in compact, isolated fibers. J Struct Biol. 1997;120:353–362. doi: 10.1006/jsbi.1997.3921. [DOI] [PubMed] [Google Scholar]
- Horowitz-Scherer RA, Woodcock CL. Organization of interphase chromatin. Chromosoma. 2006;115:1–14. doi: 10.1007/s00412-005-0035-3. [DOI] [PubMed] [Google Scholar]
- Jaeger AW, Kuenzle CC. The chromatin repeat length of brain cortex and cerebellar neurons changes concomitant with terminal differentiation. Embo J. 1982;1:811–816. doi: 10.1002/j.1460-2075.1982.tb01252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, I, Moore K, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. The location of nucleosomes in chromatin: specific or statistical. Nature. 1981;292:579–580. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kruithof M, Chien FT, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat Struct Mol Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- Kunkel GR, Martinson HG. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981;9:6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmore JP, Paulson JR. Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol. 1983;96:1120–1131. doi: 10.1083/jcb.96.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmore JP, Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–622. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- McBryant SJ, V, Adams H, Hansen JC. Chromatin architectural proteins. Chromosome Res. 2006;14:39–51. doi: 10.1007/s10577-006-1025-x. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Nickol JM, Felsenfeld G, Rau DC. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983;33:831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330– 332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JR, Langmore JP. Low angle x-ray diffraction studies of HeLa metaphase chromosomes: effects of histone phosphorylation and chromosome isolation procedure. J Cell Biol. 1983;96:1132–1137. doi: 10.1083/jcb.96.4.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson EC, Bates DL, Prospero TD, Thomas JO. Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur J Biochem. 1984;144:353–360. doi: 10.1111/j.1432-1033.1984.tb08471.x. [DOI] [PubMed] [Google Scholar]
- Razin SV. Chromosomal DNA loops may constitute basic units of the eukaryotic genome organization and evolution. Crit Rev Eukaryot Gene Expr. 1999;9:279–283. doi: 10.1615/critreveukargeneexpr.v9.i3-4.120. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci U S A. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Campos A, Shimamura A, Worcel A. Assembly and properties of chromatin containing histone H1. J Mol Biol. 1989;209:135–150. doi: 10.1016/0022-2836(89)90177-0. [DOI] [PubMed] [Google Scholar]
- Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- Schlick T, Perisic O. Mesoscale simulations of two nucleosome-repeat length oligonucleosomes. Phys Chem Chem Phys. 2009;11:10729–10737. doi: 10.1039/b918629h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- Scipioni A, Turchetti G, Morosetti S, De Santis P. Geometrical, conformational and topological restraints in regular nucleosome compaction in chromatin. Biophys Chem. 2010 doi: 10.1016/j.bpc.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Simpson RT, Stafford DW. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Siriaco G, Deuring R, Chioda M, Becker PB, Tamkun JW. Drosophila ISWI regulates the association of histone H1 with interphase chromosomes in vivo. Genetics. 2009;182:661–669. doi: 10.1534/genetics.109.102053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling L, Weiss MC. Chromatin repeat length correlates with phenotypic expression in hepatoma cells, their dedifferentiated variants, and somatic hybrids. Proc Natl Acad Sci U S A. 1980;77:3412–3416. doi: 10.1073/pnas.77.6.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, Hayes JJ, Everaers R, Angelov D, Bednar J, Dimitrov S. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A, Hiriart E, Churcher M, Caserta M, Di Mauro E. The DNA sequence-dependence of nucleosome positioning in vivo and in vitro. J Biomol Struct Dyn. 2010;27:713–724. doi: 10.1080/073911010010524942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde KE. Chromatin. Springer-Verlag; New York: 1988. [Google Scholar]
- Van Holde KE, Sahasrabuddhe CG, Shaw BR. A model for particulate structure in chromatin. Nucleic Acids Res. 1974;1:1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Fondufe-Mittendorf Y, Xi L, Tsai GF, Segal E, Widom J. Preferentially quantized linker DNA lengths in Saccharomyces cerevisiae. PLoS Comput Biol. 2008a;4:e1000175. doi: 10.1371/journal.pcbi.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008b;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978;5:1179–1188. doi: 10.1093/nar/5.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci U S A. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J, Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- Williams SP, Athey BD, Muglia LJ, Schappe RS, Gough AH, Langmore JP. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J. 1986;49:233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Dimitrov S. Higher-order structure of chromatin and chromosomes. Curr Opin Genet Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Frado LL, Rattner JB. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- Worcel A, Strogatz S, Riley D. Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci U S A. 1981;78:1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Bassett A, Travers A. A variable topology for the 30-nm chromatin fibre. EMBO Rep. 2007;8:1129–1134. doi: 10.1038/sj.embor.7401115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Lowary PT, Widom J. Linker DNA bending induced by the core histones of chromatin. Biochemistry. 1991;30:8408–8414. doi: 10.1021/bi00098a019. [DOI] [PubMed] [Google Scholar]
- Yao J, Lowary PT, Widom J. Twist constraints on linker DNA in the 30-nm chromatin fiber: implications for nucleosome phasing. Proc Natl Acad Sci U S A. 1993;90:9364–9368. doi: 10.1073/pnas.90.20.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]