Abstract
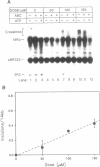
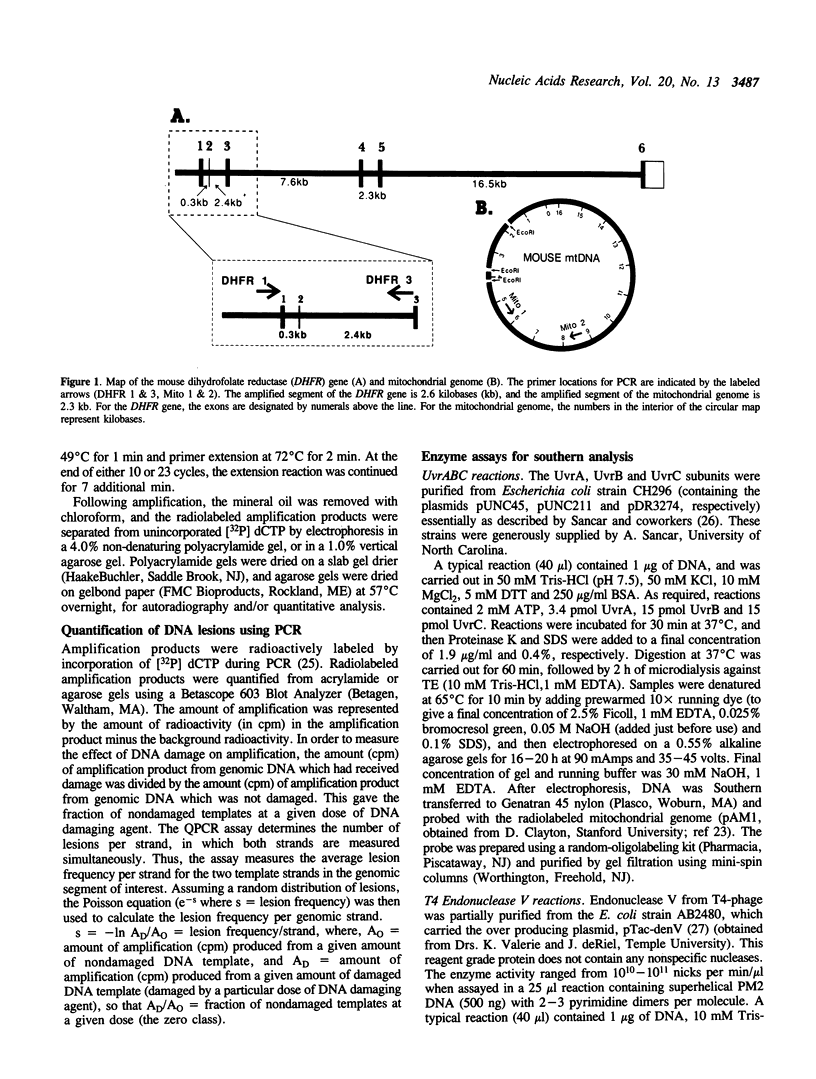
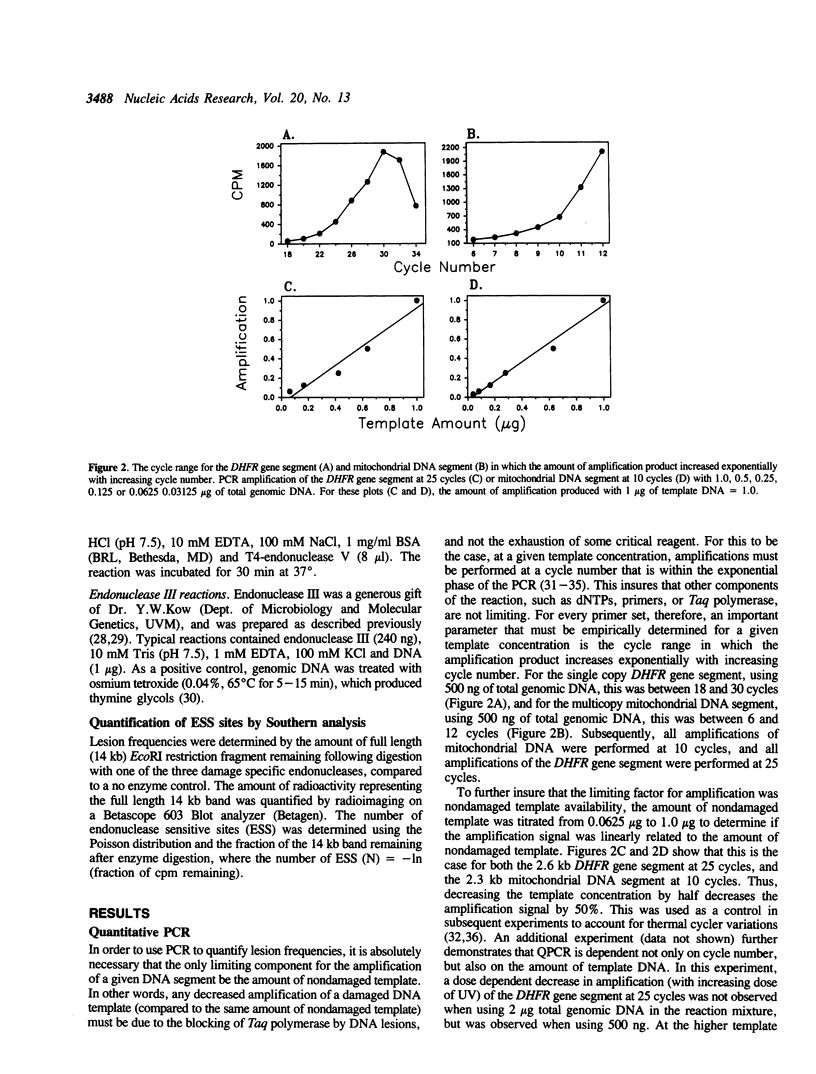
The polymerase chain reaction (PCR) represents an alternative to the current methods for investigating DNA damage and repair in specific genomic segments. In theory, any DNA lesion which blocks Taq polymerase can be measured by this assay. We used quantitative PCR (QPCR) to determine the lesion frequencies produced by cisplatin and ultraviolet light (UV) in a 2.3 kilobase (kb) segment of mitochondrial DNA and a 2.6 kb segment of the DHFR gene in mouse leukemia L1210 cells. The frequency of UV-induced lesions increased linearly with dose, and was 0.58 lesions/10 kb/10 J/m2 in the mitochondrial DNA, and 0.37 lesions/10 kb/10 J/m2 in the DHFR gene. With cisplatin, the lesion frequency also increased linearly with dose, and was 0.17 lesions/10 kb/10 microM in the DHFR gene, and 0.07 lesions/10 kb/10 microM in mitochondrial DNA. This result is contrary to that of Murata et al., 1990 (1), in which mitochondrial DNA received greater cisplatin damage than did nuclear DNA. Using PCR to measure the repair of UV-induced lesions in the DHFR gene segment, we observed that less than 10% of the lesions were removed by 4 h, but over 70% of the lesions were removed by 8 h. Repair of 43% of UV-induced lesions in mitochondrial DNA was also observed during a 24 h period.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. A., Coombs M. M. Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA. Nature. 1980 Sep 18;287(5779):244–245. doi: 10.1038/287244a0. [DOI] [PubMed] [Google Scholar]
- Axelrod J. D., Majors J. An improved method for photofootprinting yeast genes in vivo using Taq polymerase. Nucleic Acids Res. 1989 Jan 11;17(1):171–183. doi: 10.1093/nar/17.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J. M., Weinstein I. B. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980 Jul 11;209(4453):297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bohr V. A., Phillips D. H., Hanawalt P. C. Heterogeneous DNA damage and repair in the mammalian genome. Cancer Res. 1987 Dec 15;47(24 Pt 1):6426–6436. [PubMed] [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985 Feb;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Clayton D. A., Doda J. N., Friedberg E. C. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Simonsen C. C., McEwan R. N., Schimke R. T. Structure of amplified normal and variant dihydrofolate reductase genes in mouse sarcoma S180 cells. J Biol Chem. 1982 Jul 10;257(13):7887–7897. [PubMed] [Google Scholar]
- Govan H. L., 3rd, Valles-Ayoub Y., Braun J. Fine-mapping of DNA damage and repair in specific genomic segments. Nucleic Acids Res. 1990 Jul 11;18(13):3823–3830. doi: 10.1093/nar/18.13.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzel R. The trouble with 'PCR' machines. Trends Genet. 1990 Aug;6(8):237–238. doi: 10.1016/0168-9525(90)90191-8. [DOI] [PubMed] [Google Scholar]
- Jennerwein M. M., Eastman A. A polymerase chain reaction-based method to detect cisplatin adducts in specific genes. Nucleic Acids Res. 1991 Nov 25;19(22):6209–6214. doi: 10.1093/nar/19.22.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. V., Walsh M. L., Bockus B. J., Chen L. B. Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J Cell Biol. 1981 Mar;88(3):526–535. doi: 10.1083/jcb.88.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. V., Walsh M. L., Chen L. B. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci U S A. 1980 Feb;77(2):990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. C., Zhen W. P., Reed E., Parker R. J., Sancar A., Bohr V. A. Gene-specific formation and repair of cisplatin intrastrand adducts and interstrand cross-links in Chinese hamster ovary cells. J Biol Chem. 1991 Apr 15;266(11):7101–7107. [PubMed] [Google Scholar]
- Katcher H. L., Wallace S. S. Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry. 1983 Aug 16;22(17):4071–4081. doi: 10.1021/bi00286a013. [DOI] [PubMed] [Google Scholar]
- Kow Y. W., Wallace S. S. Mechanism of action of Escherichia coli endonuclease III. Biochemistry. 1987 Dec 15;26(25):8200–8206. doi: 10.1021/bi00399a027. [DOI] [PubMed] [Google Scholar]
- Kow Y. W., Wallace S. S., Van Houten B. UvrABC nuclease complex repairs thymine glycol, an oxidative DNA base damage. Mutat Res. 1990 Mar;235(2):147–156. doi: 10.1016/0921-8777(90)90068-g. [DOI] [PubMed] [Google Scholar]
- LeDoux S. P., Patton N. J., Nelson J. W., Bohr W. A., Wilson G. L. Preferential DNA repair of alkali-labile sites within the active insulin gene. J Biol Chem. 1990 Sep 5;265(25):14875–14880. [PubMed] [Google Scholar]
- Leadon S. A., Snowden M. M. Differential repair of DNA damage in the human metallothionein gene family. Mol Cell Biol. 1988 Dec;8(12):5331–5338. doi: 10.1128/mcb.8.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Bohr V. A., Hanawalt P. C. Differential DNA repair in transcriptionally active and inactive proto-oncogenes: c-abl and c-mos. Cell. 1986 May 9;45(3):417–423. doi: 10.1016/0092-8674(86)90327-2. [DOI] [PubMed] [Google Scholar]
- Martens P. A., Clayton D. A. Mechanism of mitochondrial DNA replication in mouse L-cells: localization and sequence of the light-strand origin of replication. J Mol Biol. 1979 Dec 5;135(2):327–351. doi: 10.1016/0022-2836(79)90440-6. [DOI] [PubMed] [Google Scholar]
- Mellon I., Bohr V. A., Smith C. A., Hanawalt P. C. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Hanawalt P. C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989 Nov 2;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Miyaki M., Yatagai K., Ono T. Strand breaks of mammalian mitochondrial DNA induced by carcinogens. Chem Biol Interact. 1977 Jun;17(3):321–329. doi: 10.1016/0009-2797(77)90095-3. [DOI] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Murata T., Hibasami H., Maekawa S., Tagawa T., Nakashima K. Preferential binding of cisplatin to mitochondrial DNA and suppression of ATP generation in human malignant melanoma cells. Biochem Int. 1990;20(5):949–955. [PubMed] [Google Scholar]
- Myers K. A., Saffhill R., O'Connor P. J. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis. 1988 Feb;9(2):285–292. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- Niranjan B. G., Bhat N. K., Avadhani N. G. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982 Jan 1;215(4528):73–75. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- Ponti M., Forrow S. M., Souhami R. L., D'Incalci M., Hartley J. A. Measurement of the sequence specificity of covalent DNA modification by antineoplastic agents using Taq DNA polymerase. Nucleic Acids Res. 1991 Jun 11;19(11):2929–2933. doi: 10.1093/nar/19.11.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M. S., Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989 Sep 8;58(5):1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- Schowalter D. B., Sommer S. S. The generation of radiolabeled DNA and RNA probes with polymerase chain reaction. Anal Biochem. 1989 Feb 15;177(1):90–94. doi: 10.1016/0003-2697(89)90019-5. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Sam J., Robinson M. O., Bellvé A. R., Simon M. I., Riggs A. D. Measurement by quantitative PCR of changes in HPRT, PGK-1, PGK-2, APRT, MTase, and Zfy gene transcripts during mouse spermatogenesis. Nucleic Acids Res. 1990 Mar 11;18(5):1255–1259. doi: 10.1093/nar/18.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon M. J., Thoma F. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell. 1990 May 18;61(4):675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- Tang M. S., Bohr V. A., Zhang X. S., Pierce J., Hanawalt P. C. Quantification of aminofluorene adduct formation and repair in defined DNA sequences in mammalian cells using the UVRABC nuclease. J Biol Chem. 1989 Aug 25;264(24):14455–14462. [PubMed] [Google Scholar]
- Terleth C., van Sluis C. A., van de Putte P. Differential repair of UV damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1989 Jun 26;17(12):4433–4439. doi: 10.1093/nar/17.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. C., Morton A. G., Bohr V. A., Sancar A. General method for quantifying base adducts in specific mammalian genes. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3723–3727. doi: 10.1073/pnas.85.11.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. C., Okumoto D. S., Sancar A., Bohr V. A. Preferential DNA repair of (6-4) photoproducts in the dihydrofolate reductase gene of Chinese hamster ovary cells. J Biol Chem. 1989 Oct 25;264(30):18005–18010. [PubMed] [Google Scholar]
- Tomkinson A. E., Bonk R. T., Kim J., Bartfeld N., Linn S. Mammalian mitochondrial endonuclease activities specific for ultraviolet-irradiated DNA. Nucleic Acids Res. 1990 Feb 25;18(4):929–935. doi: 10.1093/nar/18.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990 Mar;54(1):18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. M., Doyle M. V., Mark D. F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann K., Kohn K. W., Bohr V. A. Heterogeneity of nitrogen mustard-induced DNA damage and repair at the level of the gene in Chinese hamster ovary cells. J Biol Chem. 1990 Aug 15;265(23):13906–13913. [PubMed] [Google Scholar]
- Wunderlich V., Schütt M., Böttger M., Graffi A. Preferential alkylation of mitochondrial deoxyribonucleic acid by N-methyl-N-nitrosourea. Biochem J. 1970 Jun;118(1):99–109. doi: 10.1042/bj1180099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich V., Tetzlaff I., Graffi A. Studies on nitrosodimethylamine: preferential methylation of mitochondrial DNA in rats and hamsters. Chem Biol Interact. 1972 Jan;4(2):81–89. doi: 10.1016/0009-2797(72)90001-4. [DOI] [PubMed] [Google Scholar]