Abstract
Catechol O-methyltransferase (COMT) metabolizes catechol moieties by methylating a single hydroxyl group at the meta- or para- hydroxyl position. Hydrophobic amino acids near the active site of COMT influence the regioselectivity of this reaction. Our sequence analysis highlights their importance by showing that these residues are highly conserved throughout evolution. Reaction barriers calculated in the gas phase reveal a lower barrier during methylation at the meta- position, suggesting that the observed meta-regioselectivity of COMT can be attributed to the substrate itself, and that COMT has evolved residues to orient the substrate in a manner that increases the rate of catalysis.
1. Introduction
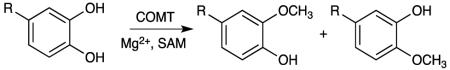
Catechol O-methyltransferase (COMT) is an enzyme that is primarily responsible for the deactivation of biologically active and toxic catechols and is found in virtually all mammalian tissues [1]. Many neurotransmitters and hormones within the body are characterized by a catechol or catecholamine structure and hence serve as substrates for COMT. Deactivation of substrates proceeds by methylation of a single hydroxyl to form the meta- or para- product [2].
The active site consists of the catechol ligand, which is mono-coordinated to Mg2+, and the co-enzyme S-adenosyl methionine (SAM) that serves as the methyl donor. Conversion to the guaiacol product proceeds via an SN2 mechanism, where Lys144 serves as a catalytic base and deprotonates a single hydroxyl to form the oxyanion that attacks a methyl group from the sulfonium. Several groups have calculated the reaction barrier for catechol to be approximately 20 kcal/mol in height [3-5].
Meta-methylation is the preferential product of COMT with the extent of regioselectivity dependent upon the catechol side-chain (Table 1) [6]. There has been no reported ligand where para-methylation is favored. It has been largely believed that the preference for meta-methylation stems from side-chain interactions with the residues comprising a hydrophobic wall near the active site. Previous molecular dynamics simulations have proposed that Trp143 forms a cation-π interaction with the substrate, conferring additional stability to the meta- position exclusively [4]. Migration of the Trp143 residue from binding to the adenosine motif of the SAM cofactor (as found in the crystal structure) to binding the side-chain of the catechol ligand demonstrates how the active site dynamics of COMT can modulate regioselectivity [7].
Table 1.
Methylation of COMT Ligands
 | ||
|---|---|---|
| R | Ligand | Meta/Para Ratio [6] |
| -CH2CH2NH3+ | Dopamine | 4.2 |
| -CH2CH(NH3+)COO− | Levodopa | 15.4 |
It is intriguing that an enzyme whose sole function is to degrade neurotransmitters and hormones displays specific regioselective behavior at the catechol moiety, a function in which site-specific chemistry seems unnecessary. Additionally, it has been assumed throughout the literature that regioselectivity has been imposed by the enzyme alone and that the ligands themselves have no preference [1,6,8]. In this Letter, we examine to see if the ligands possess intrinsic regioselective properties and whether the protein has adapted to take advantage of such properties by using regioselection as a method to enhance the rate of catalysis. Three models are constructed for this study: Model 1) a simple model composed of the catecholamine and trimethylsulfonium; Model 2) a model of catecholamine and trimethylsulfonium with four solvation shells of water; and Model 3) a model of the protein’s active site composed of the residues Trp143 and Lys144, Mg2+ coordinated to crystallographic water and surrounding residues, and the catecholamine and trimethylsulfonium. We compute the reaction barriers for methylating dopamine and levodopa at the meta- and para- positions using DFT since they represent two extremes of regioselection with meta/para ratios of approximately 4 and 16, respectively.
2. Methods
2.1. Sequence conservation of COMT
Sequence alignments of COMT were performed for 13 different species: Homo sapiens, Mus musculus, Bos taurus, Equus caballus, Sus scrofa, Rattus norvegicus, Mycobacterium vanbaalenii, Pichia stipitis, Mycobacterium gilvum, Nicotiana tabacum, Salmo salar, Populus trichocarpa, Papaver somniferum, Thalictrum tuberosum. The software ClustalX 2.0.10 was used to perform the alignment using the Neighbor Joining algorithm with default parameters.
2.2. MD simulation of ligand complexes
The crystal structure of human soluble-COMT (PDB: 3BWM) was used as a starting point for our MD simulations. A caveat of the crystal structure, as published by several groups, is that the inhibitor used to crystallize the protein has a different binding geometry compared to natural ligands. Specifically, since most inhibitors decrease the nucleophilicity of the attacking oxygen with nitrate groups, these ligands bind bidentate to the Mg2+ in the active site. Several groups have proposed that natural substrates of COMT contain a monodentate coordination number in the active site [4,5]. Additionally, the interaction between Trp143 and the ligand side chain is possible only after MD simulations [4].
Thus, we initially dock either dopamine or levodopa into the active site using the 3,5-dinitrocatechol substrate position as a reference. We adopt a similar simulation methodology as outlined by Kuhn et al in order to obtain our minimized complexes [4]. Briefly, we simulate our system using the AMBER force field (amber03) and derived constraints for our ligands and S-adenosylmethionine using Antechamber. Parameters for Mg2+ were adjusted according to Kuhn’s values; i.e., an atomic charge of +2, R*Mg = 0.7868 Å, εMg = 0.8751 kcal/mol. The crystallographic water remains coordinated to Mg2+. Additionally, the optimal nonbonded parameters between the attacking oxygen and donor sulfur were derived from Kollman (R*S = 2.0000 Å, εS = 0.2500 kcal/mol).
Charges were assigned using RESP, with two Na+ counterions to neutralize charges, and the ligand was solvated with a 20 Å sphere of TIP3P water. The counterions were over 16 Å away from the active site and were fixed to remove potential artificial long-range electrostatic effects. Simulations were performed with a nonbonding cutoff of 16 Å, a time step of 1.5 fs with SHAKE, and at a constant temperature of 300 K using the Berendsen coupling scheme.
For the first minimization, the water molecules were relaxed for 1500 steps while all other atoms remained rigid. The water was then equilibrated using MD simulation for 20 ps. Next, we progressively removed the positional restraints on the protein-ligand complex (from 25 to 0 kcal/(mol Å2)) over a period of 4000 minimization steps. The system then underwent MD equilibration for 20 ps. To avoid loss of water near the vicinity of the protein-ligand complex due to diffusion, the water shell was stripped and a new 20 Å shell of TIP3P was added, minimized, and equilibrated as described above. The protein-ligand complex was then minimized for another 4000 steps followed by an additional 20 ps of MD equilibration. A final minimization of 2000 steps was performed to yield our final structure.
2.3. QM determination of reaction barriers
We used three different models to study the methylation reaction between levodopa/dopamine and trimethylsulfonium (a substitute for S-adenosylmethionine). Model 1 was constructed by modeling only the ligand and trimethylsulfonium. Model 2 was constructed by taking Model 1 and adding four solvation layers of water, followed by minimization. Model 3 was generated from the minimized ligand pose generated using AMBER simulations of COMT with bound ligand [4]. We included residues coordinated to Mg2+ (as well as its crystallographic water), the catalytic Lys144, and the regioselective Trp144 in our final construct for Model 3. In all models, the reaction coordinate is examined along the sulfur-oxygen axis.
Using a B3LYP functional, we performed a QST2 search for Models 1 and 3 to find the transition state of each methylation reaction using Gaussian03. A mixed basis set of 6-31G for C, H and 6-311+G* for S, Mg, O, and N were used. Reactants of each methylation reaction were computed by geometry optimization. NBO analysis was used to examine the interaction energies between the two ligands. In addition, a level-of-theory study was performed on Model 1 to monitor the effects of changing density functionals on our barrier height calculations.
We calculate the energies of the transition state and reactant for Model 2 by taking the transition state for the simple model in the gas phase and solvating it with four water shells. The water was minimized using molecular mechanics, and then a single point calculation was done at the quantum level (using the same basis set) with the water treated using molecular mechanics. It is important to note that the single point calculations done in explicit solvent likely underestimate the stabilization due to the polarization of water since the dielectric response of water is primarily determined by its orientational dynamics. Nonetheless Model 2 serves as an estimate of the methylation reaction occurring in an aqueous environment.
3. Results and discussion
3.1. Residues involved in regioselection are evolutionarily conserved
Our sequence analysis shows that residues comprising the hydrophobic wall of COMT are conserved throughout evolution (Figure 1). Many residues from the hydrophobic wall are as equally conserved as those coordinated to magnesium in the active site. Trp143 is as equally conserved as the catalytic base Lys144, though Trp143 also is known to initially bind to the adenosine moiety of SAM. For comparative purposes, we also show residues found in loops of the solvent accessible surface that expectedly have poorer sequence conservation.
Figure 1.
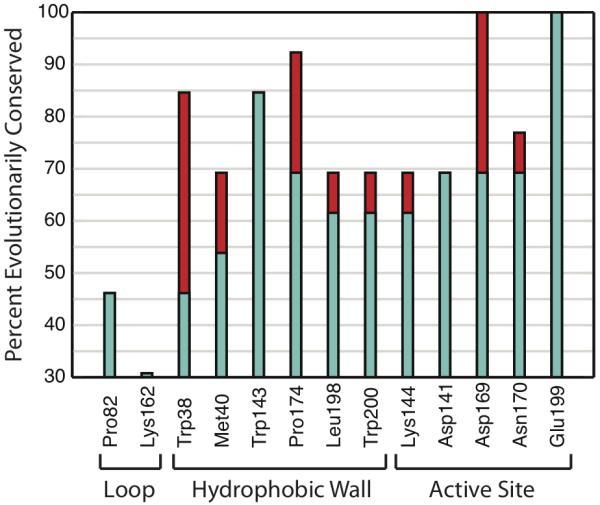
Sequence analysis of COMT shows high conservation of residues comprising hydrophobic wall. (Turquoise) Identical sequence conservation. (Red) Similar sequence conservation.
High conservation of the hydrophobic wall residues suggests that its regioselective role is important for enzymatic function. Regioselection is most commonly associated with chemical specificity, but from a biological standpoint there is no need for such specificity since the products are degraded after interactions with COMT and monoamine oxidase (in no specified order) [1]. The fundamental purpose of enzymes is to accelerate chemical reactions, and thus the conservation of regioselectivity has led us to hypothesize that COMT may have evolved regioselectivity as a method for chemical rate enhancement.
3.2. Reaction barrier differences between ligand conformations
The purpose of Models 1 and 2 is to deduce whether or not the ligands possess intrinsic regioselective properties, and what are the contributing components. Model 3 mimics the active site environment to give insight into what extent the protein contributes to regioselection. After obtaining the MD minimized structure for Model 3, we conducted DFT minimization using only the atoms described above. Our final complex after DFT minimization supports the hypothesis from Kuhn et al that Trp143 is involved in a cation-π with the catechol side-chain, as we can directly observe this interaction when modeling quantum mechanically (Figure 2).
Figure 2.
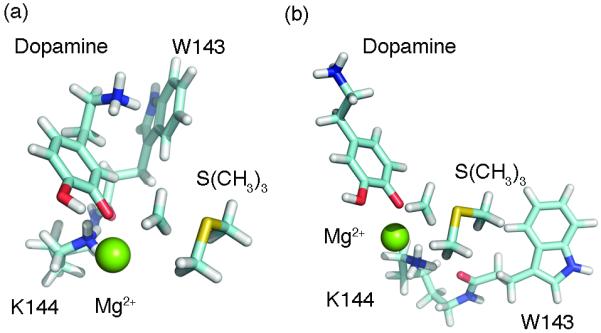
Transition state structures of COMT show Trp143 forming cation-π interaction exclusively in the meta conformation. Residues coordinated to Mg2+ removed for clarity. (a) Meta-methylation. (b) Para-methylation.
From our calculations, we observe higher reaction barriers for the para conformation of levodopa and dopamine that effectively slows its rate of methylation (Table 2, Table 3). The energies calculated between Models 1 and 2 had no significant difference, likely due to utilizing a single-point calculation. Meta-methylation becomes more favorable for levodopa when occurring in the active site, potentially underlying an important role for Trp143 in minimizing the reaction barrier.
Table 2.
Reaction Barriers and Energy Decompositions for Different COMT Ligands Conformationsa
| Ligand-Conformation (Model) | ΔEe | ΔExc | ΔTS | ΔE |
|---|---|---|---|---|
| Levodopa-Meta (1) | 11.9 | 33.2 | −27.8 | 17.3 |
| Levodopa-Para (1) | 22.0 | 27.0 | −28.9 | 20.1 |
| Dopamine-Meta (1) | 10.1 | 31.5 | −24.8 | 16.8 |
| Dopamine-Para (1) | 30.4 | 26.6 | −31.9 | 25.0 |
| Levodopa-Meta (3) | 37.9 | 36.8 | −57.4 | 17.3 |
| Levodopa-Para (3) | 33.7 | 41.2 | −51.9 | 23.0 |
| Dopamine-Meta (3) | 42.6 | 49.9 | −58.3 | 34.2 |
| Dopamine-Para (3) | 39.5 | 42.8 | −52.2 | 30.2 |
Units are in kcal/mol. Legend: ΔEe = electrostatic energy, ΔExc = exchange-correlation energy, ΔTS = kinetic energy, ΔE = reaction barrier height.
Table 3.
Performance comparison of difference density functionals on the transition state barrier height of the dopamine and levodopa reactions for the simplest model systems. Units in kcal/mol
| Dopamine | Levodopa | Dopamine | Levodopa | |||
|---|---|---|---|---|---|---|
| Meta- | Para- | Meta- | Para- | Meta-Para | Meta-Para | |
| HF | 58.57 | 66.19 | 59.41 | 62.14 | −7.62 | −2.74 |
| BP86 | 34.38 | 42.97 | 34.96 | 39.24 | −8.59 | −4.29 |
| BLYP | 36.94 | 45.04 | 37.37 | 41.29 | −8.09 | −3.92 |
| TPSSTPSS | 36.71 | 45.25 | 37.02 | 41.82 | −8.54 | −4.80 |
| MP2 | 40.67 | 47.57 | 41.06 | 45.65 | −6.89 | −4.60 |
| SVWN5 | 27.55 | 36.36 | 28.01 | 33.05 | −8.81 | −5.05 |
| VSXC | 27.40 | 31.19 | 24.99 | 35.94 | −3.79 | −10.94 |
| B3LYP | 42.18 | 50.18 | 42.70 | 46.49 | −8.00 | −3.79 |
| B3PW91 | 41.31 | 49.79 | 42.04 | 46.00 | −8.48 | −3.96 |
| B972 | 42.79 | 50.97 | 43.57 | 47.37 | −8.18 | −3.79 |
| O3LYP | 43.91 | 52.11 | 44.90 | 48.14 | −8.20 | −3.24 |
| BMK | 43.39 | 51.02 | 44.02 | 47.57 | −7.63 | −3.55 |
| BHandHLYP | 48.49 | 56.29 | 49.11 | 52.45 | −7.80 | −3.34 |
| M062X | 43.46 | 50.65 | 43.66 | 48.43 | −7.20 | −4.76 |
| tHCTHhyb | 38.95 | 47.21 | 39.55 | 43.86 | −8.26 | −4.32 |
| X3LYP | 42.06 | 49.97 | 42.55 | 46.38 | −7.91 | −3.83 |
| CAM-B3LYP | 45.33 | 53.30 | 45.94 | 49.56 | −7.98 | −3.62 |
| B2PLYP | 41.21 | 57.95 | 41.59 | 45.73 | −16.74 | −4.14 |
| mPW2PLYP | 41.75 | 49.25 | 42.13 | 46.27 | −7.51 | −4.14 |
| LC-wPBE | 47.48 | 55.75 | 48.42 | 51.99 | −8.27 | −3.57 |
| HSE2PBE | 40.34 | 48.57 | 41.00 | 45.03 | −8.23 | −4.04 |
| B1LYP | 43.68 | 51.58 | 44.20 | 47.87 | −7.90 | −3.67 |
| wB97X | 44.53 | 52.04 | 45.25 | 48.88 | −7.51 | −3.62 |
| wB97XD | 42.03 | 49.68 | 42.64 | 47.13 | −7.64 | −4.49 |
| LC-BLYP | 48.13 | 56.20 | 48.89 | 52.30 | −8.07 | −3.41 |
Our comparison of different functionals on Model 1 calculations unanimously shows that the meta-methylation reaction is more favorable (Table 3). Although there are large variations in how quantitatively accurate the barrier heights are, the qualitative trend clearly favors meta-methylation for both ligands. In our model of dopamine inside the active site of COMT (Model 3), there is actually a 4 kcal/mol preference for para-methylation which is in opposition to experimental observations. The meta/para ratio for dopamine is four times lower than the ratio for levodopa, and thus Model 3 of dopamine may be insufficient to detect such minor differences. Nonetheless, from Models 1 and 2 it can be concluded that it is the enzyme itself that is giving rise to regioselection as the ligand itself has no preference.
3.3. Chemical differences between meta- and para- nucleophiles
The differences in reaction barriers alone do not give chemical insight into why regioselectivity takes place. To investigate further, we perform an NBO analysis to calculate the interaction energies between the methyl donor and ligand using Model 1. We find that the oxyanion in the meta TS is sp3 hybridized while the para oxyanion is sp2 hybridized. As a result, the interaction energy between the methyl group and the oxyanion is significantly higher in the meta conformation (Table 4).
Table 4.
Conceptual DFT Descriptors and NBO Analysis for Model 1a
| Ligand-Conformation | Eint | χ | η | |
|---|---|---|---|---|
| Levodopa-Meta | 62.3 | 23.9 | 72.8 | 71.5 |
| Levodopa-Para | 37.9 | 2.7 | 64.0 | 55.8 |
| Dopamine-Meta | 48.9 | 26.7 | 70.9 | 70.9 |
| Dopamine-Para | 51.6 | 5.2 | 60.2 | 69.0 |
Units in kcal/mol. Legend: χ = electronegativity, η = chemical hardness, Eint = total interaction energy between SAM and ligand, = interaction energy between oxyanion and methyl group.
Chemical descriptors can describe the consequences of differences in oxyanion hybridizations [9,10]. The meta- nucleophile of each ligand is considered more electronegative and a harder base, indicative of it being a stronger nucleophile (Table 4). Notably, the meta-oxyanion of dopamine is still a more effective nucleophile than its para-oxyanion counterpart. Despite the increased nucleophilicity of the meta-oxyanion, the overall interaction energy between dopamine and trimethylsulfonium remains nearly identical for both conformations. Comparison of the total interaction energies between the ligand and methyl donor demonstrates that there is a significantly larger difference between the two conformations of levodopa as compared to dopamine (Table 4), consistent with experimental observations that levodopa has the higher meta-methylation preference (Table 1).
4. Conclusions
Our results propose that COMT utilizes regioselectivity not for the purposes of chemical specificity, but instead to enhance its catalytic rate by barrier minimization. Deprotonation of the 3′ hydroxyl creates a more effective nucleophile and enables some ligands to confer regioselectivity as demonstrated by levodopa in our work. COMT has evolved to take advantage of this property by selecting for residues that dock ligands preferentially in the meta-conformation and thereby lowering the activation barrier for methylation.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Health Grant R01GM080742 and the ARRA supplement 3R01GM080742-03S1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mannisto PT, Kaakkola S. Pharmacol. Rev. 1999;51:593. [PubMed] [Google Scholar]
- [2].Masri MS, Robbins DJ, Emerson OH, Deeds F. Nature. 1964;202:878. doi: 10.1038/202878a0. [DOI] [PubMed] [Google Scholar]
- [3].Zheng Y, Bruice TC. J. Am. Chem. Soc. 1997;119:8137. [Google Scholar]
- [4].Kuhn B, Kollman PA. J. Am. Chem. Soc. 2000;122:2586. [Google Scholar]
- [5].Roca M, Marti S, Andres J, Moliner V, Tunon I, Bertran J, Williams IH. J. Am. Chem. Soc. 2003;125:7726. doi: 10.1021/ja0299497. [DOI] [PubMed] [Google Scholar]
- [6].Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melen K, Julkunen I, Taskinen J. Biochemistry. 1995;34:4202. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- [7].Papoian GA, DeGrado WF, Klein ML. J. Am. Chem. Soc. 2003;125:560. doi: 10.1021/ja028161l. [DOI] [PubMed] [Google Scholar]
- [8].Rutherford K, Le Trong I, Stenkamp RE, Parson WW. J. Mol. Biol. 2008;380:120. doi: 10.1016/j.jmb.2008.04.040. [DOI] [PubMed] [Google Scholar]
- [9].Liu S. Acta Phys. Chim. Sin. 2009;25 [Google Scholar]
- [10].Liu S, Hu H, Pedersen LG. J. Phys. Chem. A. 2010;114:5913. doi: 10.1021/jp101329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


