Abstract
Estrogens regulate osteoblast differentiation and mineralization. We identified GATA4 as a transcription factor expressed in osteoblasts and directly regulated by 17β-estradiol in this cell type but not in breast cancer cells, another estrogen-responsive tissue. Chromatin immunoprecipitation sequencing (chromatin immunoprecipitation sequencing) reveals that estrogen receptor α (ERα) binds to chromatin near GATA4 at five different enhancers. GATA4 and ERα are both recruited to ERα binding sites near genes that are specifically expressed in osteoblasts and control osteoblast differentiation. Maximal binding of GATA4 precedes ERα binding, and GATA4 is necessary for histone 3 lysine 4 dimethylation at ERα binding sites, suggesting that GATA4 is a pioneer factor for ERα. As such, knockdown of GATA4 reduced recruitment of ERα to DNA. Our study illustrates that GATA4 is a pioneer factor for ERα recruitment to osteoblast-specific enhancers.
How estrogens exert cell-type- and disease-specific effects remains to be explained. Estrogen receptor α (ERα) and ERβ can bind to DNA at specific DNA motifs termed estrogen response elements (ERE). The classical ERE is a 13-bp inverted palindromic sequence, GGTCANNNTGACC. In addition, ERα can indirectly activate transcription by binding to other DNA binding proteins such as Sp1 and c-fos or c-jun (1). One computational prediction revealed over 70,000 EREs in the human genome (2). A second study predicted over 38,000 stringent EREs (3). However, when a subset of these was tested by chromatin immunoprecipitation (ChIP) in MCF7 breast cancer cells, only 14% of these sites could be validated (3). The unused EREs may never be used, or more likely, they are bound by ERα in other cell types or under other cellular conditions. Our recent data support a model for the cell-type-specific action of 17β-estradiol (E2) being driven primarily through specific ERα occupancy of epigenetically marked cis-regulatory regions of target genes (4).
Major advances in the understanding of estrogen biology have come from unbiased, whole-genome analysis of ERα binding sites. The location of ERα binding to DNA in MCF7 breast cancer cells, the ERα cistrome, was determined by combining ChIP with hybridization to whole-genome tiling arrays (ChIP-on-chip) (5, 6). These studies revealed more than 3600 high-confidence binding sites for ERα in MCF7 cells with the vast majority of sites located greater than 1 kb away from promoter proximal regions (5). In osteoblast-like cells, U2OS-ERα cells (4, 7), only 15% of the ERα binding sites overlapped with those seen in MCF7 cells, consistent with unique gene expression patterns and a high level of tissue specificity (4).
ERα and ERβ have each been detected by several methods in both osteoblasts and osteoclasts (8–10). One mechanism of estrogen's protective role in bone is induction of apoptosis in osteoclasts in a paracrine manner (11); E2 induces Fas ligand (FasL) specifically in osteoblasts, leading to FasL-mediated death of osteoclasts (12). E2 has also been shown to suppress the osteoclastogenic cytokines IL-1, IL-6, IL-7, and TNFα (13–17).
An important insight into cell-type-specific ERα binding patterns came from the discovery that forkhead box A1 (FoxA1) acts as a pioneer factor for recruitment of ERα to its binding sites in breast cancer cells (18). FoxA1 [hepatocyte nuclear factor 3α (HNF3α)], a member of the forkhead family of transcription factors, is involved in the development and differentiation of several organs including liver, kidney, pancreas, lung, prostate, and mammary gland (19–21). However, FoxA1 is not expressed in osteoblasts, and the forkhead motif is not enriched in ERα binding sites in U2OS-ERα cells (4), indicating that proteins other than FoxA1 act as pioneer factors for ERα in osteoblasts.
Six GATA transcription factors have been indentified in vertebrates, and they are important regulators of tissue-specific gene expression during development. GATA transcription factors are so named because they bind to the consensus DNA sequence (A/T)GATA(A/G). Mechanistically, the GATA proteins bind DNA through two highly conserved zinc finger domains and activate transcription via a basic N-terminal transcription activation domain (22). Based on their expression patterns, the six family members can be separated into two subgroups. GATA1, -2, and -3 are expressed in hematopoietic stem cells, where they regulate differentiation-specific gene expression in T lymphocytes, erythroid cells, and megakaryocytes (23). GATA3 is also expressed in the developing kidney (24) and mammary gland, where it specifies mammary cell fate (25) and regulates ERα expression (26). GATA4, -5, and -6 are expressed in mesoderm- and endoderm-derived tissues such as heart and intestines, where they also regulate tissue-specific gene expression (22). GATA4 is best characterized in the heart, where it is expressed at high levels. GATA4 is also expressed in the hypothalamus, pituitary, testis, ovary, placenta, adrenals, pancreas, liver, and intestines (23, 27, 28). No role for any GATA factor in osteoblasts has been identified until now.
Our previous microarray analysis of osteoblast-like cells (4) revealed differential E2 regulation of GATA family proteins. This allowed us to hypothesize that a GATA family member might play a role in the transcriptional regulation of osteoblast differentiation mediated by E2. Here for the first time, we identified GATA4 as a potential E2 direct target in osteoblasts. These studies further show that GATA4 is critical in the regulation of osteoblast-specific genes by functioning to recruit ERα to DNA in osteoblasts.
Results
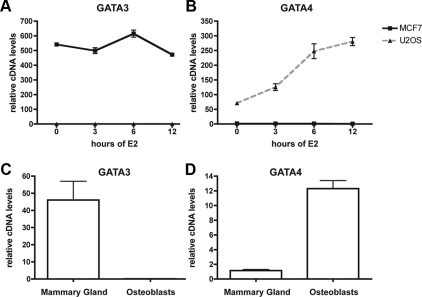
One of the genes whose expression is the most highly correlated with that of ERα in breast cancer encodes GATA3 (26). GATA3 regulates ERα in MCF7 cells, and reciprocally, ERα directly stimulates transcription of GATA3. However, in primary osteoblasts and an osteoblast-like cell line (U2OS-ERα), GATA3 is not expressed and is not regulated by E2 (Fig. 1, A and C). The U2OS-ERα cell line is an osteosarcoma cell line that was stably transfected with a doxycycline-inducible ERα (7). Upon treatment with doxycycline, the expression of ERα in U2OS-ERα cells is similar to MCF7 cells. The U2OS-ERα cells have some osteoblast-like properties, especially after ERα induction (4); however, these cells do not mineralize matrix. In search of tissue-specific GATA family members in osteoblasts, we used U2OS-ERα cells as a model and compared the gene expression profile with that obtained from MCF7 cells. We conducted microarray analysis that revealed unique ERα-mediated regulation controlling cell-type-specific gene regulation (4). Using a hierarchical clustering algorithm, we indentified GATA2 and -3 as being highly expressed basally in MCF7 cells (Supplemental Fig. 1, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). In contrast, GATA1, 4, -5, and -6 are highly expressed in U2OS-ERα cells, and GATA4, -5, and -6 are induced upon E2 treatment.
Fig. 1.
GATA4 is expressed in osteoblasts. A, U2OS-ERα and MCF-7 cells were deprived of estrogen for 3 d in phenol red-free media containing 5% CDT-FBS. They were then treated with 10 nm E2 for 0, 3, 6, or 12 h, and RNA was obtained. GATA3 mRNA was analyzed by qPCR and normalized to actin mRNA. B, Cells were treated as in A, and GATA4 mRNA was analyzed by qPCR and normalized to actin mRNA. C, Mammary gland and osteoblast mRNA was obtained and converted to cDNA. GATA3 mRNA was analyzed by qPCR and normalized to actin mRNA. D, Cells were treated as in C, and GATA4 mRNA was analyzed by qPCR and normalized to actin mRNA. Error bars in all panels represent ±1 sd.
To verify the array data, quantitative PCR (qPCR) was performed with cDNA from MCF7 and U2OS-ERα cells exposed to E2. GATA3 is expressed at a basal level over 500-fold higher in MCF7 cells than the basal level in U2OS-ERα cells (Fig. 1A). GATA3 is slightly induced by E2 after 6 h of induction (Fig. 1A) in MCF7 cells. GATA4 expression is basally very low in MCF7 cells and is not induced by E2 at 3, 6, or 12 h of E2 treatment (Fig. 1B). In contrast, GATA4 is expressed over 70-fold higher in U2OS-ERα cells than in MCF7 cells and is induced by E2 after 3 h and continues to rise after 6 and 12 h of E2 exposure (Fig. 1B). GATA5 is also expressed higher in U2OS-ERα cells than in MCF7 cells but only by approximately 3-fold (Supplemental Fig. 2). GATA6 is expressed at equal levels in MCF7 and U2OS-ERα cells (Supplemental Fig. 2).
To confirm that GATA family members are expressed in a tissue-specific manner as seen in the model cell lines, cDNA from primary calvarial osteoblasts was compared with cDNA from primary mouse mammary gland (Fig. 1, C and D). The basal level of GATA3 expression is high in the mammary gland (Fig. 1C), whereas the basal level of GATA4 is high in osteoblasts (Fig. 1D). GATA5 was expressed at equal levels in the mammary gland and osteoblasts; GATA6 was not detected in the osteoblasts (data not shown). Taken together, these results recapitulate findings in the osteoblast cell line model and led us to focus our attention on GATA4 as a tissue-specific transcription factor in osteoblasts.
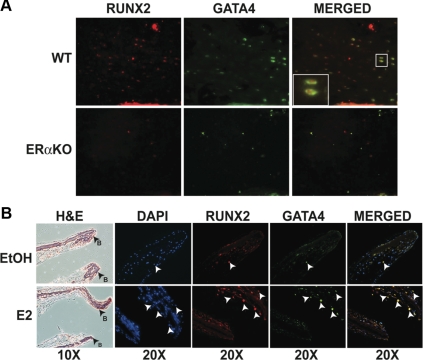
GATA4 is localized to osteoblasts in vivo and is induced by E2
To confirm that GATA4 is expressed in osteoblasts in vivo, we performed immunohistochemistry with a monoclonal antibody to GATA4. GATA4 was localized to the trabecular region of femurs from wild-type mice (Fig. 2A). To directly show that GATA4-positive cells were osteoblasts, immunofluorescence for the osteoblast-specific RUNX2 was performed. Indeed, GATA4 colocalized with nuclear RUNX2. In ERα-knockout (ERαKO) mice, fewer cells stained positive for either GATA4 or RUNX2 than in wild-type mice, suggesting that estrogen regulates both GATA4 and RUNX2. [It has been previously reported that E2 can enhance RUNX2 in osteoblasts) (29, 30).]
Fig. 2.
GATA4 is localized to osteoblasts in vivo and is induced by E2. A, Femurs from wild-type (WT) and ERαKO mice were snap-frozen and embedded in OCT. The 10-μm sections were stained with RUNX2 and GATA4. Images are of trabecular bone near the growth plate. B, Calvariae from neonatal d-2 mice were grown as organ cultures for 7 d and then treated with vehicle control [ethanol (EtOH)] or 10 nm E2 for 24 h. Calvariae were fixed and paraffin embedded. Serial sections were stained with hematoxylin and eosin (H&E) or RUNX2 and GATA4. Arrows indicate RUNX2 and GATA4 colocalization. DAPI, 4′,6-Diamidino-2-phenylindole.
To determine whether GATA4 is regulated by E2 in vivo, we used a calvarial organ culture system (31). Calvariae were isolated from neonatal d-2 wild-type mice and differentiated in the presence of β-glycerophosphate and ascorbic acid. After 1 wk of differentiation, calvariae were treated with 10 nm E2 or vehicle control. Cells were fixed and sectioned, and immunofluorescence was performed with antibodies to GATA4 and RUNX2. Vehicle-treated osteoblasts expressed a low level of GATA4 (Fig. 2B). After treatment with E2, many RUNX2-positive osteoblasts costained with GATA4. Nearby chondrocytes do not express GATA4. Together, these experiments clearly show that GATA4 is expressed in osteoblasts and is regulated by E2 through ERα.
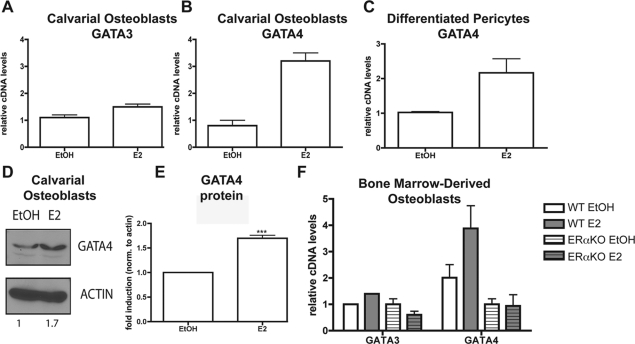
E2 induces GATA4 transcription in primary murine osteoblasts
Our data demonstrate that GATA4 is regulated by E2 in U2OS-ERα cells (Fig. 1B and Supplemental Fig. 1) and primary organ cultures (Fig. 2A). To further confirm that GATA4 is directly regulated by E2 in osteoblasts, primary calvarial osteoblasts were isolated and differentiated into mature osteoblasts. After differentiation for 10 d, treatment for 3 h with 10 nm E2 had no effect on GATA3 expression (Fig. 3A), but GATA4 mRNA was induced by 3-fold (Fig. 3B). We also verified that GATA4 is regulated by E2 in primary human osteoblasts. Perivascular cells were isolated and differentiated under osteoblastic conditions (Supplemental Fig. 3). E2 was added for 3 h, and qPCR demonstrates that GATA4 mRNA is up-regulated by E2 in human osteoblasts (Fig. 3C). To confirm that GATA4 protein is also induced by E2, primary calvarial osteoblasts were differentiated and treated with 10 nm E2 for 24 h. As shown in Fig. 3D, GATA4 protein, as detected by immunoblotting with a monoclonal antibody to GATA4, was induced 1.7-fold after E2 treatment. Three independent immunoblots were performed, and on average, GATA4 is induced 1.7-fold after E2 treatment (Fig. 3E). Gata4 mRNA was also induced by E2 in primary osteoblasts differentiated from bone marrow-derived osteoblasts (Fig. 3F). Furthermore, this induction is dependent on ERα, because GATA4 is not induced in primary osteoblasts from ERαKO mice (Fig. 3F). Interestingly, the basal level of GATA4 is lower in ERαKO osteoblasts than in wild-type mice, indicating that ERα is necessary for proper GATA4 expression. GATA3 is not regulated by E2 or ERα in bone marrow-derived osteoblasts (Fig. 3F). Therefore, GATA4 is a previously unrecognized osteoblast-specific transcription factor and is up-regulated by E2 via ERα.
Fig. 3.
GATA4 is induced by E2 in primary calvarial osteoblasts. A, Primary calvarial osteoblasts were differentiated for 10 d and then treated for 3 h with 10 nm E2. RNA was obtained, and GATA3 mRNA was analyzed by qPCR and normalized to actin mRNA. B, Cells were treated as in A, and GATA4 mRNA was analyzed by qPCR and normalized to actin mRNA. C, Human pericytes were differentiated to osteoblasts and then treated for 3 h with 10 nm E2. RNA was obtained, and GATA4 mRNA was analyzed by qPCR and normalized to actin mRNA. D, Primary calvarial osteoblasts were differentiated for 10 d and then treated for 24 h with 10 nm E2. Protein was obtained and immunoblotted for the presence of GATA4 and actin. Blots were quantitated using GeneSnap software, and the fold induction (normalized to actin) is depicted below the blots. E, Three independent immunoblots of GATA4 were performed as in D and quantitated using GeneSnap software, and the fold induction (normalized to actin) is graphed. ***, P value < 0.0003. F, Bone-marrow-derived osteoblasts from wild-type (WT) and ERαKO mice were differentiated for 10 d and then treated for 3 h with 10 nm E2. RNA was obtained, and GATA3 and GATA4 mRNA was analyzed by qPCR and normalized to actin mRNA. Error bars in all panels represent ±1 sd.
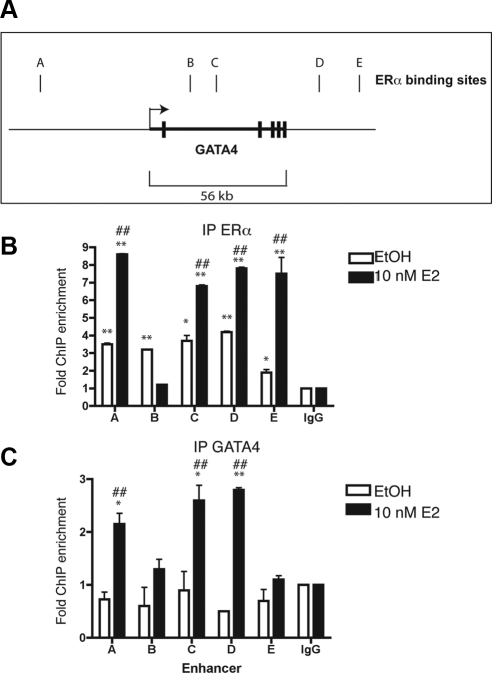
GATA4 and ERα regulate GATA4 transcriptional activity
Our results suggest that Gata4 transcription is directly activated by ERα both in vivo and in vitro. To determine whether ERα directly binds to DNA near the GATA4 gene, ChIP followed by massively parallel sequencing (ChIP-Seq, see below) identified five ERα binding sites near Gata4, and these were confirmed by directed ChIP and qPCR (Fig. 4, A and B). One of these sites is 5′ of the Gata4 coding region. Two of the sites are within Gata4 introns, and two of the sites are 3′ of Gata4 (Fig. 4A).
Fig. 4.
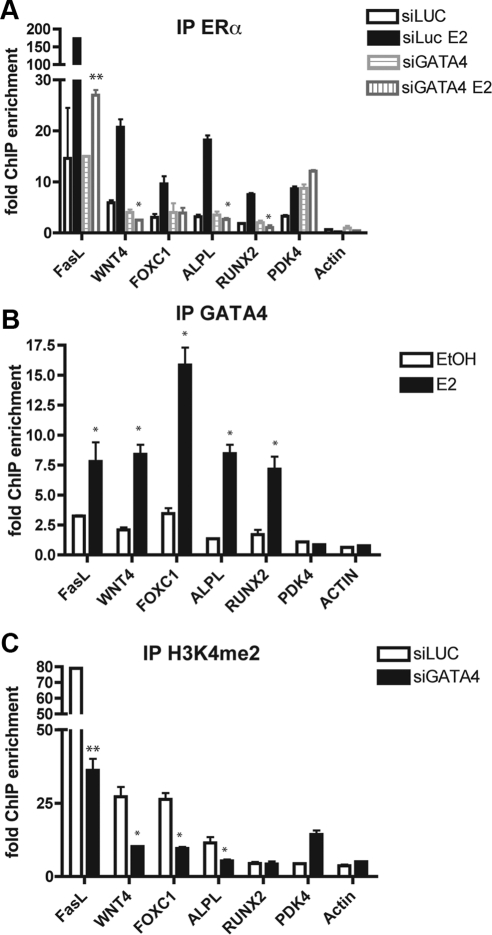
GATA4 and ERα regulate each other. Panel A, Schematic diagram of the GATA4 genomic locus. The transcriptional start site is indicated by the arrow. The exons are denoted by thick vertical lines. The ERα binding sites are named A–E and are represented by thin vertical lines. Panel B, U2OS-ERα cells were deprived of estrogen for 3 d in phenol red-free media containing 5% CDT-FBS. Cells were then treated for 45 min with 10 nm E2. ChIP was performed with antibodies to IgG or ERα, and qPCR was performed to detect the GATA4 enhancers. C, U2OS-ERα cells were treated as in panel B, and ChIP was performed with antibodies to IgG or GATA4. Quantitative PCR was performed to detect the GATA4 enhancers. Each PCR was normalized to input and represented as enrichment over a negative genomic locus [actin (ACTB) promoter]. *, P value < 0.01 vs. IgG immunoprecipitation using an unpaired t test; **, P value < 0.01 vs. IgG immunoprecipitation using an unpaired t test; # P value < 0.01 vs. ethanol (EtOH)-treated cells using an unpaired t test; ##, P value < 0.001 vs. EtOH-treated cells using an unpaired t test. Error bars in all panels represent ±1 sd.
GATA3 has been shown to regulate its own expression (26); therefore, we tested whether GATA4 also regulates itself. As demonstrated in Fig. 4C, GATA4 can be immunoprecipitated at three of the five ERα binding sites in U2OS-ERα cells to varying degrees of enrichment over a nonspecific genomic locus (the actin promoter) and ChIP performed with IgG. Taken together, the data suggest that ERα is recruited to multiple Gata4 enhancers directly and that GATA4 is recruited to its own enhancers to regulate Gata4 expression.
Genome-wide analysis of ERα binding sites reveals ERα binding near osteoblast genes
ChIP followed by deep sequencing was performed in U2OS-ERα cells with antibodies to ERα. A total of 22,151 peaks (binding sites) were identified (data not shown). Similar to other genome-wide analyses of ERα, less than 3% of binding sites are at the proximal promoter (Supplemental Fig. 4). Almost 31% of binding sites were intronic, 6.5% were in exons, and nearly 58% were considered to be distal intergenic regions (not in the gene itself). We searched the ERα binding sites for enriched transcription factor motifs by both de novo and candidate scanning approaches. This screen identified ERE half-sites as the most highly enriched transcription factor motif, as would be expected. Activation protein-1 sites were also highly enriched (data not shown).
We next searched for ERα binding sites near osteoblast genes and identified binding sites near Runx2, Alpl, and Faslg (FasL). RUNX2 is the master osteoblast transcription factor (32). There are six ERα binding sites downstream of Runx2 (Supplemental Fig. 5). Runx2 is not regulated by E2 in U2OS-ERα cells (Supplemental Fig. 8) but is regulated by E2 in primary osteoblasts (Fig. 2). Alkaline phosphatase is important for bone mineralization and is a key marker of osteoblast differentiation (33). There are four binding sites within Alpl (alkaline phosphatase) and two binding sites 3′ of Alpl (Supplemental Fig. 6). Alkaline phosphatase (ALPL) is up-regulated by E2 in U2OS-ERα cells (Supplemental Fig. 8) and primary osteoblasts (4). There are four binding sites 3′ of FasL and one site 5′ of FasL (Supplemental Fig. 7). FasL is up-regulated by E2 in U2OS-ERα cells (Supplemental Fig. 8) and osteoblasts to induce osteoclast apoptosis (12). Although these ERα binding sites are closest to the indicated genes and the genes are up-regulated by E2, it remains to be proven that the ERα binding sites are directly regulating the indicated genes.
GATA4 binding to ERα enhancers precedes ERα binding
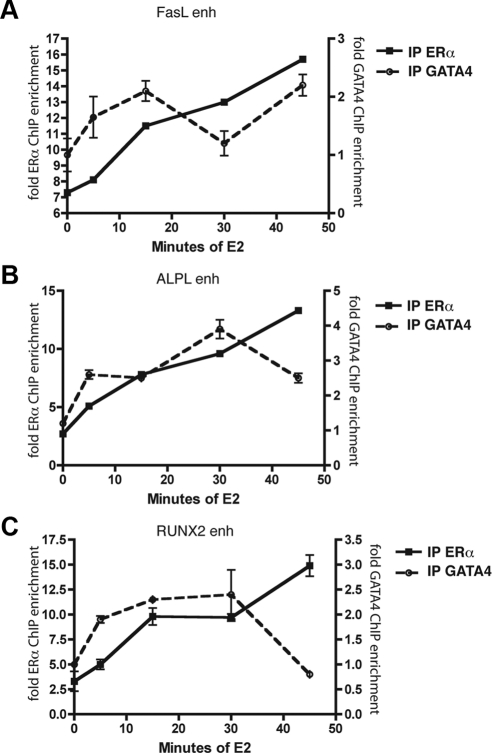
The kinetics of ERα at the pS2 promoter in MCF7 cells has been well characterized (34), with peak ERα binding occurring 45 min after E2 treatment. To determine the kinetics of ERα and GATA4 recruitment to various osteoblast genes, we performed ChIP after 5, 15, 30, and 45 min of E2 treatment in U2OS-ERα cells. Immunoprecipitation of ERα showed maximal ERα recruitment to enhancers near FasL, ALPL, and Runx2 45 min after E2 treatment (Fig. 5), in agreement with the time-course experiments in MCF7 cells. In contrast, GATA4 had maximal recruitment to these same enhancers 15–30 min after E2 treatment but preceding ERα recruitment to those same genes (Fig. 5). The data taken together illustrate that GATA4 recruitment to osteoblast genes precedes that of ERα and suggest a pioneer factor function for GATA4 for E2-responsive osteoblast-specific genes.
Fig. 5.
GATA4 binding to DNA precedes ERα binding. U2OS-ERα cells were deprived of estrogen for 3 d in phenol red-free media containing 5% CDT-FBS. Cells were then treated for 0, 5, 15, 30, or 45 min with 10 nm E2. ChIP was performed with antibodies to ERα or GATA4, and qPCR was performed to detect the enhancers (enh) near the indicated genes (A, FasL; B, ALPL; and C, RUNX2). Each PCR was normalized to input and represented as enrichment over a negative genomic locus [actin (Actb) promoter]. *, P value < 0.05 vs. time 0 for the corresponding antibody.
GATA4 is necessary for ERα binding near osteoblast-specific enhancers
Because GATA4 binding preceded ERα binding, and because GATA4 helps open chromatin in liver cells (35), we hypothesized that GATA4 opens the chromatin to help recruit ERα to ER binding sites. If GATA4 is a pioneer factor for ERα, knockdown of Gata4 should reduce the presence of ERα at the enhancers of E2-responsive osteoblast-specific genes. To test this, we knocked Gata4 down with small interfering RNA (siRNA) and performed ChIP at several ERα-binding enhancers. A reduction of Gata4 mRNA was confirmed by qPCR (Supplemental Fig. 9). Knockdown of Gata4 reduced the amount of ERα binding at enhancers near FasL, WNT4A, FOXC1, ALPL, and Runx2 (Fig. 6A). WNT4A and FOXC1 were identified in our previous microarray analysis as potential targets for ERα and are up-regulated in U2OS-ERα cells (Supplemental Fig. 8). FoxC1 is important in calvarial bone development (36), although its role in adult bone, especially in response to estrogens is unknown.
Fig. 6.
GATA4 is necessary for ERα recruitment to estrogen-responsive enhancers in osteoblasts. A, U2OS-ERα cells were transfected with either an siRNA directed at luciferase (siLUC) or GATA4 (siGATA4). At 48 h after transfection, cells were treated for 45 min with 10 nm E2. ChIP was performed with antibodies to ERα, and qPCR was performed to detect the ERα binding sites at the indicated enhancer regions. Each PCR was normalized to input and represented as enrichment over a negative genomic locus [hemoglobin (HBB) promoter]. B, U2OS-ERα cells were deprived of estrogen for 3 d in phenol red-free media containing 5% CDT-FBS. Cells were then treated for 45 min with 10 nm E2. ChIP was performed with antibodies to GATA4, and qPCR was performed to detect the enhancers near the indicated genes. Each PCR was normalized to input and represented as enrichment over a negative genomic locus [hemoglobin (HBB) promoter]. EtOH, Ethanol. C, Cells were treated as in B, and ChIP was performed with and antibody to H3K4me2. Quantitative PCR was performed to detect the indicated enhancer regions. Each PCR was normalized to input and represented as enrichment over a negative genomic locus [hemoglobin (HBB) promoter]. Error bars represent mean ± 1 sd. **, P value < 0.01; *, P value < 0.05 using an unpaired t test.
To confirm that GATA4 is recruited to these same sites, GATA4 was immunoprecipitated at the sites examined above. ERα and GATA4 were both enriched at enhancers near FasL, WNT4A, FOXC1, ALPL, and Runx2 (Fig. 6, A and B) after 45 min of E2 exposure. Not all ERα binding sites showed GATA4 recruitment, as demonstrated by the PDK4 enhancer (Fig. 6, A and B). GATA4 was not recruited to the ERα binding site near PDK4 (Fig. 6B), and siGATA4 had no effect on ERα binding to this enhancer (Fig. 6A). As a negative control, neither ERα nor GATA4 were recruited to the β-actin promoter before or after E2 treatment.
We showed previously that active ERα enhancers are enriched for histone 3 lysine 4 dimethylation (H3K4me2) (4). We therefore tested whether or not knockdown of Gata4 would reduce H3K4me2 at these ERα binding sites, thus explaining mechanistically why they are less capable of recruiting ERα. Indeed, less GATA4 correlated with a reduction in H3K4me2 at enhancers near FasL, WNT4A, FOXC1, and ALPL (Fig. 6C). H3K4me2 near Runx2 was not affected by siGATA4. Although ERα binds to the RUNX2 enhancer in U2OS-ERα cells, RUNX2 is not regulated by E2 in these cells, and thus, the region is already heterochromatic. However, RUNX2 is up-regulated by E2 in primary osteoblasts, and we would predict GATA4 would change the chromatin modifications in primary osteoblasts. There was no reduction in H3K4me2 near ACTB or PDK4 as would be expected, because these sites do not recruit GATA4. These results suggest that GATA4 binding is necessary for H3K4me2 at some ERα enhancers to recruit ERα.
Coactivator-associated arginine methyltransferase 1 (CARM1) action is critical to E2-stimulated gene expression in a tissue-specific manner (37). CARM1 dimethylates arginine residues in histones (i.e. H3R17me2) as well as other proteins in the transcriptional activation complex. E2-induced genes often have an induction of CARM1 activity (37). We tested whether knockdown of GATA4 would result in a reduction of other members of the transcriptional activation complex, in addition to ERα. Indeed, a reduction of GATA4 led to diminished E2-induced CARM1 activity (Supplemental Fig. 10). As such, it is proposed that GATA4 functions to recruit ERα to DNA at specific sites to induce cell-type-specific transcription.
Discussion
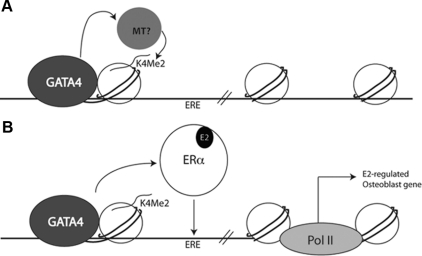
In attempts to better understand the mechanisms by which ERα is able to exert cell-type-specific regulation of the E2-responsive genes, we identified a tissue-specific transcription factor in osteoblasts. Experiments in vitro using assembled nucleosome arrays have demonstrated that GATA4 is a pioneer factor, capable of opening compacted chromatin using the albumin enhancer sequences in liver cells (35). Here we demonstrate that GATA4 acts as a pioneer factor in osteoblasts, capable of recruiting ERα to DNA. We hypothesize that GATA4 recruits a histone methyltransferase to the chromatin, allowing for H3K4 dimethylation in euchromatin (Fig. 7). As a result, ERα is recruited to ERE in open chromatin near E2-regulated osteoblast differentiation genes.
Fig. 7.
GATA4 is a pioneer factor for ERα. A, GATA4 recruits a histone methyltransferase (MT) leading to H3K4 dimethylation. B, After E2 treatment ERα is recruited to an ERE in an area of open chromatin marked by H3K4 dimethylation and GATA4. E2-regulated osteoblast genes are transcriptionally up-regulated. Pol II, Polymerase II.
GATA4 has been most extensively studied in the heart (reviewed in Ref. 38). It can be detected as early as d 7 of mouse embryogenesis where it precedes the expression of the earliest cardiac differentiation markers (39). In addition, GATA4 maintains cardiac gene expression in the adult heart. GATA4 is also expressed in the intestine, gonads, and liver, among other tissues (28). GATA4 has been shown to interact with many transcription factors, and these complexes may direct tissue-specific gene expression (22, 27). For example, in the heart, GATA4 interacts with NKX2.5 (40) to regulate cardiac α-actin (41). GATA4 expression has not been previously described in the bone, and understanding its tissue-specific function and how it achieves this bone specificity will be important to potentially understand postmenopausal osteoporosis. The GATA4-null mouse is embryonic lethal (42); thus, the bone phenotype is unknown, but further investigation of GATA4 function in bone is warranted. It would also be interesting to see whether GATA4 plays a role in pericytes undergoing the pathological process of arteriosclerosis, the calcification of blood vessels.
ERα binds only to a fraction of potential EREs in any cell type at a given time. The question then becomes what determines where ERα binds to DNA. We have shown previously that before E2 treatment, some enhancers have active chromatin marks (dimethylation at histone 3 lysine 4) or heterochromatic marks (dimethylation at histone 3 lysine 9) and that these correlate with ERα binding to DNA (4). Here we show for the first time that GATA4 is necessary for proper histone modifications to allow for active chromatin, as represented by H3K4 dimethylation, and subsequent ERα binding. Indeed, knockdown of Gata4 resulted in a reduction in H3K4me2 and a reduction in ERα binding. Therefore, tissue-specific transcription factors such as GATA4 help determine the pattern of ERα binding. There are still additional questions about how tissue specificity is determined. How does GATA4 choose between different GATA motifs? Does GATA4 perform any additional roles besides opening the chromatin? What other proteins are necessary for ERα binding in a tissue-specific manner? It is also possible that GATA4 is not a pioneer factor but a key coregulator that is necessary for ERα binding to DNA in osteoblasts.
In liver cells, GATA4 plays a role in opening the compacted chromatin in concert with HNF3 (FoxA) (35). HNF3 and GATA4 both occupy cis elements in the albumin gene enhancer in embryonic endoderm; in fact, GATA4 occupancy depends on the presence of FoxA at this enhancer, where they are both necessary for opening compacted chromatin, as demonstrated by deoxyribonuclease footprinting (43) and nucleosome arrays in vitro (35). Unlike MCF7 or liver cells, the FoxA factors are expressed at low levels in osteoblast cells, and there is no enrichment for the forkhead motif in the ERα binding sites in U2OS-ERα cells (4). Thus, whether ERα recruitment is independent of, or involves an additional factor besides, GATA4 in osteoblasts and other estrogen-responsive cell types remains to be determined.
In conclusion, we have shown that GATA4 is a tissue-specific transcription factor, with a high level of expression in osteoblasts. GATA4 is regulated by E2 and is necessary for recruitment of ERα to E2-dependent enhancers in osteoblast-like cells in a tissue-specific manner.
Materials and Methods
Reagents
E2 and doxycycline were purchased from Sigma-Aldrich Co. (St. Louis, MO). The following antibodies were used: ERα [Santa Cruz Biotechnology, Inc., Santa Cruz, CA (HC-20), and Lab Vision Corp. (Fremont, CA) (Ab-10)], GATA4 (Santa Cruz Biotechnology; G-4 and C-20), RUNX2 (Santa Cruz Biotechnology; M-70), H3K4me2 (Millipore, Billerica, MA),H3R17me2 (Millipore), and β-actin (Sigma-Aldrich).
Mice
All animal work was approved by the Animal Research Committee at University of California, Los Angeles. ERα, ERβ, and ERαβKO mice were kindly provided by Dr. Pierre Chambon (44).
Immunofluorescence
Femurs, taken from wild-type or ERαKO mice, were embedded in optimal cutting temperature compound, snap-frozen, cut into 10-μm-thick sections, and mounted onto slides coated with polylysine. The sections were briefly washed in dH2O and fixed for 15 min in 3% formaldehyde/PBS at room temperature and in methanol for 5 min at −20 C. After washing, they were incubated for 10 min in 3% hydrogen peroxide and rinsed in 1× PBS. Then they were blocked in 5% BSA/PBS for 30 min and incubated with rabbit anti-Runx2 and mouse anti-GATA4 diluted in 5% BSA/PBS overnight at 4 C. After washing, they were incubated with goat antirabbit Alexa Flour 568 and goat antimouse Alexa Flour 488 for 1 h at room temperature. As a negative control, serial sections were put through the same procedure with normal IgG as the primary antibody. All experiments were performed on femur sections from at least three different mice for each phenotype.
Primary calvarial organ culture
The calvariae of neonatal CD-1 mice were obtained 2 d after birth. The calvariae were placed on a stainless steel grid in a 12-well tissue culture dish, containing 2 ml phenol red-free DMEM supplemented with 10% charcoal dextran-treated fetal bovine serum (CDT-FBS), 5 mm β-glycerophosphate, and 100 μg/ml ascorbic acid (mineralization media) for 7 d. Calvariae were treated with 10 nm E2 for 24 h and then were fixed in 4% paraformaldehyde for 24 h, decalcified in Decal Stat (Decal Chemical Corporation, Tallman, NY) and paraffin embedded. Serial sections were stained with hematoxylin and eosin or GATA4 and RUNX2.
Cell culture
MCF7 cells were obtained from American Type Culture Collection (Manassas, VA). U2OS-ERα cells, kindly provided by Drs. Thomas Spelsberg and David Monroe, were maintained as described (7). At 24 h before treatment with E2, ERα expression in U2OS-ERα cells was induced by treatment with 100 ng/ml doxycycline.
Primary osteoblast cells
Primary osteoblasts were obtained from bone marrow mesenchymal stem cells (from wild-type or ERαKO mice) or the calvariae of wild-type mice. Bone marrow was incubated for 5 d in mesenchymal stem cell media (MesenCult Basal Media; StemCell Technologies Inc., Vancouver, BC, Canada), followed by differentiation in mineralization medium for 9 d. Neonatal BALB/c calvariae were obtained 2 d after birth and incubated for 40 min in α-MEM/1.0 mg/ml collagenase P/1.25% trypsin at 37 C. These were washed in α-MEM and transferred to α-MEM/1.0 mg/ml collagenase P/1.25% trypsin for 1 h at 37 C (32). Digestion was stopped by addition of α-MEM/10% FBS. The cells from the second digest were allowed to attach for 48 h and then differentiated in mineralization medium with media replacement every 3 d. Differentiation was confirmed by quantitation of col1A1, bone sialoprotein, and osteocalcin mRNA, alkaline phosphatase positivity, and alizarin red staining for mineralization (data not shown). Perivascular cells were obtained from human abdominal sc fat or lipoaspirate as described (45). Because specimens were obtained as anonymous and unidentifiable, the activities of the present research do not involve human subjects and therefore do not require Institutional Review Board review according to University of California, Los Angeles, Institutional Review Board medical committee standards. For in vitro bone formation, cells at 70% confluence were cultivated in DMEM, 10% FBS, 0.1 μm dexamethasone, 50 μg/ml l-ascorbic acid, and 10 mm β-glycerophosphate. RUNX2 RNA was quantitated to confirm osteoblast identity (Supplemental Fig. 7).
Mammary gland RNA
Mammary gland numbers 3 and 4 from 7- to 10-wk-old virgin C57BL/6 mice were isolated. RNA was purified using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
RNA and qPCR
Cells were hormone deprived by culture for 3 d in phenol red-free medium (Invitrogen) supplemented with 5% CDT-FBS (Omega Scientific, Tarzana, CA). Cells were treated with 10 nm E2 or ethanol as a vehicle control for 3, 6, or 12 h. Total RNA was converted to cDNA with Superscript III First Strand Synthesis Kit according to the manufacturer's instructions (Invitrogen). Primers were selected using Primer3 (46), and the sequences are listed in Supplemental Tables 1–3. cDNA was subjected to qPCR using the Applied Biosystems (Foster City, CA) SYBR Green Mastermix. Each RNA sample was collected in triplicate, and each PCR was amplified in triplicate.
Protein and immunoblotting
Cells were hormone deprived by culture for 3 d in phenol red-free medium (Invitrogen) supplemented with 5% CDT-FBS. Cells were treated with 10 nm E2 or ethanol as a vehicle control for 24 h and then lysed in EBC buffer [50 mm Tris (pH 8), 120 mm NaCl, 0.5% Nonidet P-40] supplemented with a protease inhibitor mixture (Complete; Roche Applied Science, Indianapolis, IN) for 30 min on ice. Proteins were subjected to SDS-PAGE and immunoblotting with antisera to the indicated proteins.
Chromatin immunoprecipitation
Cells were hormone deprived by culture for 3 d in phenol red-free medium (Invitrogen) supplemented with 5% CDT-FBS. Cells were treated with 10 nm E2 or ethanol as a vehicle control for 45 min, and ChIP was performed as described (5, 18). Immunoprecipitated DNA was amplified by qPCR using the Applied Biosystems SYBR Green Mastermix. Each ChIP was performed with triplicate biological replicates.
ChIP sequencing
Immunoprecipitated DNA was prepared for sequencing with the ChIP-Seq DNA Sample Prep Kit from Illumina (San Diego, CA). Samples were sequenced on an Illumina Genome Analyzer II, and 11.8 million tags were identified. Peaks were called with MACS software (http://cistrome.dfci.harvard.edu/ap/root) (47). Location analysis of binding sites was performed with CEAS (http://ceas.cbi.pku.edu.cn/submit.htm). A total of 22,151 binding sites were identified genome-wide. ERα binding sites on chromosomes 1 and 6 were compared with ChIP-on-chip data (4) with 81% overlap.
Small interfering RNA
Cells were hormone deprived by culture for 24 h in phenol red-free medium (Invitrogen) supplemented with 5% CDT-FBS before transfection with 100 nm siRNA oligonucleotide duplexes (Ambion, Austin, TX) directed against Gata4. A combination of three siRNA was used to obtain the highest level of knockdown. The sequences for the siRNAs are CGACUUCUCAGAAGGCAGAtt, GCCUCUUGCAAUGCGGAAAtt, and AGAUGGGACGGGUCACUAUtt. To confirm specificity for Gata4, a siRNA cocktail from Santa Cruz Biotechnology was also used (data not shown). siRNA directed against luciferase was used as a negative control. At 48 h after transfection, the cells were treated with 10 nm E2 for 24 h. RNA was obtained as described above or ChIP was performed as described above.
Statistical analysis
All experiments represent both biological and experimental triplicates. Error bars represent mean ± 1 sd.
Supplementary Material
Acknowledgments
This work was supported by a K12 BIRCWH Grant from the National Institutes of Health Office of Research on Women's Health (HD001400-08) and 1R56DK090231-01 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) to S.A.K. G.M.C. is currently supported by a K22 Grant from the National Cancer Institute (CA137168-01A1).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ALPL
- Alkaline phosphatase
- CARM1
- coactivator-associated arginine methyltransferase 1
- CDT-FBS
- charcoal dextran-treated fetal bovine serum
- ChIP
- chromatin immunoprecipitation
- E2
- 17β-estradiol
- ERα
- estrogen receptor α
- ERE
- estrogen response element
- ERαKO
- ERα knockout
- FasL
- Fas ligand
- FoxA1
- forkhead box A1
- H3K4me2
- histone 3 lysine 4 dimethylation
- HNF3α
- hepatocyte nuclear factor 3α
- qPCR
- quantitative PCR
- siRNA
- small interfering RNA.
References
- 1. Castro-Rivera E, Samudio I, Safe S. 2001. Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J Biol Chem 276:30853–30861 [DOI] [PubMed] [Google Scholar]
- 2. Bourdeau V, Deschênes J, Métivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S. 2004. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol 18:1411–1427 [DOI] [PubMed] [Google Scholar]
- 3. Vega VB, Lin CY, Lai KS, Kong SL, Xie M, Su X, Teh HF, Thomsen JS, Yeo AL, Sung WK, Bourque G, Liu ET. 2006. Multiplatform genome-wide identification and modeling of functional human estrogen receptor binding sites. Genome Biol 7:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M. 2008. Unique ERα cistromes control cell type-specific gene regulation. Mol Endocrinol 22:2393–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- 6. Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132:958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Monroe DG, Getz BJ, Johnsen SA, Riggs BL, Khosla S, Spelsberg TC. 2003. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERα or ERβ. J Cell Biochem 90:315–326 [DOI] [PubMed] [Google Scholar]
- 8. Bord S, Horner A, Beavan S, Compston J. 2001. Estrogen receptors α and β are differentially expressed in developing human bone. J Clin Endocrinol Metab 86:2309–2314 [DOI] [PubMed] [Google Scholar]
- 9. Braidman IP, Hainey L, Batra G, Selby PL, Saunders PT, Hoyland JA. 2001. Localization of estrogen receptor β protein expression in adult human bone. J Bone Miner Res 16:214–220 [DOI] [PubMed] [Google Scholar]
- 10. Vanderschueren D, Vandenput L, Boonen S, Lindberg MK, Bouillon R, Ohlsson C. 2004. Androgens and bone. Endocr Rev 25:389–425 [DOI] [PubMed] [Google Scholar]
- 11. Krum SA. 2011. Direct transcriptional targets of sex steroid hormones in bone. J Cell Biochem 112:401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krum SA, Miranda-Carboni GA, Hauschka PV, Carroll JS, Lane TF, Freedman LP, Brown M. 2008. Estrogen protects bone by inducing Fas ligand in osteoblasts to regulate osteoclast survival. EMBO J 27:535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pacifici R, Brown C, Puscheck E, Friedrich E, Slatopolsky E, Maggio D, McCracken R, Avioli LV. 1991. Effect of surgical menopause and estrogen replacement on cytokine release from human blood mononuclear cells. Proc Natl Acad Sci USA 88:5134–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weitzmann MN, Roggia C, Toraldo G, Weitzmann L, Pacifici R. 2002. Increased production of IL-7 uncouples bone formation from bone resorption during estrogen deficiency. J Clin Invest 110:1643–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jilka RL, Hangoc G, Girasole G, Passeri G, Williams DC, Abrams JS, Boyce B, Broxmeyer H, Manolagas SC. 1992. Increased osteoclast development after estrogen loss: mediation by interleukin-6. Science 257:88–91 [DOI] [PubMed] [Google Scholar]
- 16. Pacifici R, Rifas L, McCracken R, Vered I, McMurtry C, Avioli LV, Peck WA. 1989. Ovarian steroid treatment blocks a postmenopausal increase in blood monocyte interleukin 1 release. Proc Natl Acad Sci USA 86:2398–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krum SA, Chang J, Miranda-Carboni G, Wang CY. 2010. Novel functions for NFkappaB: inhibition of bone formation. Nat Rev Rheumatol 6:607–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- 19. Friedman JR, Kaestner KH. 2006. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci 63:2317–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. 2006. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127:1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spear BT, Jin L, Ramasamy S, Dobierzewska A. 2006. Transcriptional control in the mammalian liver: liver development, perinatal repression, and zonal gene regulation. Cell Mol Life Sci 63:2922–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Molkentin JD. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem 275:38949–38952 [DOI] [PubMed] [Google Scholar]
- 23. Viger RS, Guittot SM, Anttonen M, Wilson DB, Heikinheimo M. 2008. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol Endocrinol 22:781–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Labastie MC, Catala M, Gregoire JM, Peault B. 1995. The GATA-3 gene is expressed during human kidney embryogenesis. Kidney Int 47:1597–1603 [DOI] [PubMed] [Google Scholar]
- 25. Naylor MJ, Ormandy CJ. 2007. Gata-3 and mammary cell fate. Breast Cancer Res 9:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. 2007. Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res 67:6477–6483 [DOI] [PubMed] [Google Scholar]
- 27. Matsuda K, Kobune Y, Noda C, Ichihara A. 1994. Expression of GATA-binding transcription factors in rat hepatocytes. FEBS Lett 353:269–272 [DOI] [PubMed] [Google Scholar]
- 28. Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. 1993. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol 13:2235–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Plant A, Samuels A, Perry MJ, Colley S, Gibson R, Tobias JH. 2002. Estrogen-induced osteogenesis in mice is associated with the appearance of Cbfa1-expressing bone marrow cells. J Cell Biochem 84:285–294 [DOI] [PubMed] [Google Scholar]
- 30. McCarthy TL, Chang WZ, Liu Y, Centrella M. 2003. Runx2 integrates estrogen activity in osteoblasts. J Biol Chem 278:43121–43129 [DOI] [PubMed] [Google Scholar]
- 31. Mohammad KS, Chirgwin JM, Guise TA. 2008. Assessing new bone formation in neonatal calvarial organ cultures. Methods Mol Biol 455:37–50 [DOI] [PubMed] [Google Scholar]
- 32. Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747–754 [DOI] [PubMed] [Google Scholar]
- 33. Balcerzak M, Hamade E, Zhang L, Pikula S, Azzar G, Radisson J, Bandorowicz-Pikula J, Buchet R. 2003. The roles of annexins and alkaline phosphatase in mineralization process. Acta Biochim Pol 50:1019–1038 [PubMed] [Google Scholar]
- 34. Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F. 2003. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763 [DOI] [PubMed] [Google Scholar]
- 35. Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9:279–289 [DOI] [PubMed] [Google Scholar]
- 36. Rice R, Rice DP, Olsen BR, Thesleff I. 2003. Progression of calvarial bone development requires Foxc1 regulation of Msx2 and Alx4. Dev Biol 262:75–87 [DOI] [PubMed] [Google Scholar]
- 37. Lupien M, Eeckhoute J, Meyer CA, Krum SA, Rhodes DR, Liu XS, Brown M. 2009. Coactivator function defines the active estrogen receptor α cistrome. Mol Cell Biol 29:3413–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peterkin T, Gibson A, Loose M, Patient R. 2005. The roles of GATA-4, -5 and -6 in vertebrate heart development. Semin Cell Dev Biol 16:83–94 [DOI] [PubMed] [Google Scholar]
- 39. Charron F, Nemer M. 1999. GATA transcription factors and cardiac development. Semin Cell Dev Biol 10:85–91 [DOI] [PubMed] [Google Scholar]
- 40. Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. 1997. The cardiac transcription factors Nkx2–5 and GATA-4 are mutual cofactors. EMBO J 16:5687–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sepulveda JL, Belaguli N, Nigam V, Chen CY, Nemer M, Schwartz RJ. 1998. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol Cell Biol 18:3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Watt AJ, Battle MA, Li J, Duncan SA. 2004. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci USA 101:12573–12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. 1998. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J 17:244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dupont S, Krust A, Gansmuller A, Dierich A, Chambon P, Mark M. 2000. Effect of single and compound knockouts of estrogen receptors α (ERα) and β (ERβ) on mouse reproductive phenotypes. Development 127:4277–4291 [DOI] [PubMed] [Google Scholar]
- 45. Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, Norotte C, Teng PN, Traas J, Schugar R, Deasy BM, Badylak S, Buhring HJ, Giacobino JP, Lazzari L, Huard J, Péault B. 2008. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3:301–313 [DOI] [PubMed] [Google Scholar]
- 46. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.