Abstract
Resident stem/progenitor cells in the lung are important for tissue homeostasis and repair. However, a progenitor population for alveolar type II (ATII) cells in adult human lungs has not been identified. The aim of this study is to isolate progenitor cells from adult human lungs with the ability to differentiate into ATII cells. We isolated colony-forming cells that had the capability for self-renewal and the potential to generate ATII cells in vitro. These undifferentiated progenitor cells expressed surface markers of mesenchymal stem cells (MSCs) and surfactant proteins associated with ATII cells, such as CD90 and pro-surfactant protein-C (pro-SP-C), respectively. Microarray analyses indicated that transcripts associated with lung development were enriched in the pro-SP-C+/CD90+ cells compared with bone marrow-MSCs. Furthermore, pathological evaluation indicated that pro-SP-C and CD90 double-positive cells were present within alveolar walls in normal lungs, and significantly increased in ATII cell hyperplasias contributing to alveolar epithelial repair in damaged lungs. Our findings demonstrated that adult human lungs contain a progenitor population for ATII cells. This study is a first step toward better understanding of stem cell biology in adult human lung alveoli.
Keywords: lung repair, mesenchymal-to-epithelial transition, tissue stem cells, translational research
Resident stem/progenitor cells in the lung have important roles in tissue homeostasis and repair. The adult lung consists of four major, biologically distinct components including, the trachea, bronchi, bronchioles and alveoli, and each component has a specific stem/progenitor population.1, 2, 3, 4, 5 The alveoli are specialized terminal structures of distal airways for gas exchange. The gaseous alveolar surface is lined by two types of epithelial cells: alveolar type I (ATI) and alveolar type II (ATII) cells. ATI cells are broad- and flat-shaped cells, which cover 95% of the alveolar surface. ATII cells are cuboidal shaped and produce alveolar surfactants. ATI cells are more sensitive to injuries than ATII cells.6 When ATI cells are damaged, adjacent ATII cells are stimulated to proliferate and transdifferentiate into ATI cells.7, 8, 9, 10, 11 Therefore, ATII cells have long been considered to serve as progenitor cells in the alveoli.7, 8, 9, 10, 11
Recent studies on rodent models have identified stem/progenitor populations for alveolar epithelial cells, and have revealed that the stem/progenitor populations have important roles in lung repair and carcinogenesis.12, 13, 14, 15, 16, 17, 18 In distal lungs of adult human, to our knowledge, only two stem cell populations were isolated.19, 20 The first report showed that lung resident mesenchymal stem cells (MSCs) isolated from bronchoalveolar lavage fluid collected from lung transplant patients have the self-renewal ability and the differentiation capability into a mesodermal lineage.19 The second report demonstrated that MSCs isolated from distal lung tissues have the potential to express aquaporin 5 (AQP5; a marker for ATI cells) under the specific culture condition.20 However, a progenitor population that can differentiate into ATII cells has not been isolated from adult human lungs. Therefore, studies regarding roles of stem/progenitor cells in intractable pulmonary diseases, such as pulmonary fibrosis and lung emphysema, have been limited.
The aim of this study was to isolate progenitor cells from adult human lungs that can differentiate into ATII cells. We isolated undifferentiated MSC-like cells with expression of surfactant proteins associated with ATII cells. We found that these cells had the ability to self-renew and the capability to generate ATII cells in vitro. Microarray analyses indicated that genes encoding transcription factors important for lung development were enriched in this progenitor population, compared with bone marrow-MSCs (BM-MSCs). In addition, pathological evaluation showed that this progenitor population existed in normal alveoli, and significantly increased within ATII cell hyperplasia, which is recognized as regenerative epithelium in damaged lungs.10, 11
MATERIALS AND METHODS
Patients and Preparation of Tissue Specimens
For cell isolation, human lung tissues were obtained from patients who underwent lung resection at the Department of Thoracic Surgery at Tohoku University Hospital and the Department of Thoracic Surgery at the Ishinomaki Red Cross Hospital. Patients' characteristics are summarized in Table 1. In patients with lung tumors, lung tissues were obtained from sites away from the tumors. In patients with pulmonary fibrosis, fibrotic lungs and distant normal lungs were used. In patients with pneumothorax, normal lung tissues around the bulla were analyzed. Half of the tissues were used for cell isolation as described below, and remaining tissues were utilized for histological evaluations. The remaining tissues were inflated with formalin or a mixture of Tissue-Tek OCT compound (Sakura, Zoeterwounde, The Netherlands) and PBS, and embedded in paraffin or OCT compound. This study was approved by the Ethics Committee at Tohoku University School of Medicine and the Ishinomaki Red Cross Hospital. All subjects gave informed consent.
Table 1. Characteristics of patients (n=11)a.
| Age, years | 58.4±20.2 (20–80) |
| Gender, male/female | 8/3 |
| Height, cm | 163.5±9.3 (150–177) |
| Weight, kg | 60.4±13.4 (41–86.4) |
| Pack-years | 34.2±37.5 (0–95) |
| Indication for lung resection (number of the patients) | |
| Lung cancer | 7b |
| Open lung biopsy | 3c |
| Pneumothorax | 1 |
Data are expressed as mean±s.d. (range).
Two patients had chronic obstructive pulmonary disease (COPD). COPD severity was Stage I (mild) in both patients, based on spirometric classification by Global Initiative for COPD (GOLD).
Pathological examination showed that a patient had usual idiopathic pneumonia and two patients had organizing pneumonia.
Separation of Human Lung Cells and Cell Culture
Human lung cells were isolated as previously described with some modifications.12 We separated distal lung tissues within 3 cm from pleura. After pleura were separated bluntly, lung specimens were cut into 1 × 1 × 1 cm3 pieces. The samples were inflated with dispase II (Roche Applied Science, Mannheim, Germany; final concentration, 2.0 U/ml) and placed in conical tubes containing dispase II, collagenase/dispase (Roche Applied Science; final concentration, 1 mg/ml) and DNase I (Sigma-Aldrich, St Louis, MO, USA; final concentration, 0.1 mg/ml), and incubated for 90–120 min at 37°C with shaking. Enzymatically digested samples were minced with scissors, passed through needles, and filtered through a 100-μm mesh (BD Biosciences, San Jose, CA, USA). After treatment with red blood cell lysis buffer (Roche Applied Science), the cells were filtered through a 40-μm mesh (BD Biosciences) and resuspended in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (Invitrogen), 1% amino acids solution (Invitrogen), 100 units/ml penicillin, 100 μg/ml streptomycin (antibiotics; Sigma-Aldrich), and 2.5 μg/ml amphotericin B.
Hematopoietic cells were depleted from the lung cell suspension using the autoMACS Separator with anti-human CD45 antibody-coated micro-beads according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). Sorted CD45− lung cells were divided for primary culture, flow cytometric analysis, or preservation in liquid nitrogen.
To isolate a putative progenitor population, CD45– lung cells were seeded at a density of 1–5 × 104 cells/cm2 on mitotically inactivated mouse embryonic fibroblasts (feeders) with DMEM/10%FBS/1% amino acids solution/antibiotics in a humidified 5% CO2 incubator at 37°C. Mouse embryonic fibroblasts were isolated from C57BL/6 pregnant mice at 13 or 14 days of gestation. Mitotic inactivation was performed using mitomycin-C (Sigma-Aldrich). In CD45– lung cell cultures, amphotericin B (2.5 μg/ml) was added during the first week. For patient samples, subconfluent cells were trypsinized and plated on feeders on a 100-mm dish (TPP, Trasadingen, Switzerland) with DMEM/10%FBS/1% amino acid solution/antibiotics (passage 1). From passage 2, subconfluent cells were passaged with feeder-conditioned medium (FCM). FCM was defined as a mixture of DMEM/10% FBS/1% amino acids solution/antibiotics with supernatant collected from only feeder cultures, in the proportion of one part to one.
Human MSCs were purchased and cultured with TheraPEAK chemically defined MSC growth medium (Lonza Walkersiville, Walkersiville, MD, USA).
Flow Cytometry
Cell-surface and cytoplasmic antigens were examined by flow cytometry with the FACSCalibur flow cytometer (BD Biosciences). The data were analyzed by CellQuest Pro (BD Biosciences). We used the following antibodies: FITC-anti-human CD45, FITC-anti-human CD105, phycoerythrin (PE)-anti-human E-cadherin (Biolegend, San Diego, CA, USA), FITC-anti-human CD31, PE-anti-human CD73, allophycocyanin (APC) anti-human CD90, PE-anti-human c-kit (BD Biosciences), PE-anti-human CD133/1 (Miltenyi Biotec), FITC-anti-human CD34 (IMMUNOTECH, Praha, Czech) and PE-anti-human vascular endothelial growth factor receptor 2 (BD Biosciences). To stain E-cadherin, cultured cells were harvested using 1 mM EDTA. Isotype-matched antibodies were used as negative controls. To stain intracellular antigens, IntraPrep (Beckman Coulter, Fullerton, CA, USA) was used for fixation and permeabilization according to the manufacturer's instructions. Rabbit anti-human pro-surfactant protein-C (pro-SP-C) polyclonal antibody (Millipore Corporation, Billerica, MA, USA) was used as the primary antibody. PE-labeled goat anti-rabbit IgG (Molecular Probes, Carlsbad, CA, USA) or FITC-labeled goat anti-rabbit IgG (Vector, Burlingame, CA, USA) were used for the secondary antibodies. Normal rabbit serum (Sigma-Aldrich) and PE or FITC-labeled goat anti-rabbit IgG (Molecular Probes or Vector, respectively) were applied for negative control of pro-SP-C staining. We used primary CD45− lung cells as positive and negative controls for the staining of pro-SP-C and CD90. In addition, we optimized the concentration of rabbit anti-human pro-SP-C antibody and normal rabbit serum using primary CD45− lung cells in the flow cytometric analysis.
Immunofluorescence Staining in Cultured Cells
The cultured cells or the cytospun cells were fixed with acetone for 10 min at −20°C in staining for CD90. For staining of antigens other than CD90, cells were fixed with 4% paraformaldehyde for 10 min at 4°C. Samples were blocked and permeabilized with 0.1% Triton X/10% goat serum/1% BSA/PBS for 30 min at room temperature. The cells were incubated with primary antibodies as follows: mouse anti-human surfactant protein-A (SP-A) monoclonal antibody (1:1000, Abcam, Cambridge, MA, USA), rabbit anti-human pro-SP-C polyclonal antibody (1:1000, Millipore), mouse anti-human surfactant protein-D (SP-D) monoclonal antibody (1:20, Abcam), mouse anti-human vimentin monoclonal antibody (1:100, Dako, Glostrup, Denmark), mouse anti-human CD90 monoclonal antibody (1:50, BD Biosciences) overnight at 4°C. Alexa Fluor 546-conjugated goat anti-rabbit IgG and Alexa Fluor 488- or 647-conjugated goat anti-mouse IgG (1:100, Molecular Probes) were used as secondary antibodies. After mounting of sections and staining of nuclei using ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Molecular Probes), samples were observed via fluorescent microscopy (Leica Microsystems, Tokyo, Japan). To avoid false-positive fluorescence image development, the capture condition (exposure time: 200 ms) was fixed, and γ, contrast, or brightness of the images was not changed during the imaging processes.
Limiting Dilution Analysis and Single-Cell Sorting
The cells that we obtained using feeder culture were harvested at passage 4, and then 1, 10, or 100 cells/well were sorted and cultured on 96-well plates (Corning Incorporated, Corning, NY, USA). After 14 days, the number of wells without colonies was counted. To determine the frequency of clonogenic cells in each population, the cell number was determined from the point at which the regression line crossed 37%, which correlates with each colony provided by a single cell.12, 21 Regression analysis was performed with GraphPad Prism Version 5.0b (GraphPad Software, San Diego, CA, USA). To isolate and expand clonogenic cells, single cells that were sorted into 96-well plates (Corning Incorporated) by FACSAria II cell sorter (BD Biosciences) were cultured with FCM.
Total RNA Isolation and RT–PCR
Total RNA was extracted using TRIzol (Invitrogen). For microarray, the total RNA was cleaned up by miRNeasy Mini kit (QIAGEN, Tokyo, Japan). The isolated RNA was assessed for quantity and quality via UV spectrophotometry using a ND-100 spectrophotometer (Nano Drop Technologies, Rockland, DE, USA). cDNA was synthesized with QuantiTect Reverse Transcription Kit (QIAGEN) from 1 μg of the total RNA. One out of 20 of the single-strand cDNA products was used for each PCR reaction. Semiquantitative RT–PCR was performed with Takara Ex Taq Hot Start Version (Takara Bio, Shiga, Japan). Cycling conditions were as follows: (i) 5 min at 94°C; (ii) 30 s at 94°C; (iii) 30 s at each annealing temperature; (iv) 30 s at 72°C; and (v) 7 min at 72°C. The steps (ii–iv) were repeated in each cycle number. Human-specific intron-spanning primer pairs were used. The primer sequences, annealing temperatures, cycle numbers, and PCR product sizes are shown in Table 2. The PCR products were visualized with ethidium bromide after electrophoresis on 2% agarose gel.
Table 2. Sequences for primers and the PCR conditions.
| Gene | Sequence | Annealing temperature | Cycle number | Size | |
|---|---|---|---|---|---|
| SP-A | Forward | 5′-AGGAGCTCCAAGCCACACT-3′ | 62 | 40 | 197 |
| Reverse | 5′-TTCCTCTGGATTCCTTGGG-3′ | ||||
| SP-B | Forward | 5′-TGGGAGCCGATGACCTATG-3′ | 62 | 40 | 70 |
| Reverse | 5′-GCCTCCTTGGCCATCTTGT-3′ | ||||
| SP-C | Forward | 5′-ATCGGCTCCACTGGCCTCGT-3′ | 62 | 40 | 301 |
| Reverse | 5′-AGTAGAGCGGCACCTCGCCA-3′ | ||||
| AQP5 | Forward | 5′-ACTGGGTTTTCTGGGTAGGG-3′ | 60 | 40 | 184 |
| Reverse | 5′-GTGGTCAGCTCCATGGTCTT-3′ | ||||
| Vimentin | Forward | 5′-CCCTCACCTGTGAAGTGGAT-3′ | 60 | 30 | 241 |
| Reverse | 5′-TCCAGCAGCTTCCTGTAGGT-3′ | ||||
| α-SMA | Forward | 5′-TTCAATGTCCCAGCCATGTA-3′ | 60 | 30 | 222 |
| Reverse | 5′-GAAGGAATAGCCACGCTCAG-3′ | ||||
| Foxf1 | Forward | 5′-ATATCAAGCAGCAGCCCCTGTC-3′ | 50 | 35 | 271 |
| Reverse | 5′-ATGTCTTGGTAGGTGACCTG-3′ | ||||
| Tbx4 | Forward | 5′-CACTACCAGCACGAGAAC-3′ | 50 | 35 | 163 |
| Reverse | 5′-CCAGATAGGATCGCTTGC-3′ | ||||
| FosB | Forward | 5′-TCCTCACCCTCTGCCGAGTCTCAA-3′ | 60 | 35 | 306 |
| Reverse | 5′-ACTCGCTCCGCCACTGCTGTAG-3′ | ||||
| Laminin α5 | Forward | 5′-TGGCTGGATTATGTACTCGTGG-3′ | 54 | 38 | 226 |
| Reverse | 5′-CTGTAGCACCTACTTCGTGGCA-3′ | ||||
| DLX5 | Forward | 5′-CTACAACCGCGTCCCAAG-3′ | 52 | 35 | 223 |
| Reverse | 5′-CACCTGTGTTTGTGTCAATCC-3′ | ||||
| N-cadherin | Forward | 5′-CACTGCTCAGGACCCAGAT-3′ | 59 | 35 | 416 |
| Reverse | 5′-TAAGCCGAGTGATGGTCC-3′ | ||||
| HOXC10 | Forward | 5′-CCGACTCCAGCCCAGACAC-3′ | 54 | 38 | 101 |
| Reverse | 5′-CTTCTTCCTTCCGCTCTTTGC-3′ | ||||
| HAS1 | Forward | 5′-GGGCTTGTCAGAGCTACTT-3′ | 60 | 38 | 222 |
| Reverse | 5′-AGGGCGTCTCTGAGTAGCAG-3′ | ||||
| β-Actin | Forward | 5′-GCTCGTCGTCGACAACGGCTC-3′ | 55 | 28 | 353 |
| Reverse | 5′-CAAACATGATCTGGGTCATCTTCTC-3′ | ||||
SP-A, surfactant protein-A; SP-B, surfactant protein-B; SP-C, surfactant protein-C; AQP5, aquaporin 5; α-SMA, α-smooth muscle actin; Foxf1, Forkhead box F1; Tbx4, T-box4; FosB, FBJ murine osteosarcoma viral oncogene homolog B; DLX5, distal-less homeobox 5; HOXC10, homeobox C10; HAS1, hyaluronan synthase 1.
Assay for Mesenchymal Differentiation
Induction of adipogenic, osteogenic, and chondrogenic differentiation was performed with the Human Mesenchymal Stem Cell Functional Identification Kit (R&D systems, Minneapolis, MN, USA). For evaluating adipogenic differentiation, fatty acid binding protein 4 (FABP4) was analyzed by immunofluorescence staining. In addition, oil red O staining was performed to detect the accumulation of lipids. For osteogenic differentiation, osteocalcin was evaluated by immunofluorescence, and alizarin S staining was performed to show calcium deposition. To demonstrate chondrogenic differentiation, the expression of aggrecan was examined by immunofluorescence staining. All primary antibodies were supplied with the Human Mesenchymal Stem Cell Functional Identification Kit. Alexa Fluor 546-conjugated donkey anti-goat IgG antibody and Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (Molecular Probes) were used as secondary antibodies.
Microarray Analyses
To evaluate gene expression profiles, total RNA was extracted and cleaned up from three batches of our isolated cells at passage 4 as previously described in the Materials and Methods section. RNA targets were prepared according to the manufacturer's protocol using GeneChip 3′ IVT Express Kit (Affymetrix, Santa Clara, CA, USA). The total RNA (100 ng) was converted into double-stranded cDNA template for transcription. In vitro transcription synthesized amplified RNA (aRNA) and incorporated a biotin-conjugated nucleotide. The purified and fragmented aRNA (12.5 μg) was hybridized to GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix). This system covers over 47 000 transcripts from the GenBank, dbEST, and RefSeq database. The array was incubated for 16 h at 45°C, then automatically washed and stained with GeneChip Hybridization, Wash and Stain Kit (Affymetrix). The hybridized aRNA was detected with streptavidin-PE. The probe array was scanned using a GeneChip Scanner 3000 7G (Affymetrix) and quantified using Affymetrix GeneChip Command Console and Affymetrix Expression Console (Affymetrix).
The microarray data of our isolated cells was deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, GEO Series accession no. GSE21095). To compare the transcription profiles between our isolated cells and BM-MSCs, we downloaded the raw data set of BM-MSCs from the NCBI GEO website (GSE9451). A previous publication reported that the Affymetrix GeneChip system allows direct comparison between data sets obtained in different centers, provided the same chip and the same normalization are used.22 Expression measures were computed with GeneSpring GX software version 10.0 (Agilent Technologies, Santa Clara, CA, USA), using Microarray Suite version 5.0 (MAS 5.0) algorithm. Genes flagged as ‘present' or ‘marginal' call in each batch of the cell populations were defined as genes expressed in the populations. The area-proportional Venn diagram was generated using the BioVenn-web application (http://www.cmbi.ru.nl/cdd/biovenn/index.php).23 Calculation of fold change and unpaired t-test was conducted between our isolated cells and BM-MSCs using GeneSpring GX software version 10.0 (Agilent). Gene ontology term enrichment was analyzed by the Functional Annotation Clustering Tool in the Database for Annotation, Visualization and Integrated Discovery (version 6.7, DAVID, http://david.abcc.ncifcrf.gov).24 The major gene ontology terms associated with each group are manually summarized based on gene-term enrichment buttons provided for each functional group.25
Induction of Differentiation to ATII Cells
To induce differentiation into ATII cells, a human ATII cell culture system was adopted with some modifications.26 Cells (5 × 105) at passage 4 or 5 were plated on cell culture inserts (BD) that were previously coated with a mixture of 60% Matrigel (BD) and 40% rat tail collagen I (BD) in DMEM/5%FBS. After 4 h, the media was changed to DMEM containing 5% FBS with 10 ng/ml human recombinant keratinocyte growth factor (KGF; PeproTech, Rocky Hill, NJ, USA), 0.1 mM 8-bromo cAMP (cAMP; Sigma-Aldrich) and 0.1 mM IBMX (Sigma-Aldrich). The media were changed every other day and cells were cultured until day 7 when cells were analyzed.
For hematoxylin–eosin staining, whole cell-embedded gels were fixed with 10% formalin and embedded with paraffin. Sections were cut at 3 μm thickness and stained with hematoxylin–eosin. For immunofluorescence staining, whole gels were directly embedded in OCT compound and frozen in liquid nitrogen. Cryostat sections (6 μm) were fixed with acetone for 10 min at −20°C. The blocking procedure and following steps were performed as previously described. We used the following antibodies: rabbit anti-human pro-SP-C (1:1000, Millipore), mouse anti-human CD90 (1:50, BD Biosciences), Alexa Fluor 647-anti-mouse IgG and Alexa Fluor 488-anti-rabbit IgG (1:100, Molecular Probes). For electron microscopic analysis, whole gels were fixed in a mixture of 2% paraformaldehyde and 2.5% glutaraldehyde buffered with 0.1 M sodium cacodylate (pH 7.2) and post-fixed in 1% osmium tetroxide. The gels were dehydrated in ethanol followed by treatment with propylene oxide, and then embedded in epoxy resin. Sections were stained with uranyl acetate and lead citrate, and examined with an electron microscope (Hitachi High-Technologies Corporation 2000, Tokyo, Japan).
Immunofluorescence Staining of Pro-SP-C and CD90 in Human Lung Sections
For histological observations, cryostat sections (6 μm) were fixed with acetone for 10 min at −20°C. Formalin-fixed, paraffin-embedded specimens were cut into 3 μm sections. Antigen retrieval solution at pH 9 (Nichirei, Tokyo, Japan) was used for 5 min at 121°C for only formalin-fixed paraffin-embedded sections. The blocking procedure and following steps were performed as previously described. However, the permeabilization step was not necessary. The mouse anti-human CD90 antibody from Serotec was used for formalin-fixed paraffin-embedded sections and mouse anti-human CD90 antibody from BD Biosciences was used for OCT-compound frozen sections. Confocal images were taken using a Nikon A1Rsi imaging system (Nikon, Tokyo, Japan).
Statistics
Statistical analyses were performed with GraphPad Prism Version 5.0b (GraphPad Software). For each parameter measured, the value for individual samples were averaged for each experimental group and the s.d. was calculated. Data were compared using the unpaired t-test. Statistical significance was defined as P<0.05.
RESULTS
Isolation of a MSC-like Population with Alveolar Epithelial Phenotypes from Adult Human Lungs
We isolated colony-forming cells from adult human lungs, using a culture system consisting of feeders (mitotically inactivated mouse embryonic fibroblasts) that were previously used for deriving murine lung stem cells.12, 13 We seeded non-hematopoietic lung cells on feeders. In all specimens, spindle-shaped cells gave rise to colonies 6–10 days after the start of culture (Figure 1a). The colony-forming cells proliferated and reached to the confluency until 4 weeks. The spindle-shaped cells that formed colonies could be passaged and uniformly expanded (Figure 1b). Subsequent cultures were performed without feeders. After more than seven passages, these expanded cells became morphologically broad and flattened, suggesting that cellular senescence had occurred.27 Therefore, we analyzed the cells within 4–6 passages in this study.
Figure 1.
Morphology and protein expression of colony-forming cells derived from adult human lungs. Representative images of a colony generated from CD45− lung cells (a) and expanded spindle-shaped cells at passage 5 (b). (c) Representative immunophenotypes of the expanded cells at passage 4–6. These cells expressed mesenchymal stem cell (MSC) markers (CD73, CD90, and CD105) but not known endogenous stem cell markers (c-kit and CD133). Hematopoietic/endothelial markers (CD45, CD34, CD31, and VEGF receptor 2 (VEGFR2)) were not expressed. E-cadherin (an epithelial marker) was negative. (d) Flow cytometric analysis of pro-surfactant protein-C (pro-SP-C; an alveolar type II (ATII) cell marker) and CD90 (an MSC marker) in the expanded cells (left panel) and primary CD45− lung cells (right panel). The expanded cells from the colony-forming cells expressed CD90 with low expression of pro-SP-C. Primary CD45− lung cells were used for controls of pro-SP-C and CD90 staining. (e) Immunofluorescence staining of pro-SP-C (green) and CD90 (red) in the expanded cells (left panel) and CD45− lung cells (right panel). The expanded cells cultured on a chamber slide were fixed and stained. Primary CD45− lung cells were cytospun for immunostaining. ATII cells expressed pro-SP-C, but not CD90 (arrow). Arrowhead indicates cells expressing CD90 alone. Insets show isotype controls. (f–h) Immunofluorescence staining for SP-A (f; an ATII cell marker; green), SP-D (g; an ATII cell marker; green) and vimentin (h; a mesenchymal marker; green) in the expanded cells that were cytospun. Flow cytometric analyses and immunofluorescence staining were performed for samples from three different patients. Scale bars: (a, b) 200 μm; (e) 50 μm; (f–h) 10 μm.
The colony-forming cells co-expressed several surface markers of MSCs and surfactant proteins associated with ATII cells. The expanded cells at passage 4–6 expressed CD73, CD90, and CD105, which are defined as positive markers for human MSCs28 (Figure 1c). These expanded cells co-expressed CD90 with low expression of pro-SP-C (Figure 1d). Immunofluorescence staining confirmed that pro-SP-C and CD90 were expressed by the expanded cells (Figure 1e, left panel). Pro-SP-C expression in the expanded cells was weaker than that in the freshly isolated ATII cells (Figure 1e, right panel). At the same time, SP-A and SP-D were expressed by the expanded cells (Figure 1f and g). These cells were positive for vimentin (Figure 1h), but negative for E-cadherin (Figure 1c). In addition, these cells did not express markers of tissue stem cells (c-kit/CD133), hematopoietic cells, or endothelial cells (CD45/CD34/CD31/VEGF receptor 2) (Figure 1c). Positive controls for flowcytometric analyses were demonstrated in Supplementary Figure 1. These data demonstrate that we isolated an MSC-like cell population with the expression of surfactant proteins, that are represented as pro-SP-C+/CD90+ cells, from adult human lungs.
The Pro-SP-C+/CD90+ Cells Show the Clonogenic Ability and the Mesodermal Differentiation Capability In Vitro
The pro-SP-C+/CD90+ cells isolated from adult human lungs shared the same surface markers as BM-MSCs, therefore, we investigated whether the pro-SP-C+/CD90+ cells had the ability of self-renewal and mesodermal differentiation. First, we tested the clonogenic ability of the pro-SP-C+/CD90+ cells. As shown in Figure 2a, the pro-SP-C+/CD90+ cells generated secondary colonies from a single cell. To determine the frequency of clonogenic cells in each patient, limiting dilution analysis was performed. We found that the minimum frequency of clonogenic cells in each patient was 17.1±9.2% (mean±s.d., n=6; Figure 2b). To purify clonogenic cells, we sorted single cells out of the pro-SP-C+/CD90+ cells. The expanded clonogenic cells expressed CD73, CD90, CD105, and pro-SP-C at a proportion similar to that of the pre-sorted population (Figure 2c). In addition, these clonogenic cells co-expressed pro-SP-C and CD90 by immunostaining (Supplementary Figure 2), and SP-A and SP-D were also expressed (data not shown). RT–PCR showed that mRNA of SP-B, SP-C, vimentin, and α-smooth muscle actin (α-SMA) was expressed in these clonogenic cells as well as pre-sorted cells (Figure 2d). These data suggest that the pro-SP-C+/CD90+ cells had the clonogenic ability. Second, we examined whether the pro-SP-C+/CD90+ cells could differentiate into adipocytes, osteoblasts, and chondrocytes. In adipogenic differentiation, the pro-SP-C+/CD90+ cells differentiated into adipocytes that expressed FABP4 as shown by immunostaining and these differentiated cells contained lipid droplets as demonstrated by oil red O staining (Figure 2e). In osteogenic differentiation, the pro-SP-C+/CD90+ cells differentiated into osteoblasts that expressed osteocalcin as shown by immunostaining and these differentiated cells developed calcification as demonstrated by Arizarin S staining (Figure 2e). However, even after pellets of the cells were stimulated with transforming growth factor-β3 during chondrogenic differentiation, aggrecan, which is a specific marker for chondrocytes, was not expressed in the pellets as demonstrated by immunostaining (data not shown). These data demonstrate that the pro-SP-C+/CD90+ cells that we isolated from adult human lungs have a limited differentiation capability toward mesenchymal lineages.
Figure 2.
The pro-SP-C+/CD90+ cells have the ability of self-renew and mesodermal differentiation. (a) Representative images of proliferating cells and a secondary colony from a single cell. (b) Frequency of clonogenic cells in each patient. Limiting dilution analysis revealed that the minimum frequency of clonogenic cells in each patient was 17.1±9.2% (mean±s.d., n=6). The mean and s.d. of the frequencies are shown as crossbars. (c) Flow cytometric analyses for CD73, CD90, CD105, and pro-SP-C in pre-sorted cells (pre-sorted) and clonogenic cells (clone). Isotype controls were shown in gray. (d) Semiquantitative RT–PCR for SP-B, SP-C, vimentin, and α-smooth muscle actin (α-SMA) in pre-sorted cells and two batches of clonogenic cells (clones 1 and 2). Whole lung is used for positive control (lung) and RNA sample from whole lung without reverse transcriptase reaction is used for negative control (no-RT). (e) Representative images of adipogenic and osteogenic differentiation of the pro-SP-C+/CD90+ cells. Fatty acid binding protein 4 (FABP4)-positive cells indicated mature adipocytes (left upper panel, orange) and oil red O staining showed lipid deposition (right upper panel, red) in adipogenic differentiation. Osteocalcin-positive cells were osteoblasts (left lower panel, red) and alizarin S staining demonstrated calcium deposition (right lower panel, red) in osteogenic differentiation. Scale bars: (a) 100 μm and (e) 50 μm.
Comparison of Transcription Profiles Between Pro-SP-C+/CD90+ Cells and BM-MSCs
To compare transcription profiles between pro-SP-C+/CD90+ cells and BM-MSCs, RNA isolated from three batches of the pro-SP-C+/CD90+ cells from different subjects was analyzed with the Affymetrix array. We directly downloaded the raw data set of the transcription profiles of BM-MSCs from the GEO website (GSE9451). The self-renewal ability, multipotency, and genetic signatures of the BM-MSCs were well characterized according to a previous report.29 We found that 11 131 genes (21 560 probes) were expressed by all of three batches of the pro-SP-C+/CD90+ cells and 10 812 genes (20 512 probes) were expressed by all of three batches of the BM-MSCs. The Venn diagram demonstrated that 1909 genes (3037 probes) were specifically expressed by the pro-SP-C+/CD90+ cells and 1325 genes (1989 probes) were specifically expressed by the BM-MSCs (Figure 3a). To perform gene ontology term enrichment analysis in each gene set that was specifically expressed by pro-SP-C+/CD90+ cells or BM-MSCs, we utilized the web-based database (DAVID version 6.7, http://david.abcc.ncifcrf.gov).24 Functional annotation clustering in DAVID revealed that genes associated with transcription and lung development were enriched in pro-SP-C+/CD90+ cells, while genes associated with skeletal system development and blood vessel development were enriched in BM-MSCs (Figure 3b). We confirmed the expression of genes identified through the microarray analysis using RT–PCR. We selected four genes specifically expressed by pro-SP-C+/CD90+ cells, including Forkhead box f1 (Foxf1), T-box 4 (Tbx4), FBJ murine osteosarcoma viral oncogene homolog B (FosB) and laminin α5. At the same time, we evaluated mRNA expression of four genes specifically expressed by BM-MSCs, including distal-less homeobox 5 (DLX5), N-cadherin, homeobox C10 (HOXC10), and hyaluronan synthase 1 (HAS1). Figure 3c showed that the expression of Foxf1, Tbx4, FosB, and laminin α5 was upregulated in pro-SP-C+/CD90+ cells, while DLX5, N-cadherin, HOXC10, and HAS1 were exclusively expressed by BM-MSCs. SP-C mRNA was not expressed in BM-MSCs (data not shown).
Figure 3.
Comparison of transcription profiles between pro-SP-C+/CD90+ cells and bone marrow-mesenchymal stem cells (BM-MSCs). (a) The area-proportional Venn diagram presenting the overlap between transcripts expressed in pro-SP-C+/CD90+ cells (blue) and BM-MSCs (red). (b) Functional annotation clustering in specifically expressed gene sets in pro-SP-C+/CD90+ cells (blue) or BM-MSCs (red). Original array data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zpapfyaqumsqmvw&acc=GSE21095. (c) Semiquantitative RT–PCR for eight genes identified through the microarray analysis. Forkhead box f1 (Foxf1), T-box 4 (Tbx4), FBJ murine osteosarcoma viral oncogene homolog B (FosB), and laminin α5, that were selected out of the annotations of ‘transcription' and/or ‘lung development', were highly expressed by pro-SP-C+/CD90+ cells. At the same time, distal-less homeobox 5 (DLX5), N-cadherin, homeobox C10 (HOXC10), and hyaluronan synthase 1 (HAS1), that were chosen from the annotation of ‘skeletal system development' or ‘cell adhesion', were highly expressed by BM-MSCs. Three bathes of pro-SP-C+/CD90+ cells and three batches of BM-MSCs were examined. β-Actin was used as an endogenous control. In RT–PCR for Foxf1, Tbx4, FosB, laminin α5, and β-actin, a representative negative control (pro-SP-C+/CD90+ cells batch1 without reverse transcriptase reaction) is shown as no-RT. In RT–PCR for DLX5, N-cadherin, HOXC10, and HAS1, a representative negative control (BM-MSCs batch1 without reverse transcriptase reaction) is shown as no-RT.
Derivation of ATII Cells from the Pro-SP-C+/CD90+ Cells In Vitro
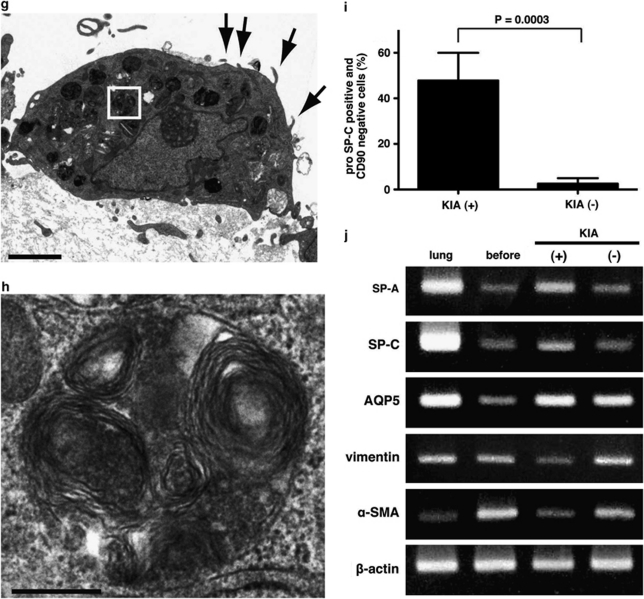
To investigate whether the pro-SP-C+/CD90+ cells could differentiate into ATII cells in vitro, we cultured these cells on a mixture of Matrigel and collagen type I gel in the presence of KGF, cAMP, and IBMX, a modified version of the culture system used by Wang et al.26 In every specimen exposed to KGF, cAMP, and IBMX (KIA(+) group), differentiated cells possessed an epithelial cell-like appearance on the gel (Figure 4a). Upon hematoxylin–eosin staining, these differentiated cells formed a monolayer and had a cuboidal shape that resembled the morphology of ATII cells (Figure 4c). Immunofluorescence staining showed that the cuboidal-shaped cells expressed pro-SP-C but not CD90 (Figure 4e), although almost all cells before differentiation expressed CD90 (Figure 1c). In addition, these differentiated cells expressed SP-A and SP-D (data not shown). Transmission electron microscopy indicated that cuboidal cells displayed lamellar bodies and apical microvilli, which are distinctive structures of ATII cells30 (Figure 4g and h). By contrast, cells cultured without any additives on Matrigel/collagen gels (KIA(−) group) showed an elongated shape and formed multiple layers (Figure 4b and d). Multilayered and flattened cells in the KIA(−) group remained positive for CD90, but lost SP-C expression (Figure 4f). To quantify ATII cell differentiation, we counted the number of pro-SP-C-positive and CD90-negative cells (ATII-like cells) as a ratio of the total population (the number of DAPI-stained nuclei). We found that the ratio of the number of pro-SP-C+/CD90− cells to the number of DAPI-stained nuclei in the KIA(+) group was 47.8±12.2%, while that in the KIA(−) group was 2.5±2.5% (n=4 in each; P=0.0003; Figure 4i). In addition, semiquantitative RT–PCR confirmed that the expression of mRNA of SP-A and SP-C was upregulated and the expression of mRNA of vimentin and α-SMA decreased in the KIA(+) group (Figure 4j). The expression of mRNA of AQP5 (an ATI cell marker) was upregulated both in the KIA(+) group and in the KIA(−) group (Figure 4j). The expression of mRNA of SP-A and SP-C in the KIA(−) group was not upregulated (Figure 4j).
Figure 4.
The pro-SP-C+/CD90+ cells differentiate into ATII cells in vitro. (a–f) Representative morphologic appearances of cells cultured on a mixture of Matrigel and rat tail collagen with keratinocyte growth factor (KGF), cAMP, and IBMX (KIA(+), left side panels) and without any additives (KIA(−), right side panels) in phase contrast images (a, b) and hematoxylin–eosin (HE) staining (c, d) and immunofluorescence staining of pro-SP-C (green) and CD90 (red) (e, f). White arrowheads indicate pro-SP-C+/CD90− cells (ATII cells) and white arrows show SP-C−/CD90+ cells (e, f). A pro-SP-C+/CD90+ cell was still observed (yellow arrowhead in e). Unidentified differentiated cells expressing neither pro-SP-C nor CD90 were seen (yellow arrows in e, f). (g, h) Transmission electron microscopic images of differentiated cells in KIA(+) group. (g) Lamellar bodies (a white-framed rectangle) and apical microvilli (arrows) were observed as distinctive structures in ATII cells. (h) A higher magnification of lamellar bodies from a white-framed rectangle in (g). (i) The ratio of the number pro-SP-C+/CD90− cells to the number of DAPI-stained nuclei was significantly higher in the KIA(+) group than that in the KIA(−) group (47.8±12.2% vs 2.5±2.5%, n=4 in each, P=0.0003). (j) Representative RT–PCR analysis of SP-A, SP-C (ATII cell markers) and aquaporin 5 (AQP5; an ATI cell marker) and vimentin and α-SMA (mesenchymal markers) before and after differentiation. Scale bars: (a, b) 200 μm; (c, d) 50 μm; (e, f) 20 μm; (g) 2 μm; (h) 300 nm.
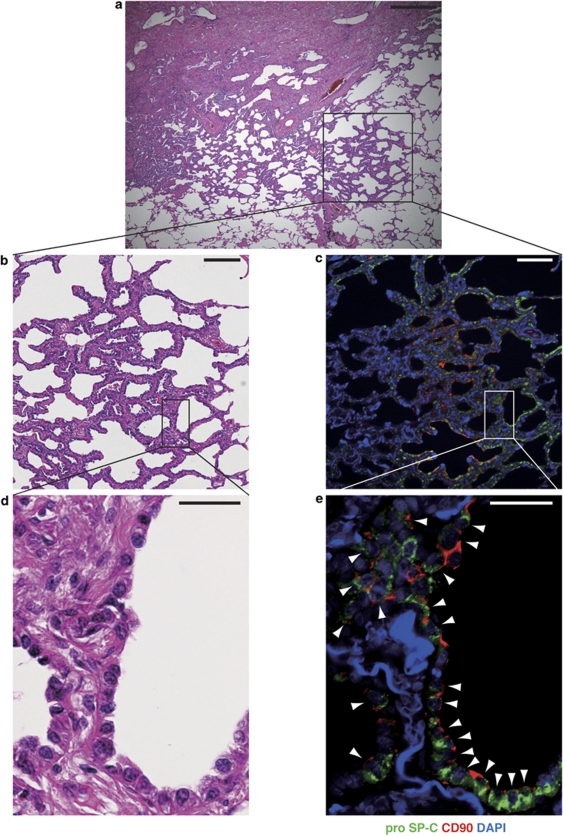
Localization of Pro-SP-C+/CD90+ Cells in Normal Lungs, Lung Fibrosis, and Lung Adenocarcinoma
Thus far, the pro-SP-C+/CD90+ cells that we isolated from adult human lungs were derived after an extended period of culture. Therefore, we sought to investigate whether cells with similar characteristics exist in situ in human lungs. In normal lung tissues, pro-SP-C+/CD90+ cells located within alveolar walls (Figure 5a and b). However, we could not find any pro-SP-C+/CD90+ cells in the bronchi, bronchioles, bronchioalveolar duct junctions, interlobular septa, blood vessels, or pleura. To demonstrate the localization of pro-SP-C+/CD90+ cells in damaged lungs, we performed immunofluorescence staining in lung sections from patients with idiopathic pulmonary fibrosis. Pro-SP-C+/CD90+ cells were localized and proliferative in hyperplasia of ATII cells, which is thought to be regenerative epithelium11 (Figure 5c–e). Flow cytometric analysis demonstrated that the ratio of pro-SP-C+/CD90+ cells to CD45− lung cells was significantly higher in fibrotic lungs than in normal lungs (1.23±0.13% vs 0.32±0.06%, mean±s.d., n=3, P=0.0003). To determine whether pro-SP-C+/CD90+ cells were involved in lung adenocarcinoma, we performed immunostaining in sections derived from lung adenocarcinomas. A subset of lung adenocarcinomas expressed both pro-SP-C and CD90 (Figure 6).
Figure 5.
Localization of pro-SP-C+/CD90+ cells in normal lung (a, b) and damaged lung (c–f). (a) Immunofluorescence staining for pro-SP-C (green) and CD90 (red) in normal alveoli. Arrowhead shows a pro-SP-C+/CD90+ cell. The pro-SP-C−/CD90+ cell was a pericyte in the alveolar wall (arrow).47, 48 (b) A confocal image from a white-framed rectangle in (a). (c, e) Low magnified images from the lung tissues from a patient with idiopathic pulmonary fibrosis (IPF). (c, e) are serial sectioned images. V, pulmonary vein. (c) HE staining showed that patchy fibrosis with remodeling of lung structure in a subpleura region. (e) Immunofluorescence staining for pro-SP-C (green) and CD90 (red) in specimen correlating to the area in (c). Positive immunostaining for CD90 (red, arrows) was observed in the wall of vascular structures (V), as previously reported.49 (d, f) Higher magnification of hyperplasia of ATII cells from the boxes in (c, e). Double-positive cells (arrowheads) for pro-SP-C (green) and CD90 (red) were seen within ATII cell hyperplasia that are thought to contribute to alveolar epithelial regeneration.11 ATII cells (arrows) are located around the pro-SP-C+/CD90+ cells. (g) The ratio of pro-SP-C+/CD90+ cells to CD45− lung cells significantly increased in fibrotic lungs (1.23±0.13%, n=3) compared with normal lungs (0.32±0.06%, n=3, P=0.0003). Data show mean±s.d. Scale bars: (a) 50 μm; (b) 10 μm; (c, e) 200 μm; (d, f) 50 μm.
Figure 6.
Representative histological analysis of lung tissues derived from an adenocarcinoma patient: HE staining (a, b, d) and immunofluorescence staining for pro-SP-C and CD90 (c, e). Images in (b, c) and (d, e) are shown from mirror sections, respectively. (a) A lower magnification of lung adenocarcinoma. (b, c) Higher magnifications from (a). Well-differentiated adenocarcinoma in (b) showed expression of pro-SP-C (green) and CD90 (red) in (c). (d, e) Higher magnifications from (b, c). Cancer cells that showed severe atypical nuclei and clear nucleoli in (d) expressed both pro-SP-C and CD90 in (e, arrowheads). Scale bars: (a–c) 400 μm (d, e) 50 μm.
DISCUSSION
In this study, we isolated progenitor cells with the ability to differentiate into ATII cells from adult human lung. We found that clonogenic cells isolated from adult human lungs expressed MSC markers combined with ATII cell-related protein expression (pro-SP-C). This cell population generated ATII cells upon differentiation-inducing stimuli in vitro. Hereafter, we refer to our isolated pro-SP-C+/CD90+ cells as alveolar epithelial type II progenitor cells (AEPCs).
AEPCs derived from adult human lungs had the clonogenic ability (Figure 2a and b). This ability of clonal expansion is one of the most essential qualifications for defining progenitor cells.12, 21, 31, 32 Flow cytometric analysis and immunofluorescence staining indicated that the undifferentiated AEPCs have already been committed to an ATII cell lineage. Indeed the undifferentiated AEPCs express both mRNA and surfactant proteins of SP-A, SP-B, pro-SP-C, and SP-D (Figures 1c–f and 2d). This feature is consistent with a previous report that adult liver progenitor cells possess undifferentiated signatures with commitment of a hepatocyte lineage.33 We demonstrated the capability that AEPCs generate ATII cells by several methods. First, AEPCs differentiated by extracellular matrix with KGF, cAMP, and IBMX exhibited cuboidal-shaped morphologies with lamellar bodies in histological examinations (Figure 4a, c, g, and h). Second, differentiated AEPCs strongly expressed surfactant proteins but were not immunopositive for CD90 (Figure 4e and i). In addition, RT–PCR confirmed that the expression of alveolar epithelial markers (SP-A, SP-C, and AQP5) was upregulated and mesenchymal markers (vimentin and α-SMA) was downregulated in differentiated AEPCs (Figure 4j).
AEPCs possess similar characteristics to BM-MSCs, but differ in the ability of chondrogenic differentiation and the global gene expression. The minimal criteria for defining MSCs were previously reported.28 In accordance with these criteria, the expression of MSC markers (CD73, CD90, and CD105) and the ability to differentiate into adipocytes and osteocytes demonstrated that AEPCs share phenotypes with MSCs (Figures 1c, 2c and e). However, we could not readily induce differentiation of AEPCs into a chondrogenic lineage under the standard culture condition used in a typical chondrogenic differentiation assay. In addition, unlike BM-MSCs, our microarray analyses and RT–PCR (Figure 3) indicated that AEPCs specifically expressed key genes encoding transcription factors or extracellular matrix associated with lung development, including Foxf1,34 Tbx4,35 FosB,36 and laminin α5.37 At the same time, AEPCs had little or no expression of crucial genes determining the phenotypes of BM-MSCs and associated with skeletal morphogenesis like DLX5,38 N-cadherin,39 HOXC10,40 and HAS1.41 Bronchoalveolar lavage (BAL)-derived MSCs expressed DLX5 and N-cadherin with the same level as BM-MSCs.19 Although, it is difficult to simply compare the expression of DLX5 and N-cadherin between our isolated AEPCs and BAL-derived MSCs, the partially shared phenotypes of AEPCs with BM-MSCs propose some ideas that AEPCs may differ from BAL-derived MSCs or AEPCs may be included in a subset of the lung MSCs.
Epithelial–mesenchymal transition and its opposite mechanism, mesenchymal–epithelial transition, have key roles in early embryonic morphogenesis, cancer stem cells, and cancer metastasis.42, 43, 44 Acquisition of a mesenchymal phenotype gives cells motility and invasive capability.44 AEPCs expressed vimentin but not E-cadherin (Figure 1c and h). In addition, AEPCs expressed mRNA of Foxf1 and Tbx4, whose expression is restricted to mesenchymal cells during lung development.45 These data suggested that AEPCs are derived from lung mesenchyme rather than epithelium. The transitional phenotypes between the mesenchyme and epithelium observed in AEPCs suggest that these cells act as lung tissue progenitor cells in tissue repair and carcinogenesis. To be involved in alveolar repair, mesenchymal properties such as anti-apoptotic activity and motility may be beneficial for functional progenitor cells.
We demonstrated that pro-SP-C+/CD90+ cells, which express the same markers as our isolated AEPCs, proliferated within ATII cell hyperplasias in idiopathic pulmonary fibrosis lungs and in a subset of lung adenocarcinomas (Figures 5c–g and 6). Positive staining of CD90 in proliferative epithelial cells in fibrotic lungs is compatible with a previous report.46 Hyperplasia of ATII cells is considered to be involved in alveolar epithelial repair in response to injury.7, 8, 9, 10, 11, 30 Our data and these previous reports suggest AEPCs possibly participate in lung fibrosis. However, a pathophysiological role of AEPCs in disease development is not yet clear. Therefore, it is possible that the pro-SP-C+/CD90+ hyperplasia and the pro-SP-C+/CD90+ cancer cells were due to the resultant of the disease progression. Further studies are needed.
We could not demonstrate a precise hierarchy between AEPCs, ATII cells, and ATI cells. According to our in vitro cellular assays and in situ histopathological evaluation, AEPCs have a potential role as precursors of ATII cells. However, the possibility of direct differentiation from AEPCs into ATI cells cannot be excluded because the expression AQP5, an ATI cell marker, increased even without upregulation of SP-C expression in the differentiation assay (Figure 4i). This result is compatible with a previous report20 and suggests that AEPCs differentiate directly into ATI cells not through ATII cells. Further studies will be necessary to establish the hierarchy among AEPCs, ATII cells, and ATI cells.
In conclusion, to our knowledge, this study is the first to report that ATII progenitors exist in adult human lungs. Impaired proliferation and differentiation, or cell senescence, in AEPCs may have an important role in the pathogenesis of severe lung diseases. Therefore, AEPCs may be a good target for drug discovery and screening.
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
Acknowledgments
We are grateful to Mr Katsuhiko Ono (Department of Pathology and Histotechnology, Tohoku University Graduate School of Medicine) for his assistance with transmission electron microscopy, and Professor Ryouichi Nagatomi (Department of Biomedical Engineering, Tohoku University Graduate School of Biomedical Engineering) for his advice on this work. This work was supported by a grant from the Japan Society for the Promotion of Science (Nos. 19390222 and 22390163) to HK.
The authors declare no conflict of interest.
Supplementary Material
References
- Liu X, Engelhardt JF. The glandular stem/progenitor cell niche in airway development and repair. Proc Am Thorac Soc. 2008;5:682–688. doi: 10.1513/pats.200801-003AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett TL, Shaheen F, Johnson A, et al. Characterization of side population cells from human airway epithelium. Stem Cells. 2008;26:2576–2585. doi: 10.1634/stemcells.2008-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangreco A, Arwert EN, Rosewell IR, et al. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci USA. 2009;106:9286–9291. doi: 10.1073/pnas.0900668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog EL, Brody AR, Colby TV, et al. Knowns and unknowns of the alveolus. Proc Am Thorac Soc. 2008;5:778–782. doi: 10.1513/pats.200803-028HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson IY, Bowden DH. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab Invest. 1974;30:35–42. [PubMed] [Google Scholar]
- Evans MJ, Cabral LJ, Stephens RJ, et al. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp Mol Pathol. 1975;22:142–150. doi: 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- Aso Y, Yoneda K, Kikkawa Y. Morphologic and biochemical study of pulmonary changes induced by bleomycin in mice. Lab Invest. 1976;35:558–568. [PubMed] [Google Scholar]
- Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med. 2000;342:1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- Kawanami O, Ferrans VJ, Crystal RG. Structure of alveolar epithelial cells in patients with fibrotic lung disorders. Lab Invest. 1982;46:39–53. [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Hegab AE, Kubo H, Fujino N, et al. Isolationand characterization of murine multipotent lung stem cells. Stem Cells Dev. 2010;19:523–535. doi: 10.1089/scd.2009.0287. [DOI] [PubMed] [Google Scholar]
- Ventura JJ, Tenbaum S, Perdiguero E, et al. p38alpha MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat Genet. 2007;39:750–758. doi: 10.1038/ng2037. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goss AM, Cohen ED, et al. A Gata6–Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet. 2008;40:862–870. doi: 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi S, Kishimoto H, Kawahara K, et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J Clin Invest. 2007;117:2929–2940. doi: 10.1172/JCI31854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey JS, Zacharek SJ, Kim CF, et al. Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc Natl Acad Sci USA. 2008;105:11857–11862. doi: 10.1073/pnas.0803574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Buckley S, Doerken M, et al. Isolation of a putative progenitor subpopulation of alveolar epithelial type 2 cells. Am J Physiol Lung Cell Mol Physiol. 2004;286:L658–L667. doi: 10.1152/ajplung.00159.2003. [DOI] [PubMed] [Google Scholar]
- Lama VN, Smith L, Badri L, et al. Evidence for tissue-resident mesenchymal stem cells in human adult lung from studies of transplanted allografts. J Clin Invest. 2007;117:989–996. doi: 10.1172/JCI29713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoubi G, Cortes-Dericks L, Breyer I, et al. Identification of mesenchymal stromal cells in human lung parenchyma capable of differentiating into aquaporin 5-expressing cells. Lab Invest. 2009;89:1100–1114. doi: 10.1038/labinvest.2009.73. [DOI] [PubMed] [Google Scholar]
- Tropepe V, Coles BL, Chiasson BJ, et al. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- Assou S, Le Carrour T, Tondeur S, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Edeen K, Manzer R, et al. Differentiated human alveolar epithelial cells and reversibility of their phenotype in vitro. Am J Respir Cell Mol Biol. 2007;36:661–668. doi: 10.1165/rcmb.2006-0410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digirolamo CM, Stokes D, Colter D, et al. Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol. 1999;107:275–281. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Kubo H, Shimizu M, Taya Y, et al. Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry. Genes Cells. 2009;14:407–424. doi: 10.1111/j.1365-2443.2009.01281.x. [DOI] [PubMed] [Google Scholar]
- Anderson WR, Thielen K. Correlative study of adult respiratory distress syndrome by light, scanning, and transmission electron microscopy. Ultrastruct Pathol. 1992;16:615–628. doi: 10.3109/01913129209023751. [DOI] [PubMed] [Google Scholar]
- Bearzi C, Rota M, Hosoda T, et al. Human cardiac stem cells. Proc Natl Acad Sci USA. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Buck DW, He D, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera MB, Bruno S, Buttiglieri S, et al. Isolation and characterization of a stem cell population from adult human liver. Stem Cells. 2006;24:2840–2850. doi: 10.1634/stemcells.2006-0114. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Gusarova GA, Kim IM, et al. Foxf1 haploinsufficiency reduces Notch-2 signaling during mouse lung development. Am J Physiol Lung Cell Mol Physiol. 2004;286:L521–L530. doi: 10.1152/ajplung.00212.2003. [DOI] [PubMed] [Google Scholar]
- Sakiyama J, Yamagishi A, Kuroiwa A. Tbx4-Fgf10 system controls lung bud formation during chicken embryonic development. Development. 2003;130:1225–1234. doi: 10.1242/dev.00345. [DOI] [PubMed] [Google Scholar]
- Millien G, Spira A, Hinds A, et al. Alterations in gene expression in T1 alpha null lung: a model of deficient alveolar sac development. BMC Dev Biol. 2006;6:35. doi: 10.1186/1471-213X-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NM, Miner JH, Pierce RA, et al. Laminin alpha 5 is required for lobar septation and visceral pleural basement membrane formation in the developing mouse lung. Dev Biol. 2002;246:231–244. doi: 10.1006/dbio.2002.0658. [DOI] [PubMed] [Google Scholar]
- Hsieh JY, Fu YS, Chang SJ, et al. Functional module analysis reveals differential osteogenic and stemness potentials in human mesenchymal stem cells from bone marrow and Wharton′s jelly of umbilical cord Stem Cells Dev 2010. doi: 10.1089/scd.2009.0485(in press). [DOI] [PubMed]
- Gao L, McBeath R, Chen CS. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells. 2010;28:564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke S, Buchheiser A, Bosch J, et al. The HOX Code as a ‘biological fingerprint' to distinguish functionally distinct stem cell populations derived from cord blood. Stem Cell Res. 2010;5:40–50. doi: 10.1016/j.scr.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Calabro A, Oken MM, Hascall VC, et al. Characterization of hyaluronan synthase expression and hyaluronan synthesis in bone marrow mesenchymal progenitor cells: predominant expression of HAS1 mRNA and up-regulated hyaluronan synthesis in bone marrow cells derived from multiple myeloma patients. Blood. 2002;100:2578–2585. doi: 10.1182/blood-2002-01-0030. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial–mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Hugo H, Ackland ML, Blick T, et al. Epithelial–mesenchymal and mesenchymal–epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- Sanders YY, Pardo A, Selman M, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39:610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Kamiyashiki T, Ito Y. Isometric contraction of microvascular pericytes from mouse brain parenchyma. Microvasc Res. 2007;73:20–28. doi: 10.1016/j.mvr.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Hagood JS, Prabhakaran P, Kumbla P, et al. Loss of fibroblast Thy-1 expression correlates with lung fibrogenesis. Am J Pathol. 2005;167:365–379. doi: 10.1016/S0002-9440(10)62982-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.