Abstract
The plasma proteome holds enormous clinical potentials, yet an in-depth analysis of the plasma proteome remains a daunting challenge due to its high complexity and the extremely-wide dynamic range in protein concentrations. Furthermore, existing antibody-based approaches for depleting high-abundance proteins are not adaptable to the analysis of animal plasma proteome, which are often essential for experimental pathology/pharmacology. Here we describe a highly-comprehensive method for the investigation of animal plasma proteomes, which employs an optimized combinatorial peptide ligand libraries (CPLL) treatment to reduce the protein concentration dynamic range and a dual-enzyme, dual-activation strategy to achieve high proteomic coverage. The CPLL-treatment enriched the lower-abundance proteins by >100-fold when loading the samples in moderately-denaturing condition with multiple loading-washing cycles. The native and the CPLL-treated plasma were digested in-parallel respectively by two enzymes (trypsin and GluC) carrying orthogonal specificities. By performing this differential proteolysis, the proteome coverage is improved where peptides produced by only one enzyme are poorly detectable. Digests were fractionated with high-resolution SCX chromatography and then resolved on a long, heated nano-LC column. MS analysis was performed on an LTQ/Orbitrap respectively with two complementary activation methods (CID and ETD). We applied this optimized strategy to investigate the plasma proteome from swine, a prominent animal model for cardiovascular diseases(CVD). This large-scale analysis results in an identification of a total 3421 unique proteins, spanning a concentration range of 9–10 orders of magnitude. The proteins were identified under a set of commonly-accepted criteria including precursor mass error<15 ppm, Xcorr cutoffs, ≥two unique peptides at the peptide probability≥95% and protein probability≥99%, and the peptide FDR of the dataset was 1.8% as estimated by searching reversed database. CPLL treatment resulted in 55% more identified proteins over these from native plasma; moreover, compared with using only trypsin and CID, the dual enzyme/activation approach enabled the identification of 2.6-fold more proteins and substantially higher sequence coverage for most individual proteins. Further analysis revealed 657 proteins as significantly associated with CVD (p<0.05), which constitute five CVD-related pathways. This study represents the first in-depth investigation of non-human plasma proteome and the strategy developed here is adaptable to the comprehensive analysis of other highly complex proteomes.
INTRODUCTION
Plasma proteome is highly valuable for clinical and biomedical investigation because it contains a broad range of markers for diseases and drug effects, and that plasma specimens can be readily collected via non-invasive procedures from a large number of individuals1. However, investigation of plasma proteomes remains highly challenging for several reasons: first, the extremely-wide dynamic range (i.e. 9–10 orders of magnitude) of concentrations of plasma proteins presents a tremendous challenge for proteomic techniques1, 2. By comparison, the dynamic ranges achievable by current proteomic methods, such as by two-dimensional electrophoresis (2-DE) and liquid chromatography mass spectrometry (LC/MS)3, 4, are far short of the range that is required for an extensive proteomic analysis of plasma. Secondly, the plasma proteome is highly complex, as it may contain proteins from all the differentiated sub-proteomes of the body, as well as exogenous proteins such as those from bacteria, viruses or fungi5, 6. These unique characteristics of plasma render it difficult to achieve a comprehensive investigation of the plasma proteome.
To address the dynamic range problem, a number of immunodepletion methods, which can reduce the dynamic range of a plasma proteome by 1–2 orders of magnitude and thereby improving the analysis of lower-abundance proteins, were employed prior to gel-based and/or LC/MS-based analysis of human plasma proteome 7–9. Although many of these depletion techniques provide considerable reproducibility, efficiency and selectivity of the depletion process7, 8, they may suffer from problems associated with the co-depletion of lower-abundance proteins and carryover among samples10, 11. More importantly, the currently developed immunodepletion methods are exclusively based on the antibodies against human plasma proteins, and thus don’t work well for specimens derived from non-human sources. This significantly limits plasma biomarker research in animal models, which are critical tools that allow us to understand human physiological and pathological processes. Furthermore, even if efficient depletion is achievable, a comprehensive analysis of the depleted plasma remains challenging because it still holds a wide protein concentration dynamic range (e.g. ~8 orders of magnitude) and is highly complex, as suggested by numerous previous studies7, 8, 12.
Here we report the development of a highly comprehensive and efficient strategy for the investigation of animal plasma proteomes, where the current approaches are not capable of providing high proteomic coverage. The strategy is comprised of two steps. Step one employs an optimized combinatorial peptide ligand libraries (CPLL) technique to reduce markedly the dynamic range of the animal plasma proteome. The CPLL utilizes a combinatorial library containing a population of millions of different hexa-peptides ligands, which are conjugated to porous beads in equal numbers. Through affinity interaction, the vastly diversified CPLL beads provide retentions of numerous proteins when incubated with a complex proteomic sample 13. As there are a limited number of each ligand, abundant proteins readily saturate the available ‘baits’ and thus most are washed away during the procedure; conversely, the low-abundance proteins, whose levels are too low to saturate their corresponding ligands, are selectively enriched 13, 14. This technique differs from the immunodepletion methods in that it is not intended to deplete any specific high-abundance protein, but rather reduces the concentration dynamic range of a proteome. Due to the fact that the CPLL method is not proteome-specific, it has been recently employed for the analyses of proteomes including plasma from various species15–18. Additionally the CPLL-treatment employs single-use columns, which avoids the sample carryover problem that is common for the multiple-use depletion columns10. In the present study, we utilized and optimized the CPLL procedure to enrich lower abundance proteins in the plasma proteome.
In step two, we devised a dual-enzyme and dual-activation strategy for comprehensive and highly confident proteomic analysis. The CPLL-treated samples were processed and digested with a highly efficient and reproducible precipitation/on-pellet digestion method we described previously19–21. For the proteolytic digestion, each sample was split, and two enzymes carrying orthogonal specificities were employed in parallel: trypsin (cuts at K and R) and GluC (cuts at D and E). By performing this differential enzyme cleavage, we produced complementary proteolytic peptide profiles, which can improve proteome coverage, e.g., where peptides produced by one enzyme are poorly detectable. Using a high-resolution nano-LC coupled to a LTQ/Orbitrap/ETD (electron transfer dissociation) analyzer, each of the parallel samples was then analyzed using two orthogonal fragmentation techniques: collisionally induced dissociation (CID) and ETD. Generally, CID can fragment singly- to triply-charged peptides with a reasonable efficiency, but not peptides with higher charge-states or labile post-translational modifications22–24. In contrast, ETD is not optimal for peptides with charge states lower than 3, but can efficiently fragment peptides having higher charges, and is indifferent to side chain chemistry, as demonstrated in several labs including ours 20, 25–28. Therefore CID and ETD provide complementary sequencing information. By integrating the data sets obtained by this dual-enzyme and dual-activation strategy, we expected to achieve significantly higher overall coverage of the target proteome, compared to that using conventional trypsin and CID alone.
As a proof of concept, we applied this strategy for the in-depth analysis of swine plasma proteome. Large animals, and in particular swine, are desirable as models of human diseases because they share much more anatomic and physiologic characteristics with humans compared to small animals29, 30. One of the most prominent uses is in experimental cardiology, where swine exhibits many similarities to man in cardiovascular-related features, such as comparable heart-to-body size ratio, coronary arterial anatomy, development of coronary collateral circulation, etc31, 32. At our lab, we have developed a prominent swine model of chronic ischemic heart disease (hibernating myocardium), which has been utilized in proteomic biomarker discovery at the tissue level19, 33. In order to facilitate the study of cardiovascular disease mechanisms in swine models, a method capable of comprehensive proteomic investigation of the CVD markers in peripheral blood is highly desirable. Nevertheless, to our knowledge, there has thus far been no method developed for the in-depth analysis of animal plasma. With an emphasis on CVD markers, we employed the comprehensive proteomic analysis strategy developed in this study to achieve an extensive and highly confident analysis of plasma proteome from healthy pigs. An additional purpose of this study was to acquire high-quality peptide information for swine plasma proteins, which are critical for the development of highly sensitive and accurate SRM quantification methods for targeted investigation of markers of high interest, as we demonstrated previously21–23.
EXPERIMENTAL
Materials
Details are specified in the Supplementary Materials.
Preparation of Swine Plasma
etails are specified in the Supplementary Materials.
CPLL treatment
The CPLL treatments of plasma samples were carried out on spin columns each packed with 0.1 mL of ProteoMiner CPLL beads (Bio-Rad, Hercules, CA). The optimal conditions we developed for loading, washing, and elution for the treatment of swine plasma are as follows: a plasma sample containing 60 mg of proteins (~0.9 mL) was mixed with 0.1 mL of 10×PBS buffer, and SDS was added to yield a final concentration of 0.1% (w/v). After the loading procedure, the column was centrifuged at 1000 g for 2 min, and then washed with 1mL PBS buffer containing 0.1% SDS and centrifuged for 2 min. Three loading-washing cycles were employed to maximize the enrichment of low-abundance proteins. Flow-through buffers were collected for BCA analysis. After removing the residual wash buffer by adding 0.8 mL of water and then centrifugation for 2 min, the proteins bound to the CPLL beads were eluted with 1 mL of elution buffer (8 M urea/2% CHAPS/5% acetic acid, pH 2.8). The yields of eluted swine plasma proteins ranged from 1.1–1.3 mg, as determined by BCA. The eluate was then subjected to the following proteomic procedure.
Estimation of the effectiveness of the CPLL treatment by 2D-Gel
By spiking dye-labeled E. Coli. proteins into the plasma samples, we evaluated the effectiveness of CPLL treatments for enriching lower-abundance proteins under different loading/washing conditions. Based on the results of this evaluation, the final CPLL procedure was developed. Details are specified in the Supplementary Materials.
Precipitation/on-pellet Digestion
Details are specified in the Supplementary Materials.
Peptide Fractionation using Strong Cation Exchange (SCX) Chromatography
A high-resolution SCX procedure was developed for the fractionation of peptide mixtures generated by each enzyme. The procedure is detailed in the Supplementary Materials.
Nano-RPLC/Mass Spectrometry analysis
Four groups of samples were analyzed and consisted of the following: trypsin digest of native plasma, GluC digest of native plasma, trypsin digest of CPLL-treated plasma and GluC digest of CPLL-treated plasma. These were fractionated by SCX and reconstituted to 25 fractions each. Each of the fractions was analyzed in duplicate by CID and ETD, respectively. Therefore, in total, 400 nano-LC/MS runs were performed. The nano-LC/MS procedure is detailed in the Supplementary Materials.
Protein identification
Individual data files were searched against the Swiss-Prot human and pig protein database containing 20,328 human entries and 1,334 pig entries using the SEQUEST program34 on a 64-node Cluster. The search parameters used were as follows: 15 ppm tolerance for precursor ion masses and 1.0 Da for fragment ion masses to process all CID and ETD data. The Charger algorithm (Thermo) was used to process the ETD data prior to database search. Two and five missed cleavages were permitted for tryptic peptides and GluC produced peptides, respectively. In terms of modifications, carbamidomethylation of cysteines was specified as a static modification and a variable modification of methionine oxidation was allowed. In order to minimize false-positive identifications, Scaffold 3.0 (Proteome Software, Portland, OR), which integrates both Protein Prophet and Peptide Prophet to minimize the false positive identifications35 and is capable of handling large-scale proteomic datasets like the one generated here, was used to merge and summarize result files. A peptide probability of 95% or higher, a protein probability of 99% or higher and at least two unique peptides per protein, are required for the identification of each protein. Furthermore, in order to minimize false-positives in such a large dataset, additional Xcorr criteria (Xcorr >2.2 for 1+ and 2+, >3 for 3+ and >3.5 for 4+ and up), were applied to filter all peptides. The same peptide identified by both CID and ETD was considered as one unique peptide. If the same protein was identified for both human and swine, the human entry was removed. Peptide FDR was estimated by searching against the decoy database.
Bioinformatics Analysis
The Physicochemical characteristics and the cardiovascular significance of the identified plasma proteins were investigated. Details are specified in the Supplemental Materials.
RESULTS AND DISCUSSION
Overall strategy for method development
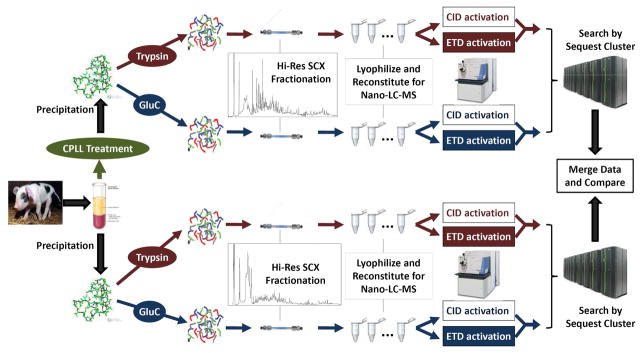
The overall strategy consists of the CPLL treatment to decrease the dynamic range of the plasma proteome, the dual-enzyme and dual-activation approach to enhance the proteomic coverage, and the high-resolution SCX and nano-LC separations to sufficiently resolve the complex plasma proteome, and thus enabling the confident detection of low-abundance proteins. The scheme of this work flow is shown in Fig. 1. As the CPLL technique may not be able to retrieve all proteins in a proteome, as suggested in previous works14 and in this study, a parallel analysis of the native plasma were also performed. With the purpose of achieving a comprehensive proteomic analysis to the maximum extent possible, each of these steps was thoroughly optimized.
Fig. 1.
The scheme of the overall strategy for the comprehensive analysis of swine plasma proteome.
Optimization of CPLL treatment procedure
Recent works demonstrated that CPLL treatment can dramatically reduce the protein concentration dynamic range of a complex proteome36. As the CPLL technique is not proteome-specific, it appears to be a logical choice for the analysis of swine plasma, where an antibody-based depletion method is not available. Nevertheless, to our knowledge, the applicability of CPLL for a comprehensive proteomic analysis of animal plasma has not been thoroughly investigated. So we set out to evaluate and optimize this technique for the treatment of swine plasma. Based upon the results from preliminary studies, we made two important modifications to the “standard” CPLL procedure described previously14.
Firstly, we employed a moderately-denaturing condition (i.e. 0.1% SDS in the loading buffer) for sample loading and washing. Our exploratory experiments indicated that unlike the antibody-based depletion approaches, the CPLL technique was well tolerant of a moderately-denaturing loading condition under a neutral pH value. Detailed data is presented in the Supplementary Materials. Though a moderately-denaturing condition could potentially weaken the binding of plasma proteins with the CPLL ligands, our pilot studies showed that loading plasma samples in a buffer containing 0.1% SDS did not cause no loss of proteins, as revealed by comparing both the numbers and intensities of 2D-Gel spots and the protein concentrations of the eluates when loading respectively under denaturing and non-denaturing (i.e. the PBS loading buffer) conditions. We speculate it is because the interactions of the CPLL ligands with plasma proteins carry considerable specificity and strength, rendering it tolerate to a mildly denaturing condition such as 0.1% SDS. By comparison, the non-specific bindings of lower-abundance proteins to abundant ones are largely non-specific and relatively weak, which may be prone to dissociation in the presence of 0.1% SDS. Consequently, the ability to load samples under denaturing conditions could be advantageous because it may help to dissociate the bindings between numerous low-abundance proteins and the abundant ones such as the albumin; such bindings could result in co-depletion of low-abundance proteins, a prevailing problem for many antibody-based depletion approaches 10. A preliminary investigation of shotgun identification indicated that loading the swine plasma sample with 0.1% SDS resulted in an average of 11% more identified proteins compared to loading under non-denaturing conditions (n=3, p<0.05).
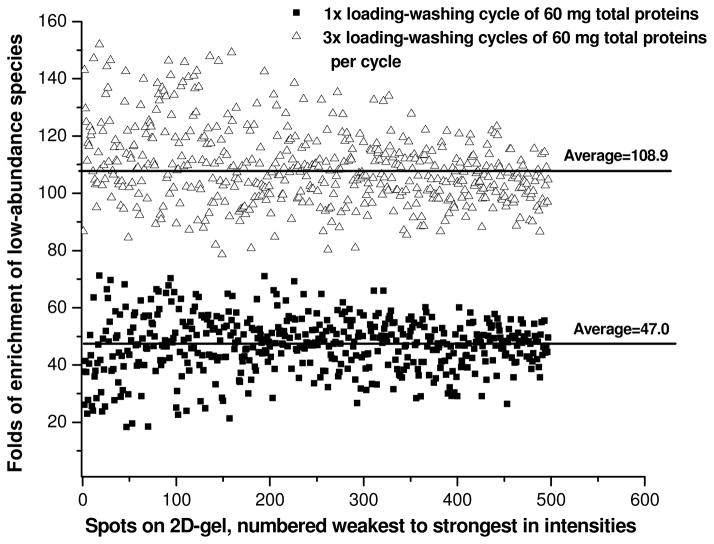
Secondly, we employed multiple sample loading-washing cycles to improve the enrichment of low-abundance plasma proteins. Based on the rationale of the CPLL technique, we hypothesized that the lower-abundance proteins that have not saturated their partner beads will be captured in progressively increasing amounts if additional plasma samples were loaded on the same column, while the high-abundance proteins would remain unbound because their corresponding partner beads have been already saturated by the first loading. To test this hypothesis, we spiked dye-labeled proteins from E. Coli., which represent a subset of lower-abundance proteins, to the non-labeled swine plasma sample at a protein mass ratio of 1:200, and then examined the efficiency of CPLL treatment for enrichment of these E. Coli. proteins using different loading-washing cycles. It was possible to load all plasma samples continuously, but the results of pilot studies indicated the residual unbound proteins from the previous loading interfered with further protein binding, and therefore should to be removed by a following washing step prior to the next loading. We compared the extents of enrichments by one single loading of 60 mg (i.e. the manufacturer suggested loading amount for the amount of CPLL beads used) of spiked plasma proteins with these by multiple loading-washing cycles, with each cycle loading 60 mg of spiked plasma proteins followed by a washing step with the loading buffer. Aliquots of mixtures were subjected to CPLL treatment respectively with single or multiple loading-washing cycles, and the relative quantities of the labeled proteins were analyzed by 2D-DIGE in each enriched preparation (details are described in the Supplementary Materials). Some typical results are shown in Fig. 2. The triple-loading of a total of 180 mg proteins resulted in an average of 110-fold enrichment of low-abundance proteins, 2.3-fold higher than by the standard single loading of 60 mg proteins (n=4, p<0.01). This indicates the triple-loading strategy will likely achieve a more comprehensive proteomic analysis. Further studies showed the trend of the increase in the enrichment of low-abundance proteins tended to level off beyond three loadings (data not shown). Based on these results, the triple loading-washing procedure that loaded 180 mg of plasma proteins was selected.
Fig. 2.
Comparison of the enrichment of low-abundance proteins by single loading-washing cycle vs. triple loading-washing cycles. Each cycle loaded 60 mg of total proteins (the mass ratio of plasma vs. E. Coli. proteins was 200:1, n=4). Quantitative values were obtained by 2D-DIGE scanning of the labeled E. Coli. The ratios for the most intensive 500 spots matched on both gels are shown.
Based on the CPLL procedures employed from previous works 14, 36, four elution buffers were evaluated for protein yield and reproducibility (SI Fig. 1). The result shows that 8 M urea/2% CHAPS/5% acetic acid (Buffer ii) provided by far the best protein yield, and thus was selected for elution.
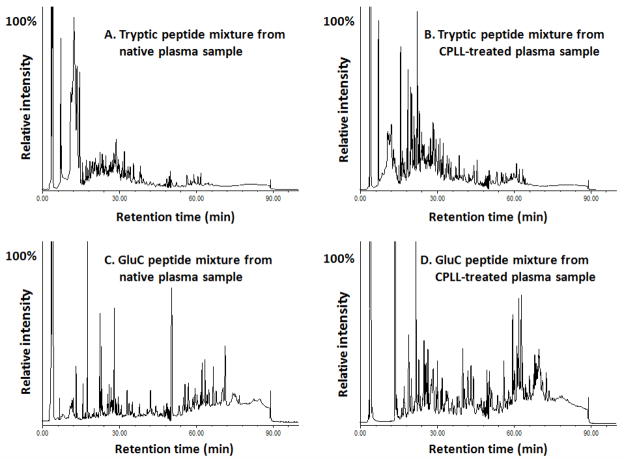
SI Fig. 2 shows some typical results of 2D electrophoresis analyses of the plasma proteins before and after the CPLL treatment. Substantially more protein spots were detected in the CPLL-treated plasma than in the native plasma, suggesting an effective enrichment of lower-abundance proteins by the CPLL beads. The high efficacy of the CPLL treatment was further supported by the comparison of the high-resolution SCX chromatographic profiles of the proteolytic peptides derived from the native and CPLL-treated swine plasma respectively (Fig. 3). Whereas the chromatograms of both the trypsin- and GluC- digests of the native plasma were dominated by scattered clusters of highly-intensive peptides (Fig. 3A and 3C), many more peptide peaks with relatively evenly-distributed intensities were observed in the CPLL-treated samples (Fig. 3B and 3D), implying a significantly reduced dynamic range in plasma protein concentrations after the CPLL treatment. These observations suggested that the CPLL treatment can improve the identification of lower-abundance plasma proteins, which has been confirmed in the following sections.
Fig. 3.
The high-resolution SCX chromatograms of (A) tryptic digest mixture of the native swine plasma, (B) tryptic digest mixture of the CPLL-treated swine plasma, (C) GluC digest mixture of the native swine plasma and (D) GluC digest mixture of the CPLL-treated swine plasma. Peptides derived from 1.2 mg of total proteins were injected for each run.
Development of the Dual-enzymes and Dual-activations Approach
Although trypsin is considered the enzyme-of-choice for most of proteomic analyses, it may not provide comprehensive proteomic coverage when used alone. For example, a large portion of tryptic peptides are fairly short, e.g. in a typical proteome, usually > 50% of tryptic peptides contain 6 or fewer residues 37. These are not optimal for either reversed-phase chromatography or tandem MS sequencing 38. Another problem is that some proteins contain long regions without lysine or arginine, rendering the resulting tryptic fragment too long to be sequenced readily by MS. Recently, our lab and others demonstrated that using multiple proteases can produce complementary proteolytic peptide profiles, significantly improving the sequence coverage for proteomic analysis where peptides produced by one enzyme are poorly detectable 20, 28, 37, 39. In this study, trypsin (cuts at the C-terminus of lysine and arginine) and GluC (cuts at the C-terminus of glutamic acid and aspartic acid) were employed to digest the swine plasma samples in parallel. As the specificities of the two enzymes are completely orthogonal, the distinct, complementary peptide profiles produced by the two enzymes can help to improve the proteomic coverage for the swine plasma.
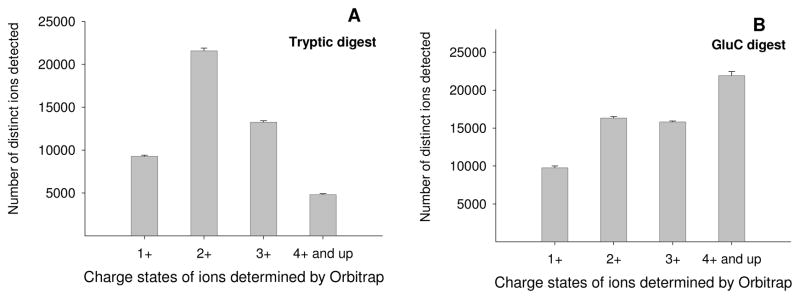
One limitation of a multiple-protease strategy is that the commonly-used CID technique may not be able to provide optimal sequencing of non-tryptic peptides. For example, peptides produced by GluC may contain multiple internal basic residues and thus may be highly charged (≥3 charges) under ESI conditions22, 27; when fragmented by CID, these highly charged precursors often fail to produce sequence-informative spectra that are required to achieve a confident identification. Moreover, as dictated by the mobile proton model22, CID may rely on a charged C-terminal basic residue (e.g. K or R) to produce a consecutive series of strong y ions. The lack of a C-terminal basic residue in a GluC-produced peptide may result in a CID spectrum that is less sequence-informative than that from a tryptic peptide. By comparison, electron-based fragmentation methods such as ECD and ETD provide extensive cleavage of the peptide backbone of highly-charged precursors, and are indifferent to the positions of the basic residues in a peptide24–26. Therefore, we hypothesize that the use of both CID and ETD with the dual-enzyme approach could markedly enhance the proteomic coverage for the analysis of swine plasma. To assess this feasibility, we mapped the distributions of peptide charge states in the tryptic and GluC digests of CPLL-treated swine plasma respectively. The nano-LC/Orbitrap was employed to measure the charge states of MS precursors (m/z range: 310–2000) detected in the digests, and the statistics of charge states were summarized using the Rawmeat (Thermofisher Scientific) software package. Given that the plasma samples underwent the protein-specific CPLL treatment followed by an efficient clean-up procedure prior to the digestion (discussed below), it is reasonable to assume the majority of the detected precursors are peptides. Some representative data is shown in Fig. 4. In the tryptic digest, the precursors were predominantly doubly-charged with only a small fraction of ions (~10%) carrying 4 or more charges (Fig. 4A). This was because trypsin cleaves at most of the K or R residues, and thus most of tryptic peptides carry only 1 basic residue, which tend to produce intensive doubly-charged ion under ESI22. Conversely, as GluC doesn’t cleave at the basic residues, it is not able to ‘regulate’ the number of charges of the produced peptides as trypsin does. Therefore, it was observed that a large portion of the GluC peptides derived from the treated plasma samples carry four or more charges (Fig. 4B), which can be sequenced with good efficiency by ETD but not by CID. Therefore, while the tryptic peptides produced from the swine plasma are relatively CID-friendly, the highly-charged precursors in the GluC digest maybe more suitable for ETD analysis.
Fig. 4.
The charge state distributions of distinct precursor ions detected respectively in (A) tryptic digest and (B) GluC digest of a CPLL-treated swine plasma sample (n=3). The nano-LC/Oribtrap was employed to measure the charge states of MS precursors (resolution: 100,000, m/z range: 310–2000) detected in the digests, and only peaks with charge states determined using isotope clusters were included. A 6-hr gradient on a heated nano-column (75 μmID × 50 cm, 3-μm particle size) was employed to resolve the mixtures efficiently. The statistics of charge states of unique ions were summarized using Rawmeat (Thermofisher Scientific) software package. Ions with differed in m/z by <0.02 and in retention times by < 2 min are grouped as one ion.
Prior to digestion, it is critical to remove the detergents and urea presenting at high concentrations in the eluate, as well as the residual non-protein matrix components, while maintaining a minimal loss of protein. To achieve this goal, a highly efficient and high-recovery precipitation/on-pellet-digestion procedure developed by our lab19, was employed. Details are specified in the EXPERIMENTAL section. A high (86–94%) and reproducible recovery was achieved for both the native and CPLL-treated swine plasma (data not shown). In order for a comprehensive proteomic analysis, an efficient proteolysis is critical 37,27. Whereas the optimal procedures for trypsin digestion have been well-established, the conditions of GluC proteolysis have not yet been thoroughly optimized. Therefore, we set out to optimize the GluC digestion procedure for the on-pellet digestion of plasma proteins. Key conditions including the composition of the digestion buffer, enzyme/substrate ratio and digestion time were optimized for the maximal number of unique peptides identified by the combination of CID and ETD on a nano-LC/LTQ/Orbitap/ETD system. It has been reported that GluC has two pH optima, respectively at pH 4.0 and pH 8.0; moreover, the composition of the digestion buffer may markedly affect its specificity40. Therefore, the effects of pH and buffer compositions were investigated and the results are presented in SI Fig. 3. It was observed that efficacy of GluC was higher at pH 8.0 than at pH 4.0, as indicated by the fact that averagely ~14% more unique peptides were identified under pH 8.0 (p<0.05); moreover, the addition of 80 mM phosphates in the buffer significantly improved the yield of the unique peptides (p<0.05), probably due to the enhanced cleavage of aspartic acid residues in the presence of phosphates40. Therefore, a buffer at pH 8.0 containing 80 mM phosphates was selected for GluC digestion. Using this buffer, we determined that the enzyme/substrate ratios for step 1 and 2 at 1:20 and 1:15 and a total digestion time of 16 h were necessary to achieve efficient cleavage. Moreover, we investigated potential benefits of using a cleavable surfactant for the on-pellet-digestion with GluC. Two acid-cleavable surfactants, the ProteasMax® and the RapiGest®, were assessed. Though the incorporation of cleavable surfactants didn’t result in significantly more unique peptides (SI. Fig. 3), both reagents provided two benefits: (i) dissolution of the pellet at the first step of digestion was accelerated, and typically required <1 h comparing with 4–5 h without a surfactant; (ii) the reproducibility of digestion was improved markedly, as indicated by the smaller variations in the numbers of peptide ID by triplicate experiments (SI Fig.3). Therefore, an acid-cleavable surfactant was used for GluC digestion. In contrast, using cleavable detergents for 2-step trypsin digestion of the swine plasma samples didn’t provide notable benefit, probably due to the higher efficacy of the trypsin. This observation agrees well with our previous observations 19. It is important to note that GluC results in a much larger number of missed cleavages than trypsin even if a fully-optimized condition is used, as we examined with both pure proteins and plasma samples (data not shown). Based on this observation, we set the tolerance of maximal missed-cleavages at 5 for the Sequest searching of GluC-produced peptides.
The conditions of MS fragmentations were optimized as well and the details are discussed in the following section.
Optimization of high-resolution SCX and nano-LC/MS
In order to enhance the proteomic coverage, an offline SCX fractionation was employed to pre-fractionate the proteolytic digests before the nano-LC/MS analysis. An SCX column with a relatively large ID (4.6-mm) was used to provide a high loading capacity (i.e. 1.2 mg total peptides were loaded per injection). Ammonium formate buffers were selected for separation because the buffer components could be readily sublimed by lyophilization, which enables facile concentration of each fraction prior to the nano-LC/MS analysis, and thus may improve the sensitivity for analyzing low abundance peptides. Chromatographic conditions, such as the composition of acetonitrile, ion strengths and the pH of each mobile phase, as well as the gradient profile were optimized extensively to achieve high-resolution separation (EXPERIMENTAL section). It is important to note that the sample loadings were performed with a very low ion-strength (i.e. 3 mM ammonium formate) in order to focus the peptides on the front end of the column, which minimized peak broadening during the sample loading, as we demonstrated previously41. Under the optimized conditions, high peak capacities ranging from 115 to 135 were achieved for the tryptic and GluC peptides (cf. Fig. 3). Such high extents of chromatographic resolution allowed the majority of the peptides to focus in the single 2-min fractions, efficiently simplifying the digest mixtures and greatly facilitating the detection of low-abundance species by the following nano-LC/MS approach.
A lab-made nano-LC/nanospray setup19, which provides high chromatographic resolution, sensitivity and separation reproducibility and low void-volume, was employed for the analysis of each fraction. A relatively long (50 cm in length) reversed-phase nano-column was used for separation; such a column, in conjunction with a long, shallow elution gradient, can help to achieve a high proteomic coverage and to improve the analysis of low-abundance peptides, as suggested by our preliminary experiments and previous works 8, 19, 42, 43. Pilot studies indicated that heating the analytical column provides significantly improved chromatographic resolution for most of the plasma-derived peptides (data not shown); as a result, an optimized separation temperature at 52 °C was employed. The chromatographic conditions were thoroughly optimized and the result is shown in the EXPERIMENTAL section. A typical chromatogram for the separation of the fractionated plasma sample was shown in SI Fig. 4. In SI Fig 4, a peptide elution window of more than 300 min and a peak capacity >650 was achieved; this high level of chromatographic separation enables extensive identifications of the swine plasma proteome (cf. SI Table I and II).
The Orbitrap was chosen as the MS analyzer because its high mass resolution/accuracy enables confident identification by both CID and ETD44. Optimal MS conditions, such as the position of the nanospray tip, ionization voltages, and dynamic exclusion conditions were experimentally identified. It is important to note that a high target ion value (8×106) was used for the Orbitrap, which improved signal-to-noise ratios for peptides by at least 10-fold over the manufacturer-default value (0.5 ×106) without compromising the accuracy and resolution for m/z measurement, as we demonstrated previously 19, 20, 43. This technical advance expanded the dynamic range of the MS detection by at least one order of magnitude, thereby enhancing the ability to identify low-abundance plasma proteins in the SCX fractions.
With the emphasis on ETD, the MS2 conditions were optimized. Successful ETD sequencing can be hampered by the production of abundant charge-reduced, non-dissociative ion species, especially for those peptide precursors carrying lower numbers of charges (e.g. the doubly-charged)45. To dissociate these charge-reduced ions, which are likely held together by intramolecular non-covalent forces such as van der Waals interactions force and/or hydrogen bonding46, we employed a short burst (~5 ms) of low-energy CID targeting the charge-reduced product populations. It was observed that this approach significantly improved the fragmentation efficiency for both the doubly- and triply- charged peptide ions (data not shown). The ETD reaction duration was optimized for all charge states and an interaction time of 110 ms was determined to be optimal. In our previous studies, we found that the ETD reaction with relatively high densities of fluoranthene anions may effectively increase the fragmentation efficiency for low-abundance peptides11b, 13c, when using a wide-band (10 amu) isolation of the anions20, 27. In this study, the results of optimization using 251 model low-abundance precursors showed that an anion number of 1E6 provided maximal Xcorr for these doubly-charged, low abundance peptides (details not shown). Therefore, an anion number of 1E6 was used for ETD analysis of all fractions.
In-depth proteomic analysis of swine plasma proteome
Using the strategy combining the CPLL-treatment and the approach of dual-enzyme and dual-activation, we conducted an in-depth proteomic analysis of the pooled plasma from healthy swine. Nano-LC/MS runs were performed in duplicate for each fraction, enzyme and activation method and totally 400 runs were performed (EXPERIMENTAL section). The following sections will discuss the method for protein identification, contributions of the CPLL treatment, enzymes and activation methods to the identification of these proteins, as well as the characterization of the identified proteins based on physiochemical properties, functions and secreted proteins, etc.
a)Strategy for protein identification and the evaluation of false-positive rates (FDR)
As currently only a small fraction of swine proteins have valid sequence information and it was speculated that swine gene sequence is highly similar to human’s47, we employed a database combining both swine and human proteins for identification. To minimize the redundancy of the final protein list, the non-redundant Uniprot-Swiss-Prot database was employed to generate the fused swine-human database. If the same protein was identified for both human and swine, the human entry was removed. Each of the 400 raw files (i.e. duplicate runs of each condition/fraction) was searched with Sequest Cluster against the combination of forward and reverse database. As this study involves a large dataset produced by the combinations of different proteases and activation methods, the FDR for identification was carefully controlled and monitored. Two different filtering strategies were employed in tandem: firstly, the search results of all runs are combined and then evaluated with Peptideprophet and ProteinProphet algorithms. The filtering threshold was set at Peptide probability of 95% or higher and protein probability of 99% or higher. Secondly, the list was further filtered by a relatively stringent set of Xcorr cutoffs (EXPERIMENTAL), a peptide precursor mass tolerances <15 ppm, and the requirement that at least two unique peptides must be identified for a protein. Under the above criteria, this large-scale investigation resulted in the identification of 3421 unique proteins. The proteins and peptides identified respectively in CPLL-treated and native swine plasma are listed in SI Table I and II. The final peptide FDR for the entire dataset was estimated by comparing the identified IDs from forward and reverse database, which is approximately 1.8%.
b) The effectiveness of CPLL treatment
Because the optimized CPLL treatment reduced the protein concentration dynamic range by at least 2 orders of magnitudes (c.f. Fig. 2), it was able to significantly improve the proteomic coverage for swine plasma, as illustrated in Fig.5A. While 1,708 unique proteins were identified in the native plasma, 2,657 were identified in CPLL-treated plasma, an increase of 55.5%. Of the 3421 total proteins identified, only 944 (27.6%) proteins were found in both native plasma and the CPLL-treated plasma samples, indicating a significant change in the distribution of plasma proteins after the CPLL treatment. In this study, we found that such change was caused mainly by the considerable reduction of protein concentration dynamic ranges by CPLL, and probably to a lesser extent, the selective enrichment/depletion of certain groups of proteins by the CPLL treatment (discussed below).
Fig. 5.
The contributions of treatment approaches, enzyme and activation methods in the identification of swine plasma proteins. (A) The distribution of proteins identified in CPLL-treated plasma vs. native plasma; (B) contributions of enzymes for CPLL-treated plasma; (C) contributions of activation methods for CPLL-treated plasma; (D) contributions of enzymes for native plasma; (E) contributions of activation methods for native plasma. Numbers in the venn diagrams denote the numbers of unique proteins identified.
c)the effectiveness of dual-enzyme, dual-activation strategy
This study demonstrated that the dual-enzyme, dual-activation strategy provided much higher proteomic coverage than using trypsin and CID alone, for the analysis of both CPLL-treated- and native swine plasma. The contributions of enzymes and activation methods to the numbers of identified proteins are illustrated in Fig. 5B-5E. In CPLL-treated plasma, 1548 proteins (58.3% of total identified) were identified with at least two unique tryptic peptides (by either CID or ETD, same below), while 1343 proteins (50.6%) were identified with at least two unique GluC-produced peptides(Fig. 5B). These two sets of proteins overlap by 555 proteins (i.e. proteins identified with at least two unique tryptic peptides and two unique GluC-peptides), accounting for only 20.9% of the total identified proteins. In addition, there are occasions where only one each unique tryptic and GluC peptide was identified for an individual protein. In these cases, a confident identification can’t be made using the dataset produced by either one of the enzymes because two unique peptides are required for a confident identification; however, the combination of the unique peptides from the two enzymes results in a confident identification of the protein. Such proteins were termed “combined” in Fig. 5. For CPLL-treated plasma, 321 proteins (12.1%) were identified by this “combined” strategy (Fig. 5B). As to the contributions by CID and ETD, 1120 (42.2%) proteins were identified in CPLL-treated plasma exclusively by CID with ≥ 2 unique peptides (produced by either trypsin or GluC, same below), while 745 (28.0%) proteins were identified exclusively by ETD with ≥2 unique peptides (Fig. 5C); in addition, 607 proteins (22.8%) were identified by both CID and ETD with at least 2 unique peptides by each. There were 185 (7.0%) proteins identified by combining one unique CID-peptide and one unique ETD-peptide. Compared with the “traditional” method using only trypsin and CID, the dual-enzyme and dual-activation strategy identified 2.6-fold more unique proteins (2657 vs 1041 proteins) and 3.3-fold more unique peptides (12,492 vs 3744 peptides) in CPLL-treated plasma. Furthermore, the dual-enzyme and dual-activation strategy markedly improved the sequence coverage for individual proteins, especially for these of lower-abundance. For example, the dual-enzyme and dual-activation strategy improved the sequence coverage of the 1041 proteins detected by the trypsin-CID by an average of 1.8 fold (SI Fig.5). In SI Fig.5, we ranked these proteins by their sequence coverage achieved by trypsin-CID, which may serve as a rough estimation of the relative abundance of these proteins. Obviously, the dual-enzyme and dual-activation strategy increased the sequence coverage to a much greater extent for the lower-abundance proteins than for the high-abundance ones, e.g. the average sequence coverage was only increased by 1.2 fold for the top 50 abundant proteins, but for the ~200 lowest abundance proteins in this dataset the sequence coverage was improved by 2.9 fold.
For native plasma, the contributions of enzymes/activations to the numbers of identified proteins as well as the improvement of protein sequence coverage by the dual-enzyme and dual-activation strategy, follow trends similar to those found for the CPLL-treated plasma, as shown in Fig. 5D-5E and SI Fig. 6.
In total, trypsin-CID identified 5706 unique peptides (excluding those peptides are not in the identified proteins) in the native and CPLL-treated plasma sample, from which we inferred 1447 unique proteins. By comparison, the dual-enzyme and dual-activation strategy enabled the identification of 3421 unique proteins and 18,495 peptides.
The above data demonstrates that the distinct, complementary peptide profiles produced by the two orthogonal enzymes substantially improved proteomic coverage for swine plasma; furthermore, the utilization of both CID and ETD with the dual-enzymes approach contributed substantially to an in-depth proteomic analysis of swine plasma.
d) Physicochemical characteristics of the identified plasma proteins
We classified the identified proteins by several physicochemical properties, including molecular weight (MW), isoelectric point (pI), hydrophobicity (as expressed by the GRAVY values) and transmembrande domains (TM domains). The large plasma protein dataset produced by the in-depth proteomic analysis in this study afforded a scrutiny of the physicochemical characteristics of swine plasma proteome (SI. Fig. 7). Key observations include: i) only 25.3% of identified plasma proteins have MW<60kDa, which is in sharp contrast to a typical tissular or cellular proteome, where generally >60% of proteins have MW <60 kDa. This phenomenon may reflect the difference between a cellular-based and a body-fluid-based proteome, and the selective renal clearance of proteins <60 kDa 48 may contribute to this difference; ii) the pattern of pI distribution of swine plasma proteins agrees well with previous data obtained with human plasma49; iii) calculation of GRAVY values of the identified proteins revealed that the vast majority (95.4%) of the identified proteins are highly hydrophilic; and iv) Approximately 18% (625 proteins) of the identified proteins contain TM domain(s), which agrees well with previous observations in human plasma2, 49. Detailed results and discussions are in the Supplementary Materials.
e) Enrichment Analysis based on Gene Ontology Annotation and the prediction of zsecreted proteins
A Gene Ontology (GO) enrichment analysis of the identified plasma proteins was performed. Within the context of the three GO term categories, 110 terms (out of a total of 2726 analyzed) were identified as significantly enriched in the plasma dataset by at least 2 fold over those in the entire database (p<0.001, SI Fig. 8). Neumerous of these enriched GO terms are well recognized as plasma-related (SI Fig 8). Interestingly, proteins in many intracellular GO terms are also enriched in the plasma dataset, which may result from different levels of selective cellular leakages. These observations agree well with the previous reports on human plasma 50, 51. Proteins containing a predicted signal peptide (SP) are considered as a secreted protein52. In this study, we identified 802 proteins with a SP, which to our knowledge is the highest number of putatively secreted proteins reported in plasma. Detailed results and discussion is specified in the Supplementary Materials.
f) Potential selective, non-abundance-dependent enrichment/loss of plasma proteins by the CPLL technique
In this study, we demonstrated the CPLL technique selectively enriched the low-MW proteins. Moreover, the data obtained in this study suggested that the CPLL enrichment resulted in a reproducible loss of certain proteins, which agreed with previous observations that certain proteins might fail to bind the CPLL beads53, 54. Detailed results and discussion is specified in the Supplementary Materials.
Candidate Biomarkers Associated with Cardiovascular Disease
For the investigations of CVD, swine constitute the preferred animal model due to the high similarities in many cardiovascular and hematologic characteristics with human55. A comprehensive identification of CVD-associated proteins in swine plasma may provide tools to enhance our understanding of the physiological interactions among plasma, heart and vasculature, and thus facilitate both mechanism studies and the discovery of novel CVD biomarkers. Here we mapped the CVD-related proteins from those identified in plasma, via two different but related approaches: classification of CVD-related proteins and pathway analysis.
Classification analysis revealed 657 unique proteins in nine classifications such as coronary artery disease, arteriosclerosis, chronic stable angina and vascular dementia, were identified as CVD-associated with high statistical significance (p<0.05). A Canonical Pathway Analysis of all the proteins identified via IPA package revealed twenty canonical pathways with high significance (p<0.05) (SI Table IV). Many of the most significant pathways are CVD-associated, such as the “Nitric Oxide Signaling in the Cardiovascular System” (20 out of 99 components identified, illustrated SI Fig. 11) and “Intrinsic Prothrombin Activation Pathway” (18 out of 35 components identified). Detailed results and discussions for the classification and pathway analysis are shown in the Supplementary Materials.
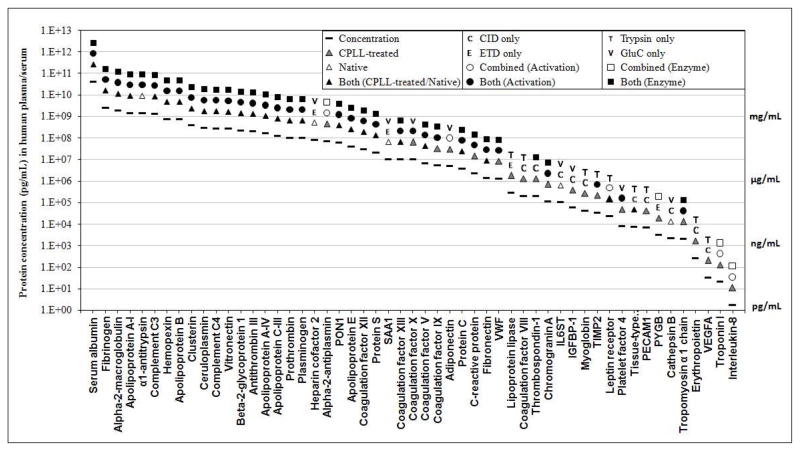
The CVD-associated proteins identified in this work span a broad dynamic range of protein concentrations. Fig.6 plots the reference levels (in healthy human plasma) 56–58 of some representative CVD-associated proteins identified in this study. As an extensive dataset of the levels of swine plasma proteins are not yet available and that existing data suggested the protein concentrations in swine plasma are consistent with those in human59, 60, we utilized levels of plasma proteins in human as reference in Fig.6. The levels of these proteins ranged from 1.7 pg/mL to 41 mg/mL, which indicates the CVD-associated proteins identified here covered a protein concentration dynamic range of at least 9–10 orders of magnitude, assuming the concentrations of proteins in swine and human plasma are similar. It is interesting to observe that most proteins at relatively high abundance (i.e. these of high-mg/mL to medium-μg/mL levels) were identified in both the CPLL-treated and native plasma, and by all methods of proteolyses/activations. Conversely, most lower-abundance proteins (i.e. these of medium ng/mL to low pg/mL) were only identified in the CPLL-treated plasma, and were either detected by a single digestion or activation method, or required the combination of peptides identified by different enzymes/activations to achieve a confident identification. This demonstrates the CPLL-treatment and the dual-enzyme, dual activation strategy contributed substantially in the identification of low-abundance proteins. Our work confidently identified some important CVD markers presenting at low abundance, such as interleukin-8 (1.7 pg/mL, average level in healthy human, same below), vascular endothelial growth factor A (32 pg/mL) and cardiac troponin I (20 pg/mL). Some representative MS/MS spectra by CID and ETD are shown in SI Fig. 12. The identified proteolytic peptides can serve as candidates for signature peptides for highly sensitive and accurate quantification of these proteins by selected reactions monitoring (SRM).
Fig. 6.
Representative CVD-associated proteins identified in this study, which span a wide protein concentration dynamic range. A total of 53 proteins were plotted. As swine data is not available, the protein concentrations data were obtained from previously reported levels in the plasma/serum of healthy humans56–58.
CONCLUSIONS
The comprehensive investigation of the plasma proteome represents a daunting challenge. In order to achieve high proteomic coverage and to improve identification of lower abundance plasma proteins, we used a suite of proteomic strategies, including the CPLL treatment, dual-enzyme and dual-activation, and high-resolution SCX and nano-LC/MS analysis.
The CPLL technique, which is a universal, non-proteome-specific approach, was employed to reduce the protein concentration dynamic ranges in swine plasma and thereby enabling a “deeper” proteomic analysis. We investigated the characteristics of CPLL-enrichment and made two important modifications to the standard CPLL procedure to enhance the effectiveness for treatment of swine plasma: first, using a moderately denaturing condition for sample loading, losses of low abundance proteins due to bindings to more abundant ones were diminished; second, the extents of reducing the dynamic range of plasma protein concentrations were markedly improved by employing multiple loading-washing cycles. Under the optimized conditions, significant enrichment of low-abundance proteins (estimated at more than 2 orders of magnitude) was observed.
In order to improve the coverage of the swine plasma proteome, we employed a dual-enzyme (trypsin and GluC) and dual-activation (CID and ETD) approach. With the emphasis on GluC, the digestion conditions were thoroughly optimized and a buffer containing phosphates and a cleavable detergent was determined to provide an optimal efficiency for GluC. Parameters for both CID and ETD were optimized to enhance the identification of low-abundance peptides. The final results showed that the dual-enzyme and dual-activation approach drastically improved proteomic coverage.
A high-resolution SCX fractionation, as well as the sufficient separation of peptides on a long, heated nano-LC column, also contributed substantially to the high proteomic coverage achieved.
The physicochemical characteristics, GO annotation enrichment and secreted proteins of the identified proteins were extensively analyzed, which led to several interesting observations. Since one of the most prominent applications of the swine models is for the study of CVD, we looked for CVD-related markers within the identified proteins via both function classification and pathway analysis. We identified 657 proteins as significantly CVD-associated (p<0.05) and constituted five canonical pathways that are closely related to CVD (p<0.05). These results indicate that the plasma proteome could be abundant in CVD-associated proteins that may be valuable to study the physiological/pathological interactions among plasma, heart and vasculature.
The authors are well cognizant of several limitations of this work. First, while this work has identified most of the reported plasma proteins down to high pg/mL levels (detailed comparison not shown), many proteins presented at low pg/mL levels, such as TNF-α, IL-1/2/7, and Oncostatin, were not detected. This indicated that the strategy may not be sufficiently robust for the proteins presenting at the concentrations of the lowest 1–2 orders of magnitude. Therefore, further fractionation or enrichment may be needed in order to extensively identify the proteins of the lowest abundance. Second, this study was not able to determine whether an identified protein is in intact or fragmented form in plasma, which is a problem common to most shotgun-based proteomic methods. In order to obtain the information on the integrity of the proteins, a MW-based fractionation strategy (such as an SDS-PAGE fractionation) before digestion may be helpful. Third, though the CPLL method effectively enriched most low abundance proteins, it appears to selectively lose certain plasma proteins, as demonstrated in this study. Therefore, it is necessary to combine the analysis of both the CPLL-treated and the native plasma in parallel to achieve comprehensive proteomic coverage. Moreover, novel CPLL-strategies such as using different pH values to improve protein capturing61 could be employed to enhance the CPLL efficiency.
This work demonstrated the synergetic effect of using CPLL treatment, dual enzyme/activation, and sufficient chromatographic separation for in-depth analysis of plasma proteome, which enabled a much more comprehensive proteomic analysis than using any of the components alone. This work also contributes to analytical sciences in that i) the CPLL loading procedures were modified to reduce significantly the protein dynamic range in swine plasma, ii) the work demonstrated the use of two orthogonal enzymes, as opposed to a collection of many enzymes with overlapping specificities, for a higher proteomic coverage, and iii) suggested that ETD may be preferable over CID for GluC-produced peptides.
To our knowledge, this study represents the first in-depth evaluation of non-human plasma proteome. The strategy used here is adaptable to other highly complex proteomes. The extensive analysis of swine plasma yielded candidates for the quantitative signature peptides that are essential for the development of an SRM-based method for the targeted quantification of proteins of interest. For example, using the sequence information, precursor m/z and fragments of the peptide candidates obtained in this study, a sensitive and selective targeted quantification method for multiple plasma proteins can be developed in a high-throughput manner using the on-the-fly Orthogonal Array Optimization strategy we described previously21.
Supplementary Material
Acknowledgments
We thank Drs. Yahao Bu, Jin Cao and Xiaotao Duan for helping with some of the experiments. This work was supported by NIH grant HL55324 and HL61610 to JMC, a UB multi-disciplinary proposal support grant to JMC and a grant by the Center of Protein Therapeutics and NIH grant DA027528 to JQ.
References
- 1.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. Mol Cell Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Shen Y, Jacobs JM, Camp DG, 2nd, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. Anal Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 4.Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang ST, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J, Steiner S. Proteomics. 2003;3:1345–1364. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 6.Anderson NL. Clin Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 7.Tu C, Rudnick PA, Martinez MY, Cheek KL, Stein SE, Slebos RJ, Liebler DC. J Proteome Res. 2010;9:4982–4991. doi: 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu T, Qian WJ, Gritsenko MA, Xiao W, Moldawer LL, Kaushal A, Monroe ME, Varnum SM, Moore RJ, Purvine SO, Maier RV, Davis RW, Tompkins RG, Camp DG, 2nd, Smith RD. Mol Cell Proteomics. 2006;5:1899–1913. doi: 10.1074/mcp.M600068-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Q, Garnham CP, Elliott ST, Bovenkamp DE, Van Eyk JE. Proteomics. 2005;5:2656–2664. doi: 10.1002/pmic.200402048. [DOI] [PubMed] [Google Scholar]
- 10.Gundry RL, White MY, Nogee J, Tchernyshyov I, Van Eyk JE. Proteomics. 2009;9:2021–2028. doi: 10.1002/pmic.200800686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandow JE. Proteomics. 2010;10:1416–1425. doi: 10.1002/pmic.200900431. [DOI] [PubMed] [Google Scholar]
- 12.Qian WJ, Kaleta DT, Petritis BO, Jiang H, Liu T, Zhang X, Mottaz HM, Varnum SM, Camp DG, 2nd, Huang L, Fang X, Zhang WW, Smith RD. Mol Cell Proteomics. 2008;7:1963–1973. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furka A, Sebestyen F, Asgedom M, Dibo G. Int J Pept Protein Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 14.Simo C, Bachi A, Cattaneo A, Guerrier L, Fortis F, Boschetti E, Podtelejnikov A, Righetti PG. Anal Chem. 2008;80:3547–3556. doi: 10.1021/ac702635v. [DOI] [PubMed] [Google Scholar]
- 15.D’Amato A, Bachi A, Fasoli E, Boschetti E, Peltre G, Senechal H, Righetti PG. J Proteome Res. 2009;8:3925–3936. doi: 10.1021/pr900221x. [DOI] [PubMed] [Google Scholar]
- 16.Beseme O, Fertin M, Drobecq H, Amouyel P, Pinet F. Electrophoresis. 2010;31:2697–2704. doi: 10.1002/elps.201000188. [DOI] [PubMed] [Google Scholar]
- 17.Fertin M, Beseme O, Duban S, Amouyel P, Bauters C, Pinet F. Proteomics Clin Appl. 2010;4:654–673. doi: 10.1002/prca.200900178. [DOI] [PubMed] [Google Scholar]
- 18.Sennels L, Salek M, Lomas L, Boschetti E, Righetti PG, Rappsilber J. J Proteome Res. 2007;6:4055–4062. doi: 10.1021/pr070339l. [DOI] [PubMed] [Google Scholar]
- 19.Duan X, Young R, Straubinger RM, Page B, Cao J, Wang H, Yu H, Canty JM, Qu J. J Proteome Res. 2009;8:2838–2850. doi: 10.1021/pr900001t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao J, Covarrubias VM, Straubinger RM, Wang H, Duan X, Yu H, Qu J, Blanco JG. Anal Chem. 2010;82:2680–2689. doi: 10.1021/ac902314m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu J, Straubinger RM. Rapid Commun Mass Spectrom. 2005;19:2857–2864. doi: 10.1002/rcm.2138. [DOI] [PubMed] [Google Scholar]
- 23.Qu J, Jusko WJ, Straubinger RM. Anal Chem. 2006;78:4543–4552. doi: 10.1021/ac0521697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coon JJ, Shabanowitz J, Hunt DF, Syka JE. J Am Soc Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Good DM, Wirtala M, McAlister GC, Coon JJ. Mol Cell Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Straubinger RM, Aletta JM, Cao J, Duan X, Yu H, Qu J. J Am Soc Mass Spectrom. 2009;20:507–519. doi: 10.1016/j.jasms.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MH, Koria P, Qu J, Andreadis ST. FASEB J. 2009;23:3874–3883. doi: 10.1096/fj.08-117804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maltaris T, Dragonas C, Hoffmann I, Mueller A, Schild RL, Schmidt W, Beckmann MW, Dittrich R. Eur J Obstet Gynecol Reprod Biol. 2006;126:56–62. doi: 10.1016/j.ejogrb.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 30.Ekser B, Rigotti P, Gridelli B, Cooper DK. Transpl Immunol. 2009;21:87–92. doi: 10.1016/j.trim.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Canty JM, Fallavollita JA. American Journal of Physiology-Heart and Circulatory Physiology. 1999;277:H417–H422. doi: 10.1152/ajpheart.1999.277.1.H417. [DOI] [PubMed] [Google Scholar]
- 32.Crick SJ, Sheppard MN, Ho SY, Gebstein L, Anderson RH. J Anat. 1998;193(Pt 1):105–119. doi: 10.1046/j.1469-7580.1998.19310105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page B, Young R, Iyer V, Suzuki G, Lis M, Korotchkina L, Patel MS, Blumenthal KM, Fallavollita JA, Canty JM., Jr Circ Res. 2008;102:103–112. doi: 10.1161/CIRCRESAHA.107.155895. [DOI] [PubMed] [Google Scholar]
- 34.Eng JK, Mccormack AL, Yates JR. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 35.Searle BC. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 36.Boschetti E, Bindschedler LV, Tang C, Fasoli E, Righetti PG. J Chromatogr A. 2009;1216:1215–1222. doi: 10.1016/j.chroma.2008.11.098. [DOI] [PubMed] [Google Scholar]
- 37.Swaney DL, Wenger CD, Coon JJ. J Proteome Res. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 39.Choudhary G, Wu SL, Shieh P, Hancock WS. J Proteome Res. 2003;2:59–67. doi: 10.1021/pr025557n. [DOI] [PubMed] [Google Scholar]
- 40.Houmard J, Drapeau GR. Proc Natl Acad Sci U S A. 1972;69:3506–3509. doi: 10.1073/pnas.69.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu J, Qu Y, Straubinger RM. Anal Chem. 2007;79:3786–3793. doi: 10.1021/ac062184r. [DOI] [PubMed] [Google Scholar]
- 42.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EA, Udseth HR, Smith RD. Anal Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 43.Qu J, Lesse AJ, Brauer AL, Cao J, Gill SR, Murphy TF. BMC Microbiol. 2010;10:162. doi: 10.1186/1471-2180-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenton AG, Godfrey AR. J Am Soc Mass Spectrom. 2010 doi: 10.1016/j.jasms.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JE, Coon JJ. Anal Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horn DM, Ge Y, McLafferty FW. Anal Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 47.Jiang Z, Rothschild MF. Int J Biol Sci. 2007;3:129–131. doi: 10.7150/ijbs.3.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schenk S, Schoenhals GJ, de Souza G, Mann M. BMC Med Genomics. 2008;1:41. doi: 10.1186/1755-8794-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu CJ, Dai J, Li SJ, Sheng QH, Deng WJ, Xia QC, Zeng R. J Proteome Res. 2005;4:1265–1273. doi: 10.1021/pr0497529. [DOI] [PubMed] [Google Scholar]
- 50.Jeong SK, Lee EY, Cho JY, Lee HJ, Jeong AS, Cho SY, Paik YK. Proteomics. 2010;10:1250–1255. doi: 10.1002/pmic.200900371. [DOI] [PubMed] [Google Scholar]
- 51.Ping P, Vondriska TM, Creighton CJ, Gandhi TK, Yang Z, Menon R, Kwon MS, Cho SY, Drwal G, Kellmann M, Peri S, Suresh S, Gronborg M, Molina H, Chaerkady R, Rekha B, Shet AS, Gerszten RE, Wu H, Raftery M, Wasinger V, Schulz-Knappe P, Hanash SM, Paik YK, Hancock WS, States DJ, Omenn GS, Pandey A. Proteomics. 2005;5:3506–3519. doi: 10.1002/pmic.200500140. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, Zhang Y, Yin Y, Gao G, Li S, Jiang Y, Gu X, Luo J. Nucleic Acids Res. 2005;33:D169–173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Righetti PG, Boschetti E, Lomas L, Citterio A. Proteomics. 2006;6:3980–3992. doi: 10.1002/pmic.200500904. [DOI] [PubMed] [Google Scholar]
- 54.Mouton-Barbosa E, Roux-Dalvai F, Bouyssie D, Berger F, Schmidt E, Righetti PG, Guerrier L, Boschetti E, Burlet-Schiltz O, Monsarrat B, de Peredo AG. Mol Cell Proteomics. 2010;9:1006–1021. doi: 10.1074/mcp.M900513-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hasenfuss G. Cardiovasc Res. 1998;39:60–76. doi: 10.1016/s0008-6363(98)00110-2. [DOI] [PubMed] [Google Scholar]
- 56.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, 2nd, Smith RD. Mol Cell Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson L. J Physiol. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Missov E, Calzolari C, Pau B. Circulation. 1997;96:2953–2958. doi: 10.1161/01.cir.96.9.2953. [DOI] [PubMed] [Google Scholar]
- 59.Carpintero R, Pineiro M, Andres M, Iturralde M, Alava MA, Heegaard PM, Jobert JL, Madec F, Lampreave F. Infect Immun. 2005;73:3184–3187. doi: 10.1128/IAI.73.5.3184-3187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muneta Y, Goji N, Tsuji NM, Mikami O, Shimoji Y, Nakajima Y, Yokomizo Y, Mori Y. J Interferon Cytokine Res. 2002;22:883–889. doi: 10.1089/107999002760274908. [DOI] [PubMed] [Google Scholar]
- 61.Fasoli E, Farinazzo A, Sun CJ, Kravchuk AV, Guerrier L, Fortis F, Boschetti E, Righetti PG. J Proteomics. 2010;73:733–742. doi: 10.1016/j.jprot.2009.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.