Abstract
Pyrano[3,2-c]pyridone and pyrano[3,2-c]quinolone structural motifs are commonly found in alkaloids manifesting diverse biological activities. As part of a program aimed at structural simplification of bioactive natural products utilizing multicomponent synthetic processes, we developed compound libraries based on these privileged heterocyclic scaffolds. The selected library members display low nanomolar antiproliferative activity and induce apoptosis in human cancer cell lines. Mechanistic studies reveal that these compounds induce cell cycle arrest in the G2/M phase and block in vitro tubulin polymerization. Because of the successful clinical use of microtubule-targeting agents, these heterocyclic libraries are expected to provide promising new leads in anticancer drug design.
Introduction
Natural products are a rich source of new medicinal leads and, therefore, preparation of natural product-based libraries of compounds is an important area of research in modern drug discovery.1–6 Such efforts, however, are often complicated by the structural complexity of natural products, which often contain a large number of stereogenic centers and intricate ring systems.7 We have recently initiated a research program aimed at structural simplification of natural products, specifically by utilizing multicomponent synthetic processes. We showed that the stereochemically complex structure of an important anticancer lead podophyllotoxin can be efficiently simplified to a dihydropyridopyrazole scaffold, which is accessible via a one-step multicomponent reaction (MCR).a The resulting library of compounds retains a significant portion of podophyllotoxin's cytotoxic potency and apoptosis inducing potential.8,9
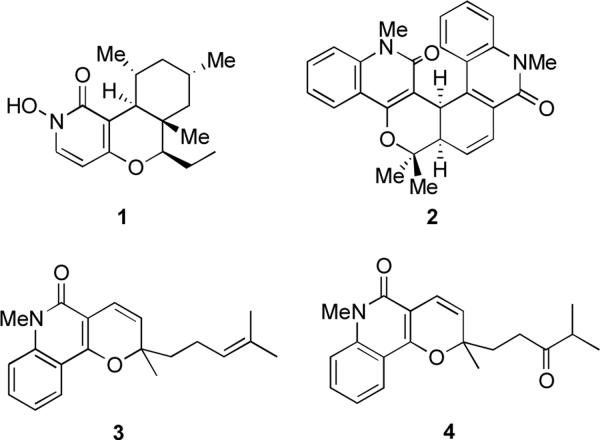
In continuation of these efforts we have been investigating compound libraries based on pyrano[3,2-c]pyridone and pyrano[3,2-c]quinolone scaffolds. This structural motif is broadly represented by pyranopyridone and pyranoquinolone alkaloids manifesting diverse biological activities. The latter include antibacterial,10–12 antifungal and antialgal,13 antiinflammatory14 and antimalarial15 properties as well as inhibition of calcium signaling,16 platelet aggregation17 and nitric oxide production.18 Furthermore, many of these alkaloids exhibit cancer cell growth-inhibitory activity and are investigated as potential anticancer agents. Examples include cytotoxic fusaricide (1)19 and melicobisquinolinone B (2, Figure 1).20 In addition, zanthosimuline (3) is active against multidrug resistant KB-VI cancer cells, while huajiaosimuline (4) exhibits a selective cytotoxicity profile showing the greatest activity with estrogen receptor-positive ZR-75-1 breast cancer cells.21
Figure 1.
Anticancer alkaloids based on pyrano[3,2-c]pyridone and pyrano[3,2-c]quinolone scaffolds.
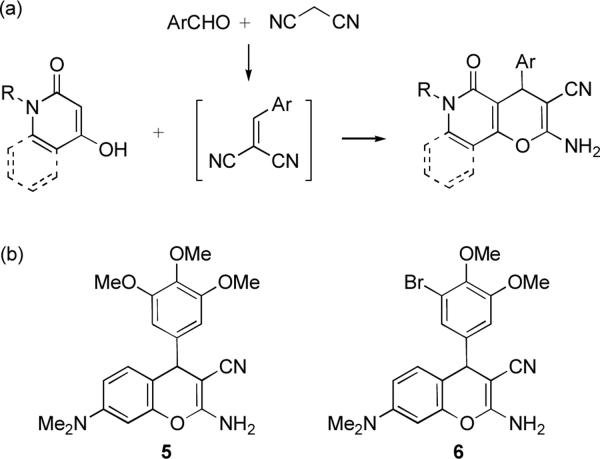
To develop compound libraries based on this fused heterocyclic scaffold we considered a reaction of 4-hydroxypyridones as well as corresponding quinolones with aldehydes and malononitrile (Figure 2a). Although we found only two literature examples pointing to the feasibility of such an MCR for pyranopyridone and pyranoquinolone library preparations,22 the stepwise version of this process involving the synthesis of the intermediate Knoevenagel adducts and their subsequent reactions with 4-hydroxypyridones and quinolones has been well studied.23 Inspection of this literature reveals that the biology of compounds prepared using this process has not been investigated with the exception of antibacterial properties reported for some of these heterocycles.24 Importantly, a number of recent publications and patents have described promising anticancer activity associated with a structurally related chromene scaffold exemplified by compounds 5 and 6 (Figure 2b).25 These compounds, shown to inhibit tubulin polymerization and induce apoptosis in cancer cells, exhibit high potency against taxoland vinblastine-resistant P-glycoprotein overexpressing cell types.25d Furthermore, chromenes 5 and 6 disrupt tumor vasculature in a number of human solid tumor xenografts and they are currently in development as anticancer agents.25b.c
Figure 2.
(a) Proposed MCR for the preparation of pyrano[3,2-c]pyridone and pyrano[3,2-c]quinolone libraries. (b) Chromenes 5 and 6 investigated as anticancer agents.
Results and Discussion
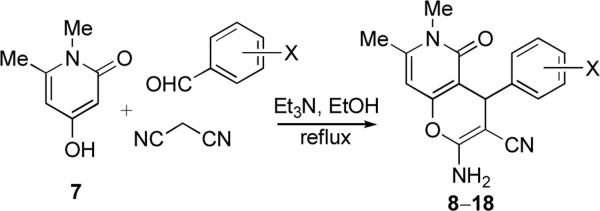
Prompted by the encouraging literature precedent we embarked on the synthesis and anticancer evaluation of the proposed pyrano[3,2-c]pyridone and pyrano[3,2-c]quinolone libraries.26 The synthesis of the pyranopyridone library is shown in Figure 3. Pyridone 7 was selected on the basis of its ready availability by a one-step synthesis involving treatment of the corresponding commercially available pyrone with aqueous MeNH2 following a literature procedure.27 A three-component reaction of pyridone 7 with malononitrile and various aromatic aldehydes in a 1:1:1 ratio proceeds smoothly in refluxing ethanol containing a small quantity of Et3N. Pyranopyridones 8–18 precipitate directly from the refluxing reaction mixtures and require no further purification. The product yields are given in Table 1.
Figure 3.
Three-component synthesis of pyrano-[3,2-c]-pyridones.
Table 1.
Synthetic yields and antiproliferative activity of pyrano-[3,2-c]-pyridones.
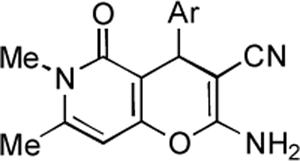 |
synthesis | cell viabilitya | |
|---|---|---|---|
| analogue | Ar | % yield | GI50, μM |
| 8 |
|
83 | 0.33 ± 0.06 |
| 9 |

|
87 | 0.58 ± 0.14 |
| 10 |
|
88 | 1.1 ± 0.8 |
| 11 |
|
75 | 2.7 ± 1.1 |
| 12 |
|
83 | 3.5 ± 1.3 |
| 13 |
|
84 | 6.4 ± 1.1 |
| 14 |
|
97 | 6.5 ± 1.3 |
| 15 |
|
98 | 18.3 ± 2.9 |
| 16 |
|
97 | 43.3 ± 5.1 |
| 17 |
|
81 | > 100 |
| 18 |
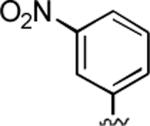
|
97 | 35.0 ± 21.8 |
| 19 |
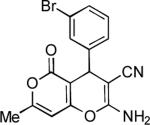
|
87 | > 100 |
| 20 |
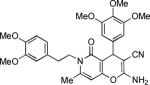
|
80 | 22.7 ± 6.4 |
Concentration required to reduce the viability of HeLa cells by 50% after 48 h of treatment with indicated compounds relative to DMSO control ± SD from three independent experiments, each performed in 4 replicates, determined by MTT assay.
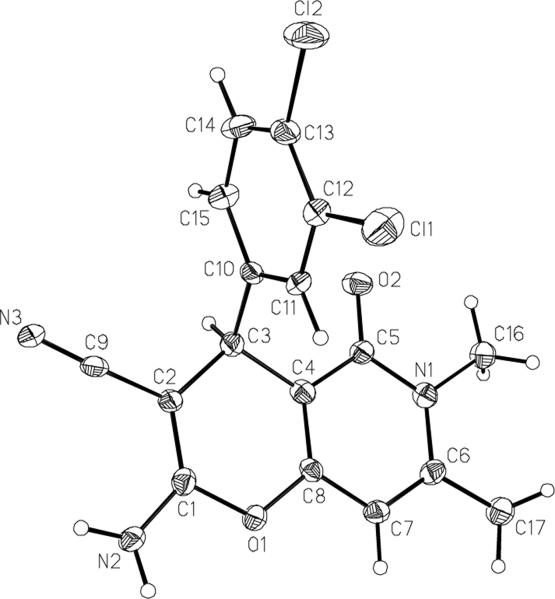
The structures of the pyranopyridones were established by 1H and 13C NMR, MS techniques as well as an X-ray structure of the library member 15 (Figure 4).28
Figure 4.
X-ray structure of pyranopyridone 15 (50% probability thermal ellipsoids).
The analogue library was tested for antiproliferative activity using the HeLa cell line as a model for human cervical adenocarcinoma. The cells were treated with respective compounds for 48 h and cell viability was assessed through measurements of mitochondrial dehydrogenase activity using the MTT method (Table 1).29 It is noteworthy that all potent analogues have a 3-bromo substituent on the aromatic ring at position C4 of the pyranopyridone skeleton (compounds 8–14) and this preference is uniform irrespective of the substitution pattern of this aromatic moiety. The 3-chloro (15) and other variously substituted analogues (16–18) are significantly less potent or are totally inactive. Further, the substitution of the nitrogen in the pyridone ring by oxygen, as in pyranopyranone 19, abolishes the activity as well. Moderate potency of the N-(β-arylethyl)pyridone 20, synthesized in a manner analogous to the rest of the library, warrants further investigation of compounds having a bulky moiety on the pyridone nitrogen. Efforts to prepare a library of such compounds are underway in our laboratories.
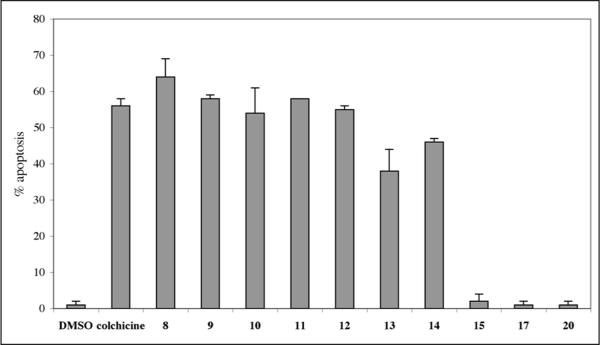
Since many clinically used anticancer agents induce apoptosis in cancer cells,30 we tested the pyranopyridone analogues for their ability to induce apoptosis in Jurkat (model for human T-cell leukemia) cells using the flow cytometric annexin-V/propidium iodide assay (Figure 5).31 Compounds 8–14, exhibiting submicromolar or low micromolar potencies for the inhibition of proliferation of HeLa cells, were found to be strong inducers of apoptosis in Jurkat cells at 5 μM concentrations. The magnitude of apoptosis induction is comparable to the known antimitotic agent colchicine used at the same concentration. In contrast, compounds 15, 17, 20, which are much less potent or totally inactive in the HeLa MTT assay, do not induce apoptosis in Jurkat cells at this concentration.
Figure 5.
Induction of apoptosis in Jurkat cells treated for 36 h with DMSO control, colchicine (5 μM) and selected pyridone library analogues (5 μM) in the flow cytometric annexin-V/propidium iodide assay. Error bars represent data from four replicates of a single experiment repeated twice with similar results.
Encouraged by the promising results obtained with pyrano-[3,2-c]-pyridone library of compounds, we investigated the corresponding pyrano-[3,2-c]-quinolones. The synthesis of analogues 21–45, utilizing commercially available 4-hydroxy-1-methylquinolin-2(1H)-one in lieu of pyridone 7, turned out to be equally facile and the yields of the library members are given in Table 2.
Table 2.
Synthetic yields and antiproliferative activity of pyrano-[3,2-c]-quinolones.
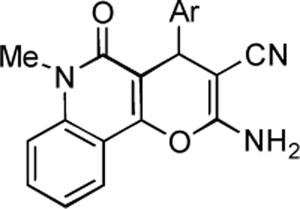 |
synthesis | cell viabilitya GI50, μM | ||
|---|---|---|---|---|
| analogue | Ar | % yield | HeLa | MCF-7 |
| 21 |
|
78 | 0.39 ± 0.01 | 3.38 ± 0.53 |
| 22 |
|
86 | 0.24 ± 0.02 | 1.0 ± 0.3 |
| 23 |
|
82 | > 50 | > 50 |
| 24 |
|
84 | 0.63 ± 0.05 | 0.5 ± 0.14 |
| 25 |
|
89 | 0.63 ± 0.02 | 0.71 ± 0.12 |
| 26 |
|
77 | 0.32 ± 0.02 | 1.1 ± 0.1 |
| 27 |
|
73 | 5.0 ± 1.4 | 3.5 ± 0.3 |
| 28 |
|
79 | > 50 | 32 ± 2 |
| 29 |
|
81 | 2.0 ± 0.1 | > 50 |
| 30 |
|
75 | 22 ± 1 | 35 ± 2 |
| 31 |
|
78 | 3.9 ± 0.2 | 5.1 ± 0.1 |
| 32 |
|
95 | 1.8 ± 0.2 | > 50 |
| 33 |
|
88 | > 50 | 36 ± 2 |
| 34 |
|
97 | 0.3 ± 0.03 | 2.5 ± 0.3 |
| 35 |
|
91 | 0.15 ± 0.04 | 2.2 ± 0.2 |
| 36 |
|
90 | 0.30 ± 0.01 | 1.0 ± 0.3 |
| 37 |
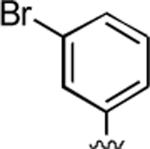
|
93 | 0.74 ± 0.03 | 0.003 ± 0.001 |
| 38 |
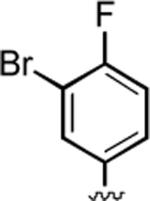
|
95 | 0.27 ± 0.03 | 0.81 ± 0.08 |
| 39 |
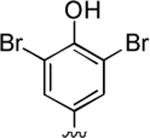
|
92 | 0.27 ± 0.02 | 0.43 ± 0.01 |
| 40 |
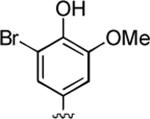
|
94 | 0.047 ± 0.010 | 0.39 ± 0.16 |
| 41 |
|
95 | 0.014 ± 0.003 | 0.38 ± 0.03 |
| 42 |
|
82 | 0.077 ± 0.006 | 0.075 ± 0.007 |
| 43 |
|
64 | 0.41 ± 0.04 | 0.5 ± 0.1 |
| 44 |
|
85 | 0.013 ± 0.003 | 0.015 ± 0.008 |
| 45 |
|
84 | 0.18 ± 0.02 | 0.025 ± 0.06 |
| 5 | NA | NA | 0.017 ± 0.004 | 0.50 ± 0.05 |
| 6 | NA | NA | 0.007 ± 0.002 | 0.50 ± 0.14 |
Concentration required to reduce the viability of cells by 50% after 48 h of treatment with indicated compounds relative to DMSO control ± SD from two independent experiments, each performed in 8 replicates, determined by MTT assay.
Analogues 21–45 as well as chromenes 5 and 6 were evaluated for antiproliferative activity in HeLa and MCF-7 (model for breast adenocarcinoma) cell lines. The general SAR trends are very similar to the corresponding pyridone-based library and, with a few exceptions, HeLas are more responsive than MCF-7 cells to both pyranoquinolones and chromenes. Most pleasingly, however, changing the pyranopyridone scaffold to the corresponding pyranoquinolone led to low nanomolar potencies exhibited by a number of library members containing the crucial 3-bromo substituent. Furthermore, the rest of the substitution pattern of this aromatic ring has a profound influence on antiproliferative potency, including the complete reversal of the strength of the effect on the two cell lines. For example, 42 and 44 exhibit low nanomolar potencies in both cell lines, while pairs 40, 41 and 37, 45 display respective low nanomolar GI50 values only with HeLa or MCF-7 cells, but not both. Synthesis of a larger number of analogues and their evaluation in a broader panel of cell lines will be required to understand these intriguing observations, which may pave the way to compounds with highly selective toxicity profiles.
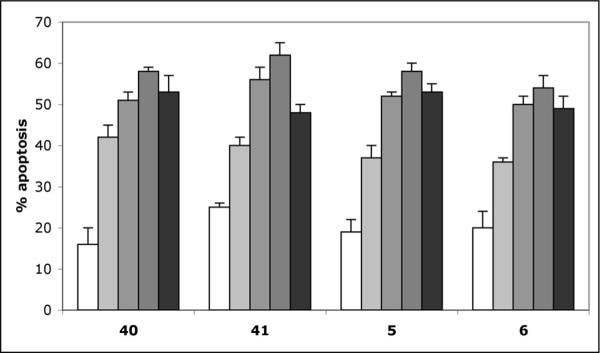
The induction of apoptosis by the potent library members was investigated using the Jurkat cell line in both time-dependent and dose-dependent manner. To this end, Jurkat cells were treated with analogues 40 and 41, as well as chromenes 5 and 6, and the proportion of cells displaying the apoptotic phenotype was determined with the flow cytometric annexin-V/propidium iodide assay every 12 h (Figure 6). For all four compounds we observed a similar, time-dependent increase in the proportion of cells undergoing apoptosis with the maximum occurring after 48 h of treatment.
Figure 6.
Induction of apoptosis in Jurkat cells treated with 40, 41, 5 and 6 (all used at 1 μM) for 12 h (open columns), 24 h (light grey columns), 36 h (medium grey columns), 48 h (dark grey columns), 60 h (black columns), determined using the flow cytometric annexin-V/propidium iodide assay. Error bars represent data from four replicates of a single experiment repeated twice with similar results.
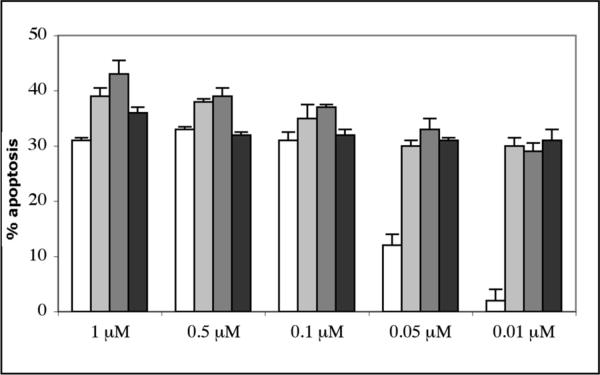
The time period of 48 h was then selected for the dose-dependent experiments to compare the apoptosis inducing power of the pyranoquinolone library members with chromenes 5 and 6 (Figure 7). While compound 40 is a strong apoptosis inducer at 100 nM, its potency drops as the concentration is reduced to 10 nM. In contrast, similarly to chromenes 5 and 6, library member 41 retains its potency at this low concentration.
Figure 7.
Dose-dependence of apoptosis induction in Jurkat cells treated for 48 h with 40 (open columns), 41 (light grey columns), 5 (dark grey columns) and 6 (black columns), determined using the flow cytometric annexin-V/propidium iodide assay. Error bars represent data from four replicates of a single experiment repeated twice with similar results.
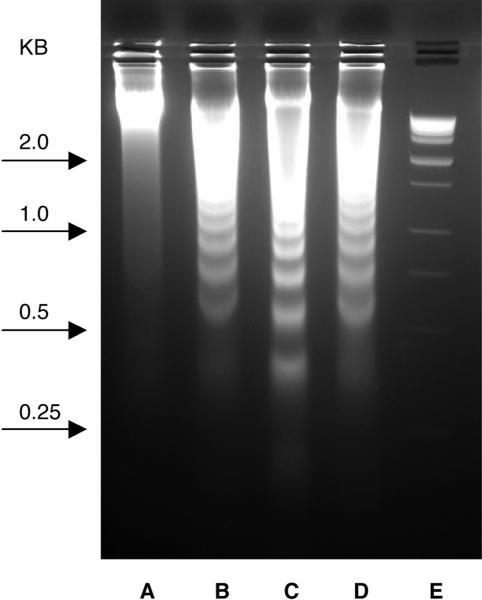
Since endonuclease-mediated cleavage of nuclear DNA resulting in formation of oligonucleosomal DNA fragments (180–200 base pairs long) is a hallmark of apoptosis in many cell types, apoptosis was further investigated with the DNA laddering assay (Figure 8). Jurkat cells were treated with DMSO (lane A), analogues 40 and 41 (lanes B and C), and paclitaxel (lane D) for 36 h. After that the cellular DNA was isolated and electrophoresed in a 1.5% agarose gel. The characteristic ladder pattern was obtained with compounds 40, 41 and paclitaxel. No laddering was observed when the cells were treated with DMSO control.
Figure 8.
DNA laddering in Jurkat cells after 36 h of treatment. (A) DMSO control, (B) 40 at 1.5 μM, (C) 41 at 1.5 μM, (D) paclitaxel at 0.4 μM, and (E) Molecular weight marker (KB).
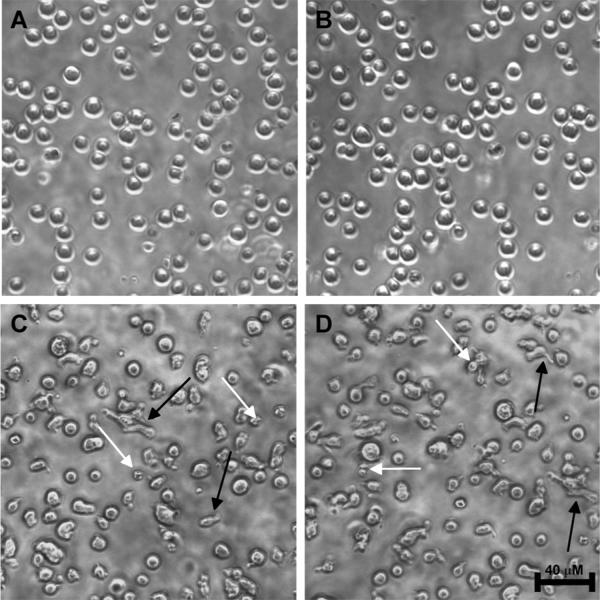
Morphological changes of cells treated with potent pyrano-[3,2-c]-quinolones can be visually observed with light microscopy (Figure 9). The phenotypic changes of Jurkat cells, such as formation of finger-like extensions and shriveling, become apparent as early as 2 h after their treatment with 40 (C), and 41 (D).
Figure 9.
Light microscopy pictures. Treatment of Jurkat cells with 40 (C) and 41 (D) for 2 hours induces formation of finger-like extensions (black arrows) and shriveling (white arrows). 0.1% DMSO (A) and 6 (B) were used as controls. All compounds are used at 1 μM. The scale bar indicates 40 μm.
Because of the structural similarity between the pyranopyridone and chromene scaffolds, we suspected that our libraries of compounds exert their antiproliferative properties through inhibition of tubulin dynamics, thereby inducing mitotic arrest and initiating apoptosis in cancer cells. Indeed, the flow cytometric cell cycle analysis, performed with pyranoquinolones 40 and 41 using the Jurkat cell line, shows a pronounced cell cycle arrest in the G2/M phase (Table 3). This effect is characteristic of antimitotic agents disrupting microtubule assembly, and is also observed with chromenes 5 and 6 that bind to or near the colchicine binding site on β-tubulin.25d
Table 3.
Flow cytometric cell cycle analysis of Jurkat cells.
| compound | % relative DNA contenta |
||
|---|---|---|---|
| G0/G1 | S | G2/M | |
| DMSO | 56 ± 2 | 21 ± 3 | 20 ± 2 |
| 40 | 27 ± 3 | 22 ± 2 | 47 ± 3 |
| 41 | 20 ± 2 | 28 ± 2 | 49 ± 2 |
| 5 | 19 ± 1 | 37 ± 3 | 42 ± 1 |
| 6 | 19 ± 2 | 36 ± 2 | 41 ± 3 |
% Relative DNA content ± SD after 24 h treatment of Jurkat cells with indicated compounds from two independent experiments each performed in triplicate. Compounds 40, 41, 5 and 6 are used at 1 μM, obtained using the flow cytometric Vybrant Orange staining assay. The remaining % DNA content is found in sub G0/G1 region.
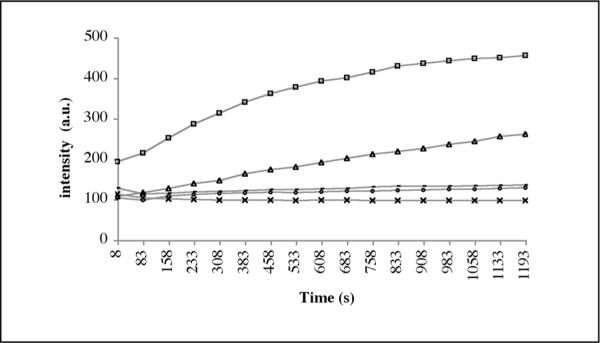
To obtain further support for the proposed antitubulin mechanism of action for our heterocycles, we assessed the effect of pyranoquinolones on in vitro tubulin polymerization.32 In this assay microtubule formation is monitored by the increase in fluorescence intensity of the reaction mixture. Paclitaxel exhibited potent enhancement of microtubule formation relative to the effect of DMSO control (Figure 10). In contrast, library members 40 and 41 displayed potent microtubule destabilizing effect in a manner similar to the known tubulin polymerization inhibitor podophyllotoxin.
Figure 10.
Effect of pyranoquinolones on in vitro tubulin polymerization. Paclitaxel (3 μM, square markers) promotes microtubule formation relative to 0.05% DMSO control (triangle markers). 40 (25 μM, dash markers), 41 (25 μM, circle markers) and podophyllotoxin (25 μM, cross markers) completely suppress tubulin polymerization. Each data point is a mean of two independent experiments producing similar results.
Conclusions
The utilization of multicomponent synthesis led to the development of compound libraries based on pyrano[3,2-c]pyridone and pyrano[3,2-c]quinolone scaffolds, which are commonly found in structurally complex alkaloids manifesting diverse biological activities. Many of the synthesized analogues exhibit low nanomolar (down to 3 nM) antiproliferative properties in HeLa and MCF-7 human cancer cell lines. Furthermore, the antiproliferative effect results from the potent apoptosis inducing ability of these heterocycles, as confirmed by the flow cytometric annexin-V staining and DNA laddering assays. The potent apoptosis inducing power of a selected library member is pronounced at concentrations as low as 10 nM. The structural similarity of these heterocycles with compounds based on a related chromene scaffold prompted investigations of a possible mode of action based on inhibition of tubulin dynamics and cell cycle arrest in the G2/M phase. Indeed, the flow cytometric detection of large populations of cells with the 4N DNA content and potent inhibition of the in vitro tubulin polymerization by library analogues in a manner similar to the well-established antitubulin agents, such as colchicine and podophyllotoxin, support this hypothesis. Because the antitubulin agents are some of the most effective drugs in cancer chemotherapy33 and many of them are now investigated as possible inhibitors of angiogenesis in cancer tissues,34 we believe that further investigation of compound libraries based on pyrano[3,2-c]pyridone and pyrano[3,2-c]quinolone scaffolds will result in important new leads in anticancer drug design.
Experimental Section
All aldehydes, malononitrile, and 4-hydroxy-1-methylquinolin-2(1H)-one were purchased from commercial sources and used without purification. 4-Hydroxy-1,6-dimethylpyridin-2(1H)-one was prepared following a literature procedure.27 Triethylamine (Et3N) was distilled from CaH2. All reactions were performed in a reaction vessel open to the atmosphere and monitored by thin layer chromatography (TLC) on pre-coated (250 μm) silica gel 60F254 glass-backed plates. Visualization was accomplished with UV light and aqueous ceric ammonium molybdate solution or potassium permanganate stain followed by charring on a hot-plate. 1H and 13C NMR spectra were recorded on JEOL 300 MHz spectrometer. MS analyses were performed at the Mass Spectrometry Facility, University of New Mexico. All compounds 8–45 decompose at temperatures exceeding 260 °C without melting.
General procedure for the synthesis of pyrano-[3,2-c]-pyridones 8–16 and pyrano-[3,2-c]-quinolones 21–45
A mixture of 4-hydroxy-1,6-dimethylpyridin-2(1H)-one (0.8 mmol) or 4-hydroxy-1-methylquinolin-2(1H)-one, malononitrile (0.8 mmol), triethylamine (0.05 mL) and a corresponding aldehyde (0.8 mmol) in EtOH (96% aqueous solution, 3 mL) was refluxed for 50 minutes. The reaction mixture was allowed to cool to room temperature, the precipitated product was collected by filtration and washed with EtOH (5 mL). In most cases the product was > 98% pure as judged by 1H NMR analysis. When necessary the products were recrystallized from DMF.
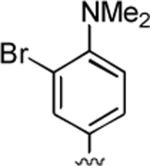
2-Amino-4-[3-bromo-4-(dimethylamino)phenyl]-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (8)
83%; 1H NMR (DMSO-d6) δ 7.27 – 7.08 (m, 5H), 6.06 (s, 1H), 4.28 (s, 1H), 3.33 (s, 3H), 2.66 (s, 6H), 2.33 (s, 3H); 13C NMR (DMSO-d6) δ 161.7, 159.8, 155.4, 153.7, 148.8, 145.0, 143.1, 122.9, 112.7, 105.5, 97.4, 60.5, 56.6, 39.3, 37.2, 31.2, 20.8; HRMS m/z (ESI) calcd for C19H19BrN4O2 (M+Na+) 437.0589, found 437.0580.
2-Amino-4-(3-bromo-4,5-dimethoxyphenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (9)
87%; 1H NMR (DMSO-d6) δ 6.93 (d, J = 6.9 Hz, 2H), 6.87–6.85 (m, 2H), 6.03 (s, 1H), 4.37 (s, 1H), 3.80 (s, 3H), 3.73 (s, 3H), 3.35 (s, 3H), 2.35 (s, 3H); 13C NMR (DMSO-d6) δ 161.7, 159.8, 155.4, 153.6, 148.5, 145.2, 142.7, 123.2, 120.1, 116.8, 113.1, 105.6, 97.4, 60.6, 58.2, 56.8, 37.3, 31.2, 20.6; HRMS m/z (ESI) calcd for C19H19BrN3O4 (M+Na+) 454.0378, found 454.0371.
2-Amino-4-(3-bromo-4-ethoxy-5-methoxyphenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (10)
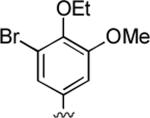
88%; 1H NMR (DMSO-d6) δ 7.09 (s, 2H), 6.90 (s, 1H), 6.79 (s, 1H), 6.07 (d, J = 2.7 Hz, 1H), 4.33 (d, J = 3.0 Hz, 1H), 3.92 (q, J = 3.0 Hz, 2H), 3.76 (s, 3H), 3.33 (s, 3H), 2.33 (s, 3H), 1.27 (t, J = 3.0 Hz. 3H); 13C NMR (DMSO-d6) δ 161.6, 159.8, 155.4, 153.8, 148.6, 144.3, 142.7, 122.9, 117.5, 112.6, 105.5, 97.42, 68.9, 57.8, 56.6, 37.1, 31.1, 20.8, 16.1; HRMS m/z (ESI) calcd for C20H20BrN3OO4 (M+Na+) 468.0535, found 468.0540.
2-Amino-4-(3-bromo-4-hydroxy-5-methoxyphenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (11)
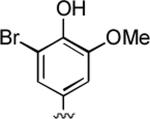
75%; 1H NMR (DMSO-d6) δ 9.32 (s, 1H), 7.04 (s, 2H), 6.82 – 6.71 (m, 2H), 6.06 (s, 1H), 4.28 (s, 1H), 3.77 (s, 3H), 3.32 (s, 3H), 2.34 (s, 3H); 13C NMR (DMSO-d6) δ 161.6, 159.6, 155.1, 148.5, 148.4, 143.0, 137.5, 123.1, 111.5, 109.6, 105.9, 97.4, 58.0, 56.7, 37.0, 31.1, 20.8; HRMS m/z (ESI) calcd for C18H16BrN3O4 (M+Na+) 440.0222, found 440.0223.
4-(2-Amino-3-cyano-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridin-4-yl)-2-bromo-6-methoxyphenyl acetate (12)
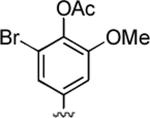
83%; 1H NMR (DMSO-d6) δ 7.02 (s, 1H), 6.97 (s, 2H), 6.92 (s, 1H) 6.05 (s, 1H), 4.44 (s, 1H), 3.77 (s, 3H), 3.34 (s, 3H), 2.34 (s, 3H), 2.30 (s, 3H); 13C NMR (DMSO-d6) δ 168.2, 161.9, 159.9, 155.6, 152.5, 148.9, 145.4, 136.5.0, 122.9, 120.2, 116.6, 112.5, 105.4, 97.4, 57.6, 56.7, 37.2, 31.1, 20.5; HRMS m/z (ESI) calcd for C20H18BrN3O5 (M+Na+) 482.0328, found 482.0322.
2-Amino-4-(3-bromo-4-fluorophenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (13)
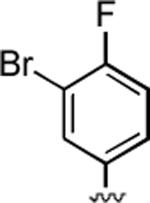
84%; 1H NMR (DMSO-d6) δ 7.42 (m, 1H), 7.29–7.15 (m, 2H), 7.07 (s, 2H) 6.05 (s, 1H), 4.37 (s, 1H), 2.33 (s, 3H), 2.30 (s, 3H); 13C NMR (DMSO-d6) δ 161.6, 159.8, 155.35, 148.7, 143.5, 132.9, 129.3, 120.2, 117.2, 116.8, 108.1, 105.3, 97.5, 57.9, 36.8, 30.9, 20.7; HRMS m/z (ESI) calcd for C17H13BrFN3O2 (M+Na+) 412.0073, found 412.0073.
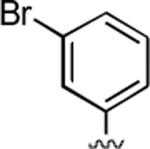
2-Amino-4-(3-bromophenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (14)
97%; 1H NMR (DMSO-d6) δ 7.39–7.11 (m, 6H), 6.07 (s, 1H), 4.35 (s, 1H), 3.30 (s, 3H), 2.34 (s, 3H); 13C NMR (DMSO-d6) δ 161.5, 159.6, 155.3, 148.7, 148.1, 131.0, 130.7, 129.9, 127.2, 121.9, 120.3, 105.3, 97.4, 57.6, 37.4, 31.1, 20.5; HRMS m/z (ESI) calcd for C17H14BrN3O2 (M+Na+) 394.0167, found 394.0159.
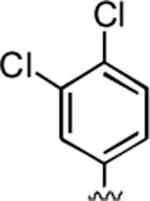
2-Amino-4-(3,4-dichlorophenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (15)
98%; 1H NMR (DMSO-d6) δ 7.54–7.49 (m, 1H), 7.38 (s, 1H), 7.18–7.16 (m, 1H), 6.83 (s, 2H), 6.03 (s, 1H), 4.43 (s, 1H) 3.35 (s, 3H), 2.35 (s, 3H); 13C NMR (DMSO-d6) δ 161.6, 159.8, 155.5, 148.7, 146.4, 131.3, 130.9, 130.0, 129.8, 128.3, 119.8, 105.1, 97.3, 58.0, 36.9, 31.1, 20.4; HRMS m/z (ESI) calcd for C17H13Cl2N3O2 (M+Na+) 384.0283, found 384.0282.
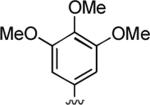
2-Amino-6,7-dimethyl-5-oxo-4-(3,4,5-trimethoxyphenyl)-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (16)
97%; 1H NMR (DMSO-d6) δ 7.01 (s, 2H), 6.42 (s, 2H), 6.06 (s, 1H), 4.35 (s, 1H), 3.69 (s, 6H), 3.60 (s, 3H), 3.32 (s, 3H), 2.34 (s, 3H); 13C NMR (DMSO-d6) δ 161.6, 159.8, 155.4, 153.3, 148.2, 140.9, 137.3, 120.2, 106.2, 105.6, 97.3, 60.6, 58.5, 56.4, 37.5, 31.0, 20.7; HRMS m/z (ESI) calcd for C20H21BrN3O5 (M+Na+) 406.1379, found 406.1379.
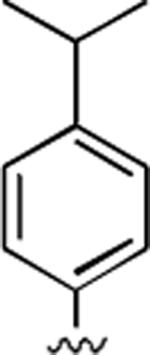
2-Amino-4-(4-isopropylphenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (17)
81%; 1H NMR (DMSO-d6) δ7.15–7.08 (m, 4H), 6.75 (s, 2H), 6.02 (s, 1H), 4.34 (s, 1H), 3.32 (s, 3H), 2.88–2.79 (m, 1H), 2.34 (s, 3H), 1.18 (d, J = 6.9 Hz, 6H); 13C NMR (DMSO-d6) δ 161.7, 159.8, 155.4, 148.0, 147.0, 142.7, 127.8, 126.5, 120.2, 106.6, 97.4, 59.3, 37.2, 33.5, 31.1, 24.3, 20.6; HRMS m/z (ESI) calcd for C20H21N3O2 (M+Na+) 358.1531, found 358.1519.
2-Amino-6,7-dimethyl-4-(3-nitrophenyl)-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (18)
97%; 1H NMR (DMSO-d6) δ8.04 (d, J = 7.7 Hz, 1H), 7.95 (s, 1H), 7.65–7.53 (m, 2H) 6.15 (s, 2H), 6.06 (s, 1H), 4.52 (s, 1H), 3.27 (s, 3H), 2.32 (s, 3H); 13C NMR (DMSO-d6) δ 161.8, 159.9, 155.6, 149.1, 148.4, 147.6, 134.9, 130.3, 122.6, 122.1, 120.1, 105.0, 97.5, 56.6, 37.4, 31.1, 20.5; HRMS m/z (ESI) calcd for C17H14N4O4 (M+Na+) 361.0913, found 361.0909.
2-Amino-4-(3-bromophenyl)-7-methyl-5-oxo-4H,5H-pyrano[4,3-b]pyran-3-carbonitrile (19)
87%; 1H NMR (DMSO-d6) δ7.43 (d, J = 8.0 Hz, 1H), 7.38 (s, 1H), 7.32–7.27 (m, 1H), 7.21 (d, J = 8.0 Hz, 1H), 7.15 (s, 2H), 6.26 (s, 1H), 4.34 (s, 1H), 2.22 (s, 3H); 13C NMR (DMSO-d6) δ 163.7, 161.9, 159.1, 158.8, 146.8, 131.3, 130.8, 130.5, 127.3, 122.1, 119.5, 100.6, 98.5, 58.1, 36.6, 19.7; HRMS m/z (ESI) calcd for C16H11BrN2O3 (M+Na+) 380.9845, found 380.9839.
2-Amino-6-(3,4-dimethoxyphenethyl)-7-methyl-5-oxo-4-(3,4,5-trimethoxyphenyl)-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carbonitrile (20)
80%; 1H NMR (DMSO-d6) δ 6.90 (s, 2H), 6.83 (d, J = 8.5 Hz, 1H), 6.69–6.63 (m, 2H), 6.52 (s, 2H),5.89 (s, 1H), 4.41 (s, 1H), 4.13–3.98 (m, 2H), 3.74 (s, 6H), 3.71 (s, 3H), 3.67 (s, 3H), 3.65 (s, 3H), 2.86–2.70 (m, 2H), 2.19 (s, 3H); 13C NMR (DMSO-d6) δ 161.6, 160.0, 155.6, 153.4, 149.5, 148.4, 147.6, 141.0, 137.4, 131.4, 121.3, 120.5, 113.6, 112.9, 106.6, 105.6, 97.6, 60.5, 58.4, 56.5, 56.3, 56.1, 40.1, 37.2, 33.3, 20.0; HRMS m/z (ESI) calcd for C29H31N3O7 (M+Na+) 556.2060, found 556.2057.
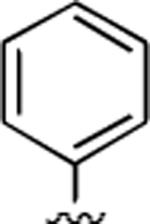
2-Amino-6-methyl-5-oxo-4-phenyl-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (21)
78%; 1H NMR (DMSO-d6) δ8.06 (d, J = 7.6 Hz, 1H), 7.70–7.66 (m, 1H), 7.52 (d, J = 7.6 Hz, 1H), 7.40–7.18 (m, 6H), 6.98 (s, 2H), 4.56 (s, 1H), 3.55 (s, 3H); 13C NMR (DMSO-d6) δ 160.5, 159.6, 150.9, 144.9, 139.4, 132.1, 128.8, 128.0, 127.2, 122.9, 122.8, 122.6, 119.9, 115.3, 113.4, 109.8, 59.2, 37.8, 29.8; HRMS m/z (ESI) calcd for C20H15N3O2 (M+Na+) 352.1062, found 352.1058.
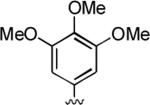
2-Amino-6-methyl-5-oxo-4-(3,4,5-trimethoxyphenyl)-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (22)
86%; 1H NMR (DMSO-d6) δ8.03 (d, J = 8.0 Hz, 1H), 7.72–7.66 (m, 1H), 7.55 (d, J = 7.6 Hz, 1H), 7.40–7.35 (m, 1H), 7.14 (s, 2H), 6.51 (s, 2H), 4.53 (s, 1H), 3.71 (s, 6H), 3.64 (s, 3H); 13C NMR (DMSO-d6) δ 160.6, 159.6, 153.4, 150.8, 140.4, 139.3, 137.5, 132.2, 122.9, 122.7, 120.1, 115.4, 113.3, 109.4, 105.9, 60.5, 58.6, 56.6, 38.2, 29.9; HRMS m/z (ESI) calcd for C23H21N3O5 (M+Na+) 442.1379, found 442.1389.
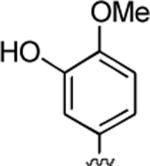
2-Amino-4-(3-hydroxy-4-methoxyphenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (23)
82%; 1H NMR (DMSO-d6) δ8.90 (s, 1H), 8.00 (d, J = 8.0 Hz, 1H), 7.72–7.67 (m, 1H), 7.55 (d, J = 7.6 Hz, 1H), 7.41–7.36 (m, 1H), 7.23 (s, 2H), 6.79 (d, J = 6.8 Hz, 1H), 6.61 (s, 2H), 4.37 (s, 1H), 3.71 (s, 3H), 3.55 (s, 3H); 13C NMR (DMSO-d6) δ 160.3, 159.3, 150.4, 147.1, 146.8, 139.1, 137.6, 132.0, 122.8, 120.4, 118.7, 115.5, 115.1, 113.2, 112.7, 110.0, 58.8, 56.4, 37.2, 29.8; HRMS m/z (ESI) calcd for C21H17N3O4 (M+Na+) 398.1117, found 398.1123.
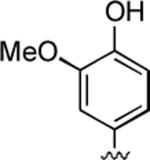
2-Amino-4-(4-hydroxy-3-methoxyphenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (24)
84%; 1H NMR (DMSO-d6) δ8.70 (s, 1H), 8.03 (d, J = 7.5 Hz, 1H), 7.70–7.65 (m, 1H), 7.53 (d, J = 7.6 Hz, 1H), 7.39–7.34 (m, 1H), 7.08 (s, 2H), 6.84 (s, 1H), 6.79 (d, J = 8.2 Hz, 1H), 6.60–6.57 (m, 1H), 4.47 (s, 1H), 3.73 (s, 3H), 3.55 (s, 3H); 13C NMR (DMSO-d6) δ 160.5, 159.5, 150.4, 147.7, 146.3, 139.3, 136.0, 132.3, 122.7, 120.4, 116.2, 115.2, 113.3, 113.1, 110.0, 59.0, 56.4, 37.1, 29.8; HRMS m/z (ESI) calcd for C21H17N3O4 (M+Na+) 398.1117, found 398.1119.
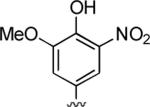
2-Amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (25)
89%; 1H NMR (DMSO-d6) δ10.20 (s, 1H), 8.03 (d, J = 8.0 Hz, 1H), 7.73–7.67 (m, 1H), 7.55 (d, J = 7.6 Hz, 1H), 7.41–7.36 (m, 1H), 7.23 (s, 4H), 4.61 (s, 1H), 3.85 (s, 3H), 3.54 (s, 3H); 13C NMR (DMSO-d6) δ 160.5, 159.7, 150.9, 149.9, 142.3, 139.3, 137.4, 135.8, 132.3, 122.7, 120.0, 117.2, 115.4, 114.8, 113.2, 108.7, 57.9, 57.4, 37.4, 29.9; HRMS m/z (ESI) calcd for C21H16N4O6 (M+Na+) 443.0968, found 443.0966.
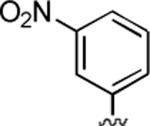
2-Amino-6-methyl-4-(3-nitrophenyl)-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (26)
77%; 1H NMR (DMSO-d6) δ8.11–8.04 (m, 3H), 7.75–7.54 (m, 4H), 7.41–7.36 (m, 1H), 7.00 (s, 2H), 4.78 (s, 1H), 3.56 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 160.0, 150.7, 149.9, 139.3, 134.0, 133.8, 132.4, 131.3, 128.4, 124.3, 122.9, 122.7, 119.4, 115.5, 113.1, 108.8, 56.9, 32.6, 29.8; HRMS m/z (ESI) calcd for C20H14N4O4 (M+Na+) 397.0913, found 397.0917.
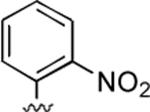
2-Amino-6-methyl-4-(2-nitrophenyl)-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (27)
73%; 1H NMR (DMSO-d6) δ8.06–8.03 (m, 1H), 7.86–7.84 (m, 1H), 7.73–7.66 (m, 1H), 7.61–7.51 (m, 2H), 7.45–7.36 (m, 3H), 7.29 (s, 2H), 5.34 (s, 1H), 3.48 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 160.1, 150.8, 150.0, 139.4, 134.0, 133.8, 132.4, 131.3, 128.5, 124.4, 122.8, 119.5, 115.3, 113.2, 108.8, 57.0, 32.6, 29.8; HRMS m/z (ESI) calcd for C20H14N4O4 (M+Na+) 397.0913, found 397.0910.
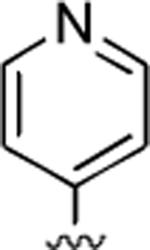
2-Amino-6-methyl-5-oxo-4-(4-pyridinyl)-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (28)
79%; 1H NMR (DMSO-d6) δ8.45 (d, J = 8.5 Hz, 2H), 8.03 (d, J = 8.0 Hz, 1H), 7.73–7.67 (m, 1H), 7.54 (d, J = 7.5 Hz, 1H), 7.41–7.22 (m, 5H), 4.55 (s, 1H), 3.52 (s, 3H); 13C NMR (DMSO-d6) δ 160.3, 159.7, 153.1, 151.2, 150.4, 150.2, 139.3, 132.3, 123.4, 122.9, 119.8, 115.3, 113.1, 107.9, 57.0, 37.6, 29.8; HRMS m/z (ESI) calcd for C19H14N4O2 (M+Na+) 353.1014, found 353.1017.
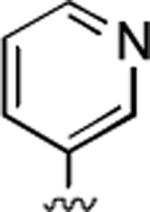
2-Amino-6-methyl-5-oxo-4-(3-pyridinyl)-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (29)
81%; 1H NMR (DMSO-d6) δ8.51 (d, J = 3.6 Hz, 1H), 8.40 (m, 1H) 8.05 (m, 1H), 7.72–7.52 (m, 3H), 7.38–7.27 (m, 2H), 7.10 (s, 2H), 4.62 (s, 1H), 3.53 (s, 3H); 13C NMR (DMSO-d6) δ 160.5, 159.8, 151.2, 149.8, 148.6, 140.1, 139.5, 135.5, 132.2, 124.4, 123.0, 122.6, 119.7, 115.3, 113.3, 108.7, 58.2, 35.7, 29.7; HRMS m/z (ESI) calcd for C19H14N4O2 (M+Na+) 353.1014, found 453.1018.
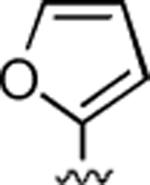
2-Amino-4-(2-furyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (30)
75%; 1H NMR (DMSO-d6) δ8.04 (d, J = 8.0 Hz, 1H), 7.73–7.67 (m, 1H), 7.56 (d, J = 7.6 Hz, 1H), 7.41–7.34 (m, 2H), 6.93 (s, 2H), 6.32 (s, 1H), 6.14 (d, J = 6.1 Hz, 1H), 4.72 (s, 1H), 3.60 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 155.9, 151.5, 142.3, 142.1, 139.4, 132.1, 122.9, 122.5, 119.5, 115.2, 113.4, 110.9, 107.2, 106.2, 56.7, 31.8, 29.9; HRMS m/z (ESI) calcd for C18H13N3O3 (M+Na+) 342.0855, found 342.0851.
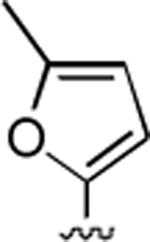
2-Amino-6-methyl-4-(5-methyl-2-furyl)-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (31)
78%; 1H NMR (DMSO-d6) δ8.03 (d, J = 8.0 Hz, 1H), 7.71–7.57 (m, 2H), 7.40–7.35 (m, 1H), 7.01 (s, 2H), 5.99–5.91 (m, 2H), 4.64 (s, 1H), 3.60 (s, 3H), 2.17 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.7, 151.5, 151.5, 145.0, 139.5, 132.5, 131.2, 129.3, 128.8, 123.1, 122.8, 119.6, 115.4, 113.1, 108.3, 56.9, 36.4, 29.9; HRMS m/z (ESI) calcd for C19H15N3O3 (M+Na+) 356.1011, found 356.1022.
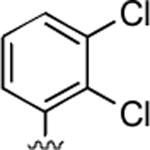
2-Amino-4-(2,3-dichlorophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (32)
95%; 1H NMR (DMSO-d6) δ8.05 (d, J = 8.0 Hz, 1H), 7.75–7.70 (m, 1H), 7.55 (d, J = 8.8 Hz, 1H), 7.48–7.38 (m, 2H), 7.27–7.14 (m, 4H), 5.15 (s, 1H), 3.52 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.6, 151.5, 145.0, 139.5, 132.5, 131.2, 129.3, 128.8, 123.1, 122.8, 122.7, 119.6, 115.6, 115.4, 113.1, 108.3, 56.9, 36.4, 29.9; HRMS m/z (ESI) calcd for C20H13Cl2N3O2 (M+Na+) 420.0283, found 420.0284.
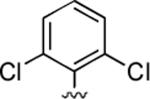
2-Amino-4-(2,6-dichlorophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (33)
88%; 1H NMR (DMSO-d6) δ8.05 (d, J = 8.0 Hz, 1H), 7.72–7.67 (m, 1H), 7.55–7.21 (m, 5H), 6.91 (bs, 2H), 5.62 (s, 1H), 3.52 (s, 3H); 13C NMR (DMSO-d6) δ 160.1, 139.6, 137.0, 132.4, 132.2, 129.6, 129.4, 122.8, 122.6, 122.4, 119.1, 115.1, 113.1, 106.7, 55.2, 34.6, 29.5; HRMS m/z (ESI) calcd for C20H13Cl2N3O2 (M+Na+) 420.0283, found 420.0280.
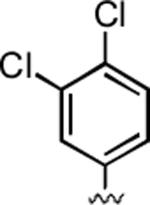
2-Amino-4-(3,4-dichlorophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (34)
97%; 1H NMR (DMSO-d6) δ8.03 (d, J = 8.0 Hz, 1H), 7.72–7.67 (m, 1H), 7.55–7.19 (m, 7H), 4.58 (s, 1H), 3.52 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.5, 150.9, 145.9, 139.4, 132.5, 131.4, 131.1, 130.2, 129.8, 128.5, 123.0, 122.7, 119.7, 115.5, 113.2, 108.4, 57.7, 37.3, 29.8; HRMS m/z (ESI) calcd for C20H13Cl2N3O2 (M+Na+) 420.0283, found 420.0274.
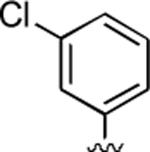
2-Amino-4-(3-chlorophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (35)
91%; 1H NMR (DMSO-d6) δ8.02 (d, J = 8.0 Hz, 1H), 7.73–7.68 (m, 1H), 7.58–7.16 (m, 8H), 4.55 (s, 1H), 3.53 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.5, 150.8, 147.4, 139.3, 133.4, 131.0, 128.0, 126.8, 122.8, 122.6, 120.1, 113.1, 108.7, 57.8, 37.3, 29.8; HRMS m/z (ESI) calcd for C20H14ClN3O2 (M+Na+) 386.0673, found 386.0677.
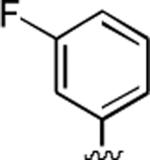
2-Amino-4-(3-fluorophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (36)
90%; 1H NMR (DMSO-d6) δ 8.05–8.02 (m, 1H), 7.72–7.67 (m, 1H), 7.54 (d, J = 7.5 Hz, 1H), 7.40–7.00 (m, 7H), 4.58 (s, 1H), 3.54 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.6, 150.9, 147.8, 139.4, 132.2, 130.8, 124.0, 122.7, 119.9, 115.5, 114.9, 114.6, 114.2, 113.9, 133.2, 108.9, 58.1, 37.7, 29.9; HRMS m/z (ESI) calcd for C20H14FN3O2 (M+Na+) 307.0968, found 370.0963.
2-Amino-4-(3-bromophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (37): 93%
1H NMR (DMSO-d6) δ 8.07 (d, J = 8.0 Hz, 1H), 7.73–7.68 (m, 1H), 7.57–7.24 (m, 7H), 6.98 (s,1H), 4.59 (s, 1H), 3.57 (s, 3H); 13C NMR (DMSO-d6) δ 160.5, 159.0, 151.8, 147.9, 139.3, 133.8, 131.0, 128.4, 122.8, 120.0, 113.0, 108.7, 105.4, 57.87 37.5, 29.7; HRMS m/z (ESI) calcd for C20H14BrN3O2 (M+Na+) 430.0167, found 430.0160.
2-Amino-4-(3-bromo-4-fluorophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (38)
95%; 1H NMR (DMSO-d6) δ 8.03 (d, J = 8.0 Hz, 1H), 7.72–7.23 (m, 8H), 4.59 (s, 1H), 3.54 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.5, 150.9, 143.0, 139.3, 133.0, 132.8, 132.3, 129.5, 123.0, 122.8, 119.9, 117.2, 115.5, 113.2, 108.6, 58.1, 36.7, 29.7; HRMS m/z (ESI) calcd for C20H13BrFN3O2 (M+Na+)448.0073, found 448.0056.
2-Amino-4-(3,5-dibromo-4-hydroxyphenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (39)
92%; 1H NMR (DMSO-d6) δ 8.03 (d, J = 8.0 Hz, 1H), 7.72–7.23 (m, 8H), 4.50 (s, 1H), 3.55 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.5, 150.9, 150.2, 139.5, 139.3, 132.2, 131.8, 123.0, 122.8, 119.9, 115.6, 113.4, 112.3, 108.7, 58.0, 37.5, 29.9; HRMS m/z (ESI) calcd for C20H14Br2N3O3 (M+Na+) 523.9221, found 523.9210.
2-Amino-4-(3-bromo-4-hydroxy-5-methoxyphenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (40)
94%; 1H NMR (DMSO-d6) δ 9.16 (s, 1H), 8.02 (d, J = 8.0 Hz, 1H), 7.71–7.53 (m, 2H), 7.39–7.34 (m, 1H), 7.15 (s, 2H), 6.86 (s, 1H), 6.82 (s, 1H), 4.47 (s, 1H), 3.77 (s, 3H), 3.54 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.5, 150.6, 148.8, 143.3, 139.2, 136.9, 132.2, 123.5, 123.0, 122.7, 120.1, 115.5, 113.3, 111.8, 109.8, 109.3, 58.5, 57.0, 37.2, 29.9; HRMS m/z (ESI) calcd for C21H16BrN3O4 (M+Na+) 476.0222, found 476.0226.
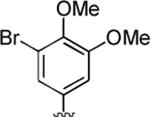
2-Amino-4-(3-bromo-4,5-dimethoxyphenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (41)
95%; 1H NMR (DMSO-d6) δ 8.03 (d, J = 8.0 Hz, 1H), 7.72–7.67 (m, 1H), 7.55 (d, J = 8.0 Hz, 1H), 7.40–7.35 (m, 1H), 7.21 (s, 2H), 6.98 (d, J = 1.7 Hz, 1H), 6.92 (d, J = 1.7 Hz, 1H) 4.56 (s, 1H), 3.79 (s, 3H), 3.71 (s, 3H), 3.56 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.6, 153.8, 150.9, 145.3, 142.2, 139.3, 132.3, 123.3, 123.0, 122.5, 120.1, 117.0, 115.5, 113.3, 113.0, 108.9, 60.5, 58.2, 56.5, 37.4, 29.8; HRMS m/z (ESI) calcd for C22H18BrN3O4 (M+Na+) 490.0379, found 490.0371.
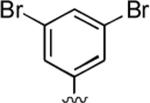
2-Amino-4-(3,5-dibromophenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (42)
82%; 1H NMR (DMSO-d6) δ 8.05 (d, J = 8.3 Hz, 1H), 7.65–7.57 (m, 2H), 7.48–7.38 (m, 4H), 7.29 (s, 2H), 4.60 (s, 1H), 3.56 (s, 3H); 13C NMR (DMSO-d6) δ 160.4, 159.7, 151.2, 149.6, 132.6, 130.1, 122.8, 115.5, 113.2, 107.8, 57.5, 37.6, 29.7; HRMS m/z (ESI) calcd for C20H13Br2N3O2 (M+Na+) 507.9272, found 507.9257.
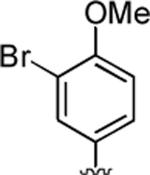
2-Amino-4-(3-bromo-4-methoxyphenyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (43)
64%; 1H NMR (DMSO-d6) δ 8.02 (d, J = 8.0 Hz, 1H), 7.72–7.67 (m, 1H), 7.57–7.54 (d, J = 7.5 Hz, 1H), 7.38 (s, 2H), 7.22 (s, 3H), 7.01 (d, J = 7.0 Hz, 1H), 4.05 (s, 1H), 3.79 (s, 3H), 3.54 (s, 3H); 13C NMR (DMSO-d6) δ 160.3, 159.3, 154.7, 150.5, 139.1, 138.6, 132.4, 128.7, 122.8, 122.6, 120.2, 115.6, 113.1, 110.7, 109.1, 58.1, 56.8, 37.0, 29.8; HRMS m/z (ESI) calcd for C21H16BrN3O3 (M+Na+) 460.0273, found 460.0287.
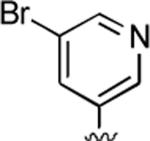
2-Amino-4-(5-bromo-3-pyridinyl)-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile (44)
85%; 1H NMR (DMSO-d6) δ 8.53 (d, J = 2.2 Hz, 1H), 8.50 (d, J = 1.6 Hz, 1H), 8.07 (d, J = 8.0 Hz, 1H), 7.84 (m, 1H), 7.74–7.69 (m, 1H), 7.56 (d, J = 7.6 Hz, 1H), 7.41–7.36 (m, 1H), 7.05 (s, 2H), 4.68 (s, 1H), 3.57 (s, 3H); 13C NMR (DMSO-d6) δ 160.7, 159.8, 151.3, 149.2, 145.8, 142.4, 139.7, 132.4, 122.8, 115.5, 108.0, 58.2, 36.0, 29.8; HRMS m/z (ESI) calcd for C19H13BrN4O2 (M+Na+) 431.0120, found 431.0139.
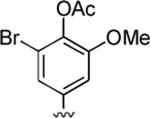
4-(2-Amino-3-cyano-6-methyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinolin-4-yl)-2-bromo-6-methoxyphenyl acetate (45)
84%; 1H NMR (DMSO-d6) δ 8.04 (d, J = 8.0 Hz, 1H), 7.74–7.69 (m, 1H), 7.57 (d, J = 7.6 Hz, 1H), 7.42–7.37 (m, 1H), 7.25 (s, 2H), 7.03 (s, 1H), 7.00 (s, 1H), 4.61 (s, 1H), 3.76 (s, 3H), 3.57 (s, 3H), 2.29 (s, 3H); 13C NMR (DMSO-d6) δ 168.3, 160.5,159.4, 153.2, 149.6, 142.3, 139.5, 132.8, 122.5, 122.0, 121.7, 120.4, 115.5, 108.1, 57.6, 36.8, 29.6, 20.4; HRMS m/z (ESI) calcd for C23H18BrN3O5 (M+Na+) 518.0328, found 518.0334.
Cell Culture
Human T-cell leukemia cell line Jurkat (ATCC TIB-152, E6-1 clone) and human cervical cancer cell line HeLa (ATCC S3) were cultured in RPMI-1640 (Invitrogen) and supplemented with 10% FBS (Invitrogen), 100 mg/L penicillin G, 100 mg/L streptomycin, 1.0 mM sodium pyruvate, 1.5 g/L sodium bicarbonate, and 4.5 g/L glucose (all from Sigma) at 37 °C in a humidified atmosphere with 10% CO2. MCF-7 (human mammary carcinoma) were cultured in a mixture of DMEM and HAMF12 (50/50) (Invitrogen) supplemented with 250 IU/mL penicillin, 100 μg/mL streptomycin and 10% FBS.
Annexin V Apoptosis Assay
2 × 105 Jurkat cells/mL were plated in 24 well plates, treated with the required compounds and incubated for the necessary number of hours. The cells were centrifuged at 400 G for 1 min. The supernatant was discarded and the cells were resuspended in 100 μL per sample of Annexin-V-FITC/ propidium iodide solution in HHB (3 μL CaCl2 (1.5 M) per mL HHB, 2 μL (10 mg/mL) propidium iodide (Sigma) per mL HHB and 20 μL Annexin-V-FITC (Southern Biotech, Birmingham, AL) per mL HHB). The samples in the labeling solution were transferred into Falcon tubes and incubated in a water bath at 37 °C for 20 min. The samples were then analyzed using a Becton Dickinson FACscan flow cytometer with CellQuest software. The results were tabulated as % of Annexin-V-FITC positive apoptotic cells.
DNA Laddering
Approximately 1 × 106 Jurkat cells were treated with the required compounds for 36 h. Cells were collected and centrifuged at 400 G for 1 min. The supernatant was discarded and the cells were gently resuspended in 20 μL of lysis buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, 0.5% SDS and 0.5 mg/mL proteinase K (Sigma)) on ice. The suspension of lysed cells was first incubated at 55 °C for 60 min and then 5 μL of RNAse (Sigma) was added and incubated at 55 °C for another 60 min. The debris was collected by centrifugation and the supernatant collected. The lysate was heated to 70 °C and loading buffer (20 μL TE buffer (10 mM Tris-HCl, 1 mM EDTA) and 10 μL FOG) was added. The samples were loaded on a 1.5% agarose gel containing 0.5 μg/mL ethidium bromide and run at 80V for approximately 1 h and visualized under UV light.
MTT Assay
HeLa and MCF-7 cells were transferred to microtiter plates in 100 μL of medium at a concentration of 2 × 104 cells/mL and then incubated for 24 h before treating with the required compounds to allow proper adhesion. Cells were further treated with the panel of test compounds and incubated for 48 h. 20 μL of MTT reagent (5 mg/mL) was added to each well and incubated for 2 h. The resulting formazan crystals were dissolved in 100 μL of DMSO and the OD was determined at a wavelength of 490 nm. The experiments were repeated at least three times for each compound per cell line.
Cell Cycle Analysis
Approximately 2 × 105 Jurkat cells/mL were treated with the respective compounds and incubated for 15 h. Cells were collected into microcentrifuge tubes and centrifuged at 400 G for 1 min. The supernantant was discarded and the cells were resuspended in 200 μL of HHB, Vybrant DyeCycle Orange stain (Invitrogen) solution (1 μL dye per 1 mL HHB). Samples were then transferred to Falcon tubes and incubated at 37 °C for 30 minutes while being protected from light. Samples were analyzed using a Beckon Dickinson FACscan flow cytometer with CellQuest software.
In Vitro Tubulin Polymerization Assay
The in vitro tubulin polymerization assay was conducted as described by the manufacturer (Cytoskeleton Inc.). In brief, paclitaxel, DMSO, 40, 41 and podophyllotoxin were incubated with purified bovine tubulin and buffer containing 10% glycerol and 1 mM GTP at 37 °C each in a separate experiment. The effect of each agent on tubulin polymerization was monitored kinetically using a fluorescent plate reader.
Supplementary Material
Acknowledgment
This work is supported by the Russian Foundation for Basic Research (grant 07-03-00577 to I.V.M) and US National Institutes of Health (grants RR-16480 and CA-99957 to A.K.) under the BRIN/INBRE and AREA programs. P. T. and M. Yu. A. are grateful to NSF/DMR (Grant 0420863) for the acquisition of X-ray single crystal diffractometer and to the Distributed Nanomaterials Characterization Network in the framework of New Mexico NSF EPSCoR Nanoscience initiative. Sophia C. Sigstedt and Manika C. Callewaert are acknowledged for their assistance with the MTT assays. We thank Dr. Olivia George and Professor Charles B. Shuster for their generous help with the tubulin polymerization assay, Professor Tatiana V. Timofeeva for her kind assistance with X-ray crystallography and Professor Scott T. Shors for stimulating discussions.
Footnotes
Abbreviations: DMEM, Dulbecco's modified Eagle's medium; DMF, dimethylformamide; DMSO, dimethyl sulfoxide; EDTA, diaminoethanetetraacetic acid; EtOH, ethanol; FBS, fetal bovine serum; FITC, fluorescein isothiocyanate; FOG, Ficoll Orange G; HEPES, 4-(2-hydroxyethyl)-1-piperazinethanesulfonic acid; HHB, Heinz-HEPES buffer; HRMS, high resolution mass spectrometry; MCR, multicomponent reaction; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; SAR, structure-activity relationship; TLC, thin layer chromatography; SD, standard deviation.
Supporting Information Available: X-ray data for compound 15 (CIF and PDF). This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 2.Arve L, Voigt T, Waldmann H. Charting Biological and Chemical Space: PSSC and SCONP as guiding principles for the development of compound collections based on natural product scaffolds. QSAR Comb. Sci. 2006;25:449–456. [Google Scholar]
- 3.Breinbauer R, Vetter IR, Waldmann H. From protein domains to drug candidates - Natural products as guiding principles in the design and synthesis of compound libraries. Angew. Chem. Int. Ed. 2002;41:2879–2890. doi: 10.1002/1521-3773(20020816)41:16<2878::AID-ANIE2878>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 4.Tan DS. Current progress in natural product-like libraries for discovery screening. Comb. Chem. High Throughput Screen. 2004;7:631–643. doi: 10.2174/1386207043328418. [DOI] [PubMed] [Google Scholar]
- 5.Boldi AM. Libraries from natural product-like scaffolds. Curr. Opin. Chem. Biol. 2004;8:281–286. doi: 10.1016/j.cbpa.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Liao Y, Hu Y, Wu J, Zhu Q, Donovan M, Fathi R, Yang Z. Diversity oriented synthesis and branching reaction pathway to generate natural product-like compounds. Curr. Med. Chem. 2003;10:2285–2316. doi: 10.2174/0929867033456738. [DOI] [PubMed] [Google Scholar]
- 7.Feher M, Schmidt JM. Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 8.Magedov IV, Manpadi M, Rozhkova E, Przheval'skii NM, Rogelj S, Shors ST, Steelant WFA, Van slambrouck S, Kornienko A. Structural Simplification of Bioactive Natural Products with Multicomponent Synthesis: Dihydropyridopyrazole Analogues of Podophyllotoxin. Bioorg. Med. Chem. Lett. 2007;17:1381–1385. doi: 10.1016/j.bmcl.2006.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magedov IV, Manpadi M, Van Slambrouck S, Steelant WFA, Rozhkova E, Przhevalískii NM, Rogelj S, Kornienko A. Discovery and Investigation of Antiproliferative and Apoptosis-Inducing Properties of New Heterocyclic Podophyllotoxin Analogues Accessible by a One-Step Multicomponent Synthesis. J. Med. Chem. 2007;50:5183–5192. doi: 10.1021/jm070528f. [DOI] [PubMed] [Google Scholar]
- 10.Kumar NV, Rajendran SP. One-pot Synthesis and the Biological Activities of 4-Methylpyrano[3,2-C]-quinolin-2,5-[6H]-diones. Asian J. Chem. 2004;16:1911–1914. [Google Scholar]
- 11.Hanawa F, Fokialakis N, Skaltsounis AL. Photo-activated DNA binding and antimicrobial activities of Furoquinoline and Pyranoquinolone alkaloids from Rutaceae. Planta Med. 2004;70:531–535. doi: 10.1055/s-2004-827153. [DOI] [PubMed] [Google Scholar]
- 12.Fujita Y, Oguri H, Oikawa H. The relative and absolute configuration of PF1140. J. Antibiot. 2005;58:425–427. doi: 10.1038/ja.2005.56. [DOI] [PubMed] [Google Scholar]
- 13.Cantrell CL, Schrader KK, Mamonov LK, Sitpaeva GT, Kustova TS, Dunbar C, Wedge DE. Isolation and identification of antifungal and antialgal alkaloids from Haplophyllum sieversii. J. Agric. Food Chem. 2005;53:7741–7748. doi: 10.1021/jf051478v. [DOI] [PubMed] [Google Scholar]
- 14.Chen JJ, Chen PH, Liao CH, Huang SY, Chen IS. New phenylpropenoids, bis(1-phenylethyl)phenols, bisquinolinone alkaloid, and anti-inflammatory constituents from Zanthoxylum integfifoliolum. J. Nat. Prod. 2007;70:1444–1448. doi: 10.1021/np070186g. [DOI] [PubMed] [Google Scholar]
- 15.Isaka M, Tanticharoen M, Kongsaeree P, Thebtaranonth Y. Structures of cordypyridones A-D, antimalarial N-hydroxy- and N-methoxy-2-pyridones from the insect pathogenic fungus Cordyceps nipponica. J. Org. Chem. 2001;66:4803–4808. doi: 10.1021/jo0100906. [DOI] [PubMed] [Google Scholar]
- 16.Koizumi F, Fukumitsu N, Zhao J, Chanklan R, Miyakawa T, Kawahara S, Iwamoto S, Suzuki M, Kakita S, Rahayu ES, Hosokawa S, Tatsuta K, Ichimura M. YCM1008A, a novel Ca2+-Signaling inhibitor, produced by Fusarium sp. YCM1008. J. Antibiot. 2007;60:455–458. doi: 10.1038/ja.2007.58. [DOI] [PubMed] [Google Scholar]
- 17.Chen IS, Tsai IW, Teng CM, Chen JJ, Chang YL, Ko FN, Lu MC, Pezzuto JM. Pyranoquinoline alkaloids from Zanthoxylum simulans. Phytochemistry. 1997;46:525–529. [Google Scholar]
- 18.Ito C, Itoigawa M, Furukawa A, Hirano T, Murata T, Kaneda N, Hisada Y, Okuda K, Furukawa H. Quinolone alkaloids with nitric oxide production inhibitory activity from Orixa japonica. J. Nat. Prod. 2004;67:1800–1803. doi: 10.1021/np0401462. [DOI] [PubMed] [Google Scholar]
- 19.McBrien KD, Gao Q, Huang S, Klohr SE, Wang RR, Pirnik DM, Neddermann KM, Bursuker I, Kadow KF, Leet JE. Fusaricide, a new cytotoxic N-hydroxypyridone from Fusarium sp. J. Nat. Prod. 1996;59:1151–1153. doi: 10.1021/np960521t. [DOI] [PubMed] [Google Scholar]
- 20.Kamperdick C, Van NH, Van Sung T, Adam G. Bisquinolinone alkaloids from Melicope ptelefolia. Phytochemistry. 1999;50:177–181. [Google Scholar]
- 21.Chen IS, Wu SJ, Tsai IL, Wu TS, Pezzuto JM, Lu MC, Chai H, Suh N, Teng CM. Chemical and Bioactive Constituents from Zanthoxylum Simulans. J. Nat. Prod. 1994;57:1206–1211. doi: 10.1021/np50111a003. [DOI] [PubMed] [Google Scholar]
- 22.(a) Rodinovskaya LA, Shestopalov AM, Gromova AV, Shestopalov AA. Substituted 4-(3-cyanopyridin-2-ylthio)acetoacetates: new convenient reagents for the synthesis of heterocycles. Synthesis. 2006:2357–2370. [Google Scholar]; (b) Rodinovskaya LA, Shestopalov AM, Gromova AV. 4-(3-Cyanopyridin-2-ylthio)acetoacetates in synthesis of heterocycles. Russ. Chem. Bull., Int. Ed. 2003;52:2185–2196. [Google Scholar]; (c) Wang XS, Zeng ZS, Shi DQ, Wei XY, Zong ZM. One-step synthesis of 2-amino-3-cyano-4-aryl-1,4,5,6-tetrahydropyrano[3,2-c]quinolin-5-one derivatives using KF-Al2O3 as catalyst. Synth. Commun. 2004;34:3021–3027. [Google Scholar]
- 23.For recent examples, see Dodia N, Shah A. New heterocycles from substituted 2-amino-5-oxo-4-phenyl-6-hydro-4H-pyrano[3,2-c]quinoline-3-carbonitriles. Indian J. Heterocycl. Chem. 2000;10:155–156.. Stoyanov EV, Ivanov IC, Heber D. General method for the preparation of substituted 2-amino-4H,5H-pyrano[4,3-b]pyran-5-ones and 2-amino-4H-pyrano[3,2-c]pyridine-5-ones. Molecules. 2000;5:19–32.. Dodia N, Shah A. Synthesis of some tricyclic and tetracyclic ring systems built on 4-hydroxy-2-quinolones. Heterocycl. Commun. 2001;7:289–294.. Stadlbauer W, Badawey ES, Hojas G, Roschger P, Kappe T. Malonates in cyclocondensation reactions. Molecules. 2001;6:338–352.. Mulwad VV, Lohar MV. One pot synthesis of novel spiro pyranoquinolones. Indian J. Heterocycl. Chem. 2002;12:57–60.. Rodinovskaya LA, Shestopalov AM, Shestopalov AA, Litvinov VP. Cross reactions of cyanoacetic acid derivatives with carbonyl compounds 3. One-step synthesis of substituted 2-amino-5-oxo-4,5-dihydropyrano[3,2-c]chromenes. Russ. Chem. Bull., Int. Ed. 2005;54:992–996.. El-Taweel FMAA. Studies with quinolines: New synthetic routes to 4H,5H,6H,9H-benzo[ij]pyrano[2,3-b]quinolizine-8-one, 4H-pyrano[2,3-b]pyridine, 2H-pyran-2-one and pyranopyridoquinoline derivatives. J. Heterocyclic Chem. 2005;42:943–946.
- 24.(a) Choudhari BP, Mulwad VV. Synthesis of biologically active 3,8-dioxo-10-hydroxypyrano[2,3-f]quinoline and its reactions. Indian J. Chem. Sect B-Org. Chem. Incl. Med. Chem. 2003;42:2080–2085. [Google Scholar]; (b) Balasubramanian C, Sekar M, Mohan PS. Synthesis and biological activity of novel pyranobisbenzopyrans and benzopyranopyranoquinolines. Indian J. Chem. Sect BO-rg. Chem. Incl. Med. Chem. 1996;35:1225–1227. [Google Scholar]
- 25.(a) Kemnitzer W, Kasibhatla S, Jiang S, Zhang H, Zhao J, Jia S, Xu L, Crogan-Grundy C, Denis R, Barriault N, Vaillancourt L, Charron S, Dodd J, Attardo G, Labrecque D, Lamothe S, Gourdeau H, Tseng B, Drewe J, Cai SX. Discovery of 4-aryl-4H-chromenes as a new series of apoptosis inducers using a cell- and caspase-based high-throughput screening assay. 2. Structure-activity relationships of the 7- and 5-, 6-, 8-positions. Bioorg. Med. Chem. Lett. 2005;15:4745–4751. doi: 10.1016/j.bmcl.2005.07.066. [DOI] [PubMed] [Google Scholar]; (b) Gourdeau H, Leblond L, Hamelin B, Desputeau C, Dong K, Kianicka I, Custeau D, Boudreau C, Geerts L, Cai S-X, Drewe J, Labrecque D, Kasibhatla S, Tseng B. Antivascular and antitumor evaluation of 2-amino-4-(3-bromo-4, 5-dimethoxy-phenyl)-3-cyano-4H-chromenes, a novel series of anticancer agents. Mol. Cancer Ther. 2004;3:1375–1383. [PubMed] [Google Scholar]; (c) Kasibhatla S, Gourdeau H, Meerovitch K, Drewe J, Reddy S, Qiu L, Zhang H, Bergeron F, Bouffard D, Yang Q, Herich J, Lamothe S, Cai SX, Tseng B. Discovery and mechanism of action of a novel series of apoptosis inducers with potential vascular targeting activity. Mol. Cancer Ther. 2004;3:1365–1373. [PubMed] [Google Scholar]; (d) Kemnitzer W, Drewe J, Jiang S, Zhang H, Wang Y, Zhao J, Jia S, Herich J, Labreque D, Storer R, Meerovitch K, Bouffard D, Rej R, Denis R, Blais C, Lamothe S, Attardo G, Gourdeau H, Tseng B, Kasibhatla S, Cai SX. Discovery of 4-aryl-4H-chromenes as a new series of apoptosis inducers using a cell- and caspase-based high-throughput screening assay. 1. Structure-activity relationships of the 4-aryl group. J. Med. Chem. 2004;47:6299–6310. doi: 10.1021/jm049640t. [DOI] [PubMed] [Google Scholar]; (e) Kemnitzer W, Drewe J, Jiang SC, Zhang H, Zhao JH, Crogan-Grundy C, Xu LF, Lamothe S, Gourdeau H, Denis R, Tseng B, Kasibhatla S, Cai SX. Discovery of 4-Aryl-4H-chromenes as a new series of apoptosis inducers using a cell- and caspase-based high-throughput screening assay. 3. Structure-activity relationships of fused rings at the 7,8-positions. J. Med. Chem. 2007;50:2858–2864. doi: 10.1021/jm070216c. [DOI] [PubMed] [Google Scholar]; (f) Cai SX, Jiang S, Kemnitzer WE, Zhang H, Attardo G, Denis R. PCT Int. Appl. WO 2003097806 A2 20031127. 2004 [Google Scholar]; (g) Cai SX, Jiang S, Attardo G, Denis R, Storer R, Rej R. PCT Int. Appl. WO 2003096982 A2 20031127. 2003 [Google Scholar]; (h) Cai SX, Zhang H, Jiang S, Storer R. PCT Int. Appl. WO 2002092594 A1 20021121. 2002 [Google Scholar]; (i) Cai SX, Xu L, Storer R, Attardo G. PCT Int. Appl. WO 2002092083 A1 20021121. 2002 [Google Scholar]; (j) Cai SX, Zhang H, Kemmitzer WE, Jiang S, Drewe JA, Storer R. PCT Int. Appl. WO 2002092076 A1 20021121. 2002 [Google Scholar]; (k) Drewe JA, Cai SX, Wang Y. PCT Int. Appl. WO 2001034591 A2 20010517. 2001 [Google Scholar]
- 26.For preliminary communication describing part of this work, see: Magedov IV, Manpadi M, Evdokimov NM, Elias EM, Rozhkova E, Ogasawara MA, Bettale JD, Przeval'skii NM, Rogelj S, Kornienko A. Antiproliferative and apoptosis inducing properties of pyrano[3,2-c]pyridones accessible by a one-step multicomponent synthesis. Bioorg. Med. Chem. Lett. 2007;17:3872–3876. doi: 10.1016/j.bmcl.2007.05.004.
- 27.Castillo S, Ouadahi H, Herault V. Reaction of 4-hydroxy-6-methyl-2-pyrone with various primary amines - synthesis of N-substituted 2-pyridones and aliphatic intermediates. Bull. Soc. Chim. Fr. 1982:257–261. [Google Scholar]
- 28.X-ray data for compound 15 is available in the Supporting Information(CIF and PDF).
- 29.Mosmann T. Rapid colorimetric assay for cellular growth and survival - application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 30.See for example: Vial JP, Belloc F, Dumain P, Besnard S, Lacombe F, Boisseau MR, Reiffers J, Bernard P. Study of the apoptosis induced in vitro by antitumoral drugs on leukaemic cells. Leukemia Res. 1997;21:163–172. doi: 10.1016/s0145-2126(96)00102-6.
- 31.(a) Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis -flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein-labeled annexin-V. J. Immunol. Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]; (b) Fadok VA, Voelkler DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 32.Hamel E. Evaluation of antimitotic agents by quantitative comparisons of their effects on the polymerization of purified tubulin. Cell Biochem. Biophys. 2003;38:1–21. doi: 10.1385/CBB:38:1:1. [DOI] [PubMed] [Google Scholar]
- 33.Jackson JR, Patrick DR, Dar MM, Huang PS. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat. Rev. Cancer. 2007;7:107–117. doi: 10.1038/nrc2049. [DOI] [PubMed] [Google Scholar]
- 34.Kesisis G, Broxterman H, Giaccone G. Angiogenesis inhibitors. Drug selectivity and target specificity. Curr. Pharm. Design. 2007;13:2795–2809. doi: 10.2174/138161207781757033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.