Abstract
Clonorchis sinensis is the causative agent of the life-threatening disease endemic to China, Korea, and Vietnam. It is estimated that about 15 million people are infected with this fluke. C. sinensis provokes inflammation, epithelial hyperplasia, and periductal fibrosis in bile ducts, and may cause cholangiocarcinoma in chronically infected individuals. Accumulation of a large amount of biological information about the adult stage of this liver fluke in recent years has advanced our understanding of the pathological interplay between this parasite and its hosts. However, no developmental gene expression profiles of C. sinensis have been published. In this study, we generated gene expression profiles of three developmental stages of C. sinensis by analyzing expressed sequence tags (ESTs). Complementary DNA libraries were constructed from the adult, metacercaria, and egg developmental stages of C. sinensis. A total of 52,745 ESTs were generated and assembled into 12,830 C. sinensis assembled EST sequences, and then these assemblies were further categorized into groups according to biological functions and developmental stages. Most of the genes that were differentially expressed in the different stages were consistent with the biological and physical features of the particular developmental stage; high energy metabolism, motility and reproduction genes were differentially expressed in adults, minimal metabolism and final host adaptation genes were differentially expressed in metacercariae, and embryonic genes were differentially expressed in eggs. The higher expression of glucose transporters, proteases, and antioxidant enzymes in the adults accounts for active uptake of nutrients and defense against host immune attacks. The types of ion channels present in C. sinensis are consistent with its parasitic nature and phylogenetic placement in the tree of life. We anticipate that the transcriptomic information on essential regulators of development, bile chemotaxis, and physico-metabolic pathways in C. sinensis that presented in this study will guide further studies to identify novel drug targets and diagnostic antigens.
Author Summary
Clonorchis sinensis is a significant pathogen that causes clonorchiasis, which is endemic to East Asian countries. This fluke provokes acute inflammation and chronic hyperplasic changes in the biliary tracts. C. sinensis promotes cholangiocarcinoma, and has been classified as a Group 1 biological carcinogen, alongside Opisthorchis viverrini, by the World Health Organization. Recently, transcriptomes for adult liver flukes have been reported with the molecular functionalities predicted on the bases of their transcriptomic data sets. We generated the developmental C. sinensis transcriptome for three different developmental stages, revealing that most functional genes were differentially expressed in each developmental stage; only a small proportion of the expressed genes were shared between the three stages. The developmental transcriptome describes the gene expression landscapes of C. sinensis adults, metacercariae, and eggs, and provides insight into how this fluke adapts to the distinctly different environments provided by its various hosts. We anticipate that the transcriptome will contribute significantly to the identification of intervention points along the developmental stages and allow the exploitation of novel potential targets for diagnostic, drug, and vaccine development purposes.
Introduction
Clonorchis sinensis causes clonorchiasis, which is endemic to Korea, China, Taiwan and Vietnam; approximately 15 million people are estimated to be infected [1]–[3]. C. sinensis is a significant pathogen both from an epidemiological and clinical perspective, as people who develop clonorchiasis are debilitated, thereby negatively impacting socio-economic activities. In C. sinensis endemic areas, inhabitants become infected by eating raw or inappropriately cooked fresh water fish caught from water bodies near their villages [4]. Fresh water fish are the hosts of C. sinensis metacercariae, which is the infective stage to humans.
Once C. sinensis eggs reach a fresh water body, they develop into miracidiae. When ingested by freshwater snails, the miracidia escapes from the egg and transforms into sporocysts, and then into rediae within several weeks. The cercariae emerge into fresh water and swim in search of freshwater fish, the second intermediate host. The cercaria penetrates the skin of a freshwater fish and its body becomes encysted by a cyst wall, followed by transformation into a metacercaria. Almost all freshwater fish can serve as the second intermediate hosts, with the highest infection rate and metacercarial burden found in the topmouth gudgeon, Pseudorasbora parva. When ingested by humans and other mammals, the metacercariae are retained for a while in the stomach and then passed down to the duodenum. The metacercariae excyst there and the hatched, juvenile C. sinensis migrate up into the bile duct. The juvenile flukes grow to adults that produce eggs in the biliary passages of the mammalian host.
C. sinensis flukes in the biliary tracts lacerate and apply pressure to the epithelia, and excrete waste products from their excretory bladder and regurgitate residual digests from their intestinal ceca. In addition, ovigerous C. sinensis adults excrete uterine fluid with high protein content when they ovulate. These various excretory and secretory products act as chemical irritants that provoke inflammation, epithelial hyperplasia, and periductal fibrosis in the biliary tracts. In human clonorchiasis patients, frequent symptoms are epigastric discomfort and dull pain, mild fever, loss of appetite, diarrhea, and jaundice [5]. Moreover, clonorchiasis has epidemiologically been reported to be associated with cholangiocarcinoma [6]–[8]. Furthermore, experimental studies have shown that C. sinensis infection induces the differentiation of liver oval cells into a bile duct cell lineage and promotes the development of cholangiocarcinoma in golden hamsters [9], [10]. Recently, C. sinensis was officially classified along with Opisthorchis viverrini as a Group 1 biological carcinogen by the World Health Organization [11]. Among the excretory-secretory products produced by liver flukes, granulin was identified as a mitogenic agent capable of stimulating cell proliferation and epithelial hyperplasia [12]. By binding to Toll-like receptors, the excretory-secretory products of adult flukes activate the NF-kB pathway resulting in increased expression of the pro-inflammatory cytokine, IL-6 [13], which in turn leads to the production of reactive oxygen radicals. The endogenous reactive radicals damage DNA and could initiate carcinogenesis [14].
Expressed sequence tags (ESTs) generated from cDNA libraries cover a large proportion of functional mRNAs and can be assembled into overlapping contigs coding for almost complete open reading frames [15]. Schistosoma mansoni and S. japonicum were the first flukes for which transcriptome data was published [16], [17]; these studies stimulated the generation of ESTs and functional cataloging of these ESTs from the human-infecting liver flukes, C. sinensis, O. viverrini, and Fasciola hepatica [18]–[22]. The recent widespread availability of next-generation sequencing technology has also stimulated high-throughput analyses of the transcriptomes of liver flukes with a focus on the pathobiological characteristics of these adult liver fluke transcriptomes [23], [24]. Given that C. sinensis adults are considered carcinogenic agents, they are predicted to express genes encoding proteins that are also known to be involved in cancer development [25].
To further elucidate the pathogenesis and carcinogenesis provoked by C. sinensis infection, comprehensive molecular and genetic information covering the different developmental stages of this parasite is required. In this study, we generated and sequenced transcriptome-scale ESTs from three developmental stages of C. sinensis and investigated the biological properties, growth, host adaptations, and pathogenic features of these different developmental stages.
Materials and Methods
Ethics statement
Rabbits were handled in an accredited Korea Food and Drug Administration animal facility in accordance with the AAALAC International Animal Care policies (Accredited Unit, Korea FDA; Unit Number 000996). Approval for animal experiments was obtained from Korea FDA animal facility (NIH-06-15, NIH-07-16 and NIH-08-19).
Parasite resources, culture and RNA extraction
C. sinensis metacercariae were collected from naturally infected P. parva caught in Jinju, Korea, and Shenyang, China. The fish were ground and digested artificially in gastric juice for 1 hr at 37°C. Particulate material was filtered out using a sieve with 0.15 mm mesh and washed several times with 0.85% saline. C. sinensis metacercariae were identified and collected under a dissecting microscope. Male New Zealand White rabbits, 1.5–3.0 kg (Samtaco Inc., Korea) were infected with 500 metacercariae each, and adult flukes were recovered from the bile ducts of these experimental rabbits 2 months after the infection. Bile juice collected from C. sinensis-infected rabbits was centrifuged at 2,000 g for 10 min and C. sinensis eggs were collected from the sediment. To extract total RNA, adult flukes, metacercariae, and eggs of C. sinensis were put into liquid nitrogen in a pre-chilled mortar on dry ice and pulverized using a Mixer Mill MM301 (Retsch GmbH, Haan, Germany). Total RNA was extracted from the ground tissues using TRI reagent (MRC, Inc., Cincinnati, OH, USA). Poly(A+) mRNA was selected from the total RNA using the Absolutely mRNA Purification Kit (Stratagene, La Jolla, CA, USA) according to the manufacturer's instruction. The amounts of total RNA and mRNA were determined by measuring the absorbance at 260 nm and the degree of protein contamination was assessed by calculating the ratio of the absorbance at 260 nm to that at 280 nm. RNA integrity was assessed by examining ribosomal RNA bands on 1% RNA agarose gels stained with ethidium bromide.
Construction of cDNA libraries
Using the poly(A+) mRNAs generated as described above, cDNA libraries of the three developmental stages of C. sinensis (adult, metacercaria, and egg) were constructed using the directional λ ZAP cDNA synthesis/Gigapack Ш Gold cloning kit (Stratagene, La Jolla, CA, USA). First stand cDNAs were synthesized from mRNAs primed at the poly-A tail using reverse transcriptase and an oligo-dT linker-primer containing an XhoI restriction enzyme site. Following second strand synthesis, an EcoR I linker was ligated to the 5′-termini followed by digestion with the restriction enzyme XhoI. These synthesized and assembled double strand cDNAs were size-fractionated using Sepharose® CL-2B gel filtration column chromatography. cDNA fractions longer than 500 bp were ligated into the ZAP Express vector pBK-CMV and the ligation products were packaged in vitro into cDNA libraries using the ZAP Express cDNA Gigapack Ш Gold cloning Kit (Stratagene). cDNAs were directionally cloned into the pBK-CMV vector, which allows both prokaryotic and eukaryotic expression of large sequences and in vivo excision into a phagemid vector. The adult, metacercaria, and egg cDNA libraries were plated onto LB-kanamycin plates, 23.5 cm×23.5 cm, coated with X-gal/IPTG for blue/white selection. White colonies were randomly picked and inoculated into each well of a 384-well plate (Corning Co., Cortland, NY, USA) containing 40 µl Terrific Broth/kanamycin, followed by incubation for 16 hr at 37°C. For storage, the culture media in the 384-well plates were mixed with an equal volume of glycerol solution (65% glycerin, 0.1 M MgSO4, 0.025 M Tris-HCl, pH 8.0) and stored at −80°C. To assess cDNA quality, additional cDNA libraries were constructed in the pBluescript SK(+) vector for the adult and in the pAD-GAL42.1 vector for the metacercaria.
cDNA sequencing
A total of 60,768 colonies were picked: 30,144 from the adult cDNA library, 20,256 from the metacercaria cDNA library, and 10,368 from the egg cDNA library. Single plasmid colonies were transferred into 540 µl of Terrific Broth medium supplemented with 50 µg/ml kanamycin in a 96-deep well plate and incubated at 37°C overnight with gentle rotation (550 rpm). Plasmids containing C. sinensis cDNA were extracted using an alkaline lysis method [26], [27]. The sequences of the cloned C. sinensis cDNAs were determined using the BigDye Terminator Cycle Sequencing Kit, ver. 3.1 (Applied Biosystems, Foster City, CA, USA). Sequencing reactions were performed in a 3-µl volume containing 250 ng plasmid DNA, 0.5 pmole universal primer, 0.87 µl of 5X Sequencing buffer, and 1.38 µl of distilled water. The cycling profile consisted of 35 cycles of denaturation at 96°C for 10 seconds, annealing at 50°C for 5 seconds, and extension at 60°C for 4 min. Sequencing products were purified via ethanol precipitation and read on an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA). The T3 forward primer and T7 reverse primer were used as sequencing primers.
DNA sequence trimming and assembly
Nucleotide sequences of the 60,768 clones were read once from the 5′-end. The vector and adapter sequences were trimmed off all reads, as well as nucleotide stretches with a Phred score of 20 or less and poly A/T stretches [28], [29]. Reads shorter than 100 bp were then filtered out of the analyses. A total of 52,745 reads that survived these quality control filters were assembled into clusters using the TGICL and CAP3 programs with the following parameters: an offset of 40 bp overlap, 95% minimum identity, and a maximum mismatched overhang of 30 bp [30], [31]. Nucleotide sequences of the reads reported in this paper are registered in the DDBJ/EMBL/GenBank databases under the accession numbers FS126466-FS179210.
Bioinformatic analyses of ESTs
To annotate the assembled EST clusters, the nucleotide sequences of the clusters were translated into putative polypeptide sequences and these sequences were blasted against the NCBI non-redundant nucleotide and protein databases using the parameters of more than 30 matched amino acids, an identity greater than 25%, and an E-value less than 1e−4 using BLASTX. The domain structures of the translated polypeptides were predicted using InterProScan (data version v14.0) and an E-value of less than 1e−4 [32] and their potential function was assessed by gene ontology (GO) analysis. Moreover, we manually curated the gene descriptions in the databases by selecting known genes with a significant E-value to prevent incorrect assignment of annotated genes. To further enhance the reliability of the data and provide more accurate gene predictions for C. sinensis, we chose the species closest to C. sinensis for which data were available.
Signal peptides in the EST clusters were searched and categorized based on their function. The open reading frame of a conceptual polypeptide was first translated from the cluster sequence using OrfPredictor [33] (https://fungalgenome.concordia.ca/tools/OrfPredictor.html). The signal peptide sequences of the conceptual polypeptides were predicted using the SignalP 3.0 server [34] (http://www.cbs.dtu.dk/services/SignalP) and the subcellular localizations of proteins were analyzed using PSORTb [35] (http://psort.nibb.ac.jp). Secondary structure features of the peptides such as alpha helices and intervening loops were predicted using TMHMM version 2.0 [36] (http://www.cbs.dtu.dk/services/TMHMM/). Putative B-cell epitopes in C. sinensis EST-encoding proteins were predicted using the ABCpred database [37] (http://www.imtech.res.in/raghava/abcpred/ABC_submission.html).
To generate developmental gene expression profiles of C. sinensis, the number of ESTs in each contig was counted according to developmental stage and analyzed using Fisher's exact test using a significance level of P<0.01 at IDEG6 [38] (http://telethon.bio.unipd.it/bioinfo/IDEG6_form/). Putative SNPs in the EST sequences were determined using the AutoSNP program [39].
To gain insight into the evolutionary history of C. sinensis and to investigate parasitism-related genes, the global similarity of C. sinensis whole ESTs at the amino acid sequence level were compared to those of other parasites and free-living platyhelminthes using the SimiTri program (cut-off score: 50) [40]. Whole EST sequences of comparator organisms were retrieved from the NCBI protein database. The relative similarities of the gene sequences of C. sinensis to those of other species were analyzed using TBLASTX and the SimiTri viewer. Bulk sequences of the comparator species were downloaded from the GenBank EST databases. Sequences with BLAST scores (bit score values) higher than 50 were collected from each large dataset. Nucleotide versus nucleotide comparisons of the dataset of interest to the different databases were performed using the TBLASTX algorithm. The primary data consisted of the similarity values of each sequence from the chosen database. The primary data was transformed into input for the SimiTri viewer. A gene is indicated as a square tile, and these tiles are colored by similarity scores to other datasets. Genes that were similar to genes in only one other database are not shown. Genes that showed similarity to genes in only two databases are shown as lines joining the two databases.
URL
More detailed information and raw data are accessible at http://grc.kribb.re.kr/pipeline2/.
ID of genes
The ID numbers for genes and proteins mentioned in the text refer to tables. The others are as follow: K+-channel, CL3811; Ca+2-channels, CL6309, CSM01492; Na+-Channel, CSA10278; Cl− channel, CL2019; Sodium transporters, CL5256, CL25, CL6319, CL420, CSA00901, CSA10105, CSA10217, CSA19634; Na+/K+-ATPases, CL2552, CSA23629; Glucose transporters, CL272, CL25; Amino acid transporters, CL1075, CL1111; Zinc transporter, CL1482; Phosphate transporters, CL5278, CSA12737; Fatty acid transporter, CL384. Apoptosis, CL632, CL635, CL621; Cell proliferation, CL1924, CL134, CL6028, CL894; cancer development, CL644, CL557, CL3820, CL1644, CL1801, CL5519, CL5976, CL1379, CL2881, CL2147, CL684, CL894, CL25, CL1289, CL1087, CL545, CL1679, CL421, CL111; Neuroreceptors, CL1876, CSM05821, CSM11397; Neurotransmitter-related proteins, CL4260, CSA10291, CL1504, CL5536, CSM14836, CL5972.
Results and Discussion
The C. sinensis transcriptome
To generate transcriptomes and evaluate developmental gene expression in C. sinensis, 60,768 clones were selected randomly from cDNA libraries of the adult, metacercaria, and egg developmental stages. After stringent quality filtering through base calling, vector sequence trimming, repeat masking, and contaminant screening, 52,745 high quality reads remained (success index of 86.8%) consisting of 27,070 reads from the adult stage, 15,872 reads from the metacercaria stage, and 9,803 reads from the egg stage ( Table 1 ). The high quality reads were assembled and clustered into 12,830 C. sinensis assembled EST sequences (CsAEs) comprising 7,184 contigs and 5,646 singletons [31]. The length of the CsAEs ranged from 100–3,328 bp with an average length of 724 bp. Over 50% of the CsAEs were between 500–799 bp. More than 70% of the CsAEs consisted of less than 30 EST members, while the largest single CsAE had 643 EST members.
Table 1. Transcriptome feature of three developmental stages from Clonorchis sinensis.
| Adults | Metacercariae | Eggs | All | |
| Total sequence reads | 30,144 | 20,256 | 10,368 | 60,768 |
| Total analyzed reads + | 27,070 | 15,872 | 9,803 | 52,745 |
| Average EST size (bp after trimming) | 528 | 524 | 569 | 522 |
| Total number of assembled sequences | 7,779 | 5,398 | 2,660 | 12,830‡ |
| Contigs | 3,921 | 2,728 | 1,523 | 7,184 |
| Singlets | 3,858 | 2,670 | 1,137 | 5,646 |
| Average contigs size (bp) | 720 | 671 | 758 | 724 |
| Total known genes * | 3,993 | 2,718 | 1,630 | 6,413 |
| Unigene number | 3,488 | 2,427 | 1,504 | 5,269 |
| Unigenes matched to S. mansoni or S. japonicum | 1,674(48%) | 1,138(47%) | 868(58%) | 2,356(45%) |
| Unigenes matched to other organisms | 1,814(52%) | 1,289(53%) | 636(42%) | 2,913(55%) |
| Total unknown genes | 3,786 | 2,680 | 1,030 | 6,417 |
+: : Total analyzed reads were determined using the stringent quality filtering and contained reads with a minimum of 100 bases and a phred quality≥20.
*: Total known genes were determined by homology ≥25% and length of minimum exact match ≥30 amino acids by using BLASTX homology search.
‡: : Each stage (column) was generated from its own dataset. Independently, data of all stage were assembled from the combination of adults, metacercariae and eggs.
Developmental gene expression
A total of 52,745 reads collected from the adult, metacercaria, and egg stages were assembled into 7,184 contigs. Of these contigs, 1,887 (26.3%) were shared by two developmental stages; 648 contigs between the adult and egg stages, 974 between the adult and metacercaria stages, and 261 between the metacercaria and egg stages. A small portion of the transcriptome (564 contigs; 7.9%) occurred in all three developmental stages, suggesting that some of these are housekeeping genes expressed constitutively across the life stages of C. sinensis. A large number of the contigs (4,733; 65.9%) occurred in one of the three developmental stages (Figure S1). This finding suggests that genes associated with growth can be identified from the C. sinensis developmental transcriptome. The expression levels of these genes, as determined by the number of ESTs contained within a cluster, were compared to investigate stage-specific patterns of transcription based on an arbitrary cut-off as well as the statistical significance of the expression differences. In C. sinensis, 119 contigs in the adult stage, 48 in the metacercaria stage, and 134 in the egg stage were significantly differentially expressed. The majority of CsAEs obtained from each developmental stage were non-annotated or hypothetical transcripts. The unknown CsAEs were broadly distributed at each stage, indicating that C. sinensis may have a unique developmental mechanism compared to other parasites.
Functional annotation
Our manual curation of the search reports generated by BLASTX and InterPro increased the annotation rate and accuracy of the functional notations of the CsAEs. Of the 12,830 CsAEs, 7,132 (55.6%) were found to have significant sequence similarities to sequences in the NCBI NR database and/or in InterPro [41], while the remaining 5,698 (44.4%) CsAEs had no homolog (Figure S2). One-half of the proteins translated from the CsAEs were annotated by BLASTX and found to be similar to proteins found in Schistosoma japonicum. Among them, the sequences of hsp90, RNA binding protein, and actin were highly conserved between C. sinensis and S. japonicum. Another 44% of the CsAEs were unique gene transcripts with no homolog in the databases searched.
Gene ontology
To predict the functions of the C. sinensis genes, we independently classified a total of 12,830 CsAEs into functional categories by analyzing automated gene ontology assignments [42]. The most accurate method to identify new members of known gene function among gene transcripts is to retrieve a sequence-based homology of the translated transcripts using domains extracted from a multiple alignment of gene members with known functions [43]. To functionally categorize CsAEs using domain homology searches, we translated the 12,830 CsAEs in six reading frames and recruited them into InterProScan [32], which aligned 6,106 CsAEs to InterPro entries (E-value≤1e−4). Among these, 2,889 CsAEs were assigned to 23,178 GO accession numbers. The 23,178 accession numbers further generated 2,349 distinguished GO mappings in two major ontologies, molecular functions and biological processes. We assigned 2,679 CsAEs (20.9%) to 17 main molecular functional subcategories and 2,195 CsAEs (17.1%) to 23 main biological process subcategories ( Figure 1 ). The most abundant groups represented under the molecular function category were assigned to the GO categories of nucleotide binding (9.1%), nucleic acid binding (6.2%), ion binding (5.7%), transferase activity (6.0%), and hydrolase activity (7.4%). The most abundant groups represented under the biological process category corresponded to the GO categories of cellular component organization and biogenesis (2.3%), transport (2.9%), localization (3.0%), biosynthetic processes (2.4%), metabolic processes of nucleobases, nucleosides, nucleotides and nucleic acids (2.7%), and protein metabolism (5.2%). The apparent discrepancies between these values may be due to the fact that one InterProScan number can be assigned to more than one GO accession number, and one GO accession number can be mapped to multiple parental categories and CsAEs [33].
Figure 1. Predicted functions of C. sinensis transcripts based on gene ontology analysis.
Distribution of molecular functional categories (a) and biological process categories (b) according to homology to genes in Uniprot with a gene ontology classification.
Putative single nucleotide polymorphisms (SNPs)
SNPs are the most common and abundant type of genetic variation between individuals and are used in population and evolutionary biology studies [44]. In addition, SNPs can be used as markers for physical and genetic mapping. SNP discovery based on large-scale EST datasets has proven an efficient technique for identifying large numbers of SNPs. Among the 7,184 CsAE contigs, SNPs were discovered in 2,896 contigs (40%). A large majority (77%) of the SNPs were detected from contigs consisting of 2–4 ESTs, while the remainder of the SNPs were discovered in contigs consisting of more than 5 ESTs ( Table 2 ). A total of 9,077 SNPs were detected from the 7,184 contigs, which is an average of 0.37 SNPs per 100 bp. The putative SNP frequency varied from 0.22 to 0.50 SNPs per 100 base pairs among contigs of different sizes. The genetic diversity of C. sinensis SNPs was similar to that reported for S. japonicum: an average of 0.35 SNPs per 100 bp [21]. In eukaryotic organisms, SNPs occur every 500 to 1,000 bp, at frequencies higher than those found in non-eukaryotic organisms [45]. Almost all of the predicted CsAEs with >0.19 SNPs/100 bp were no-match genes, and only several were homologous to S. mansoni genes of unknown function. In trematodes, SNPs in the first codon position can result in an amino acid substitution, which may lead to structural changes in the respective proteins and affect the formation of functional domains, antigenic epitopes, or drug binding sites [44].
Table 2. SNPs of C. sinensis identified using AutoSNP software.
| No. of sequences in each contig | No. of contigs with SNPs | No. of total SNPs | Total consensus length (bp) | SNP frequency (per 100 bp) |
| 2 | 950 | 2,398 | 689,883 | 0.35 |
| 3 | 760 | 2,396 | 604,004 | 0.40 |
| 4 | 520 | 2,267 | 451,608 | 0.50 |
| 5 | 128 | 305 | 112,313 | 0.27 |
| 6 | 123 | 300 | 117,116 | 0.26 |
| 7–10 | 108 | 212 | 96,884 | 0.22 |
| 11–20 | 108 | 279 | 108,014 | 0.26 |
| 21–30 | 59 | 171 | 78,977 | 0.22 |
| 31–50 | 64 | 226 | 104,073 | 0.22 |
| >50 | 76 | 523 | 112,516 | 0.46 |
| Total | 2,896 | 9,077 | 2,475,388 | 0.37 |
Abundantly expressed gene transcripts in the different developmental stages
The 30 genes most abundantly expressed were investigated further (Table S1). Genes with significant p-values were analyzed by one-to-one comparison with the number of reads per the same gene in each stage. Eighteen genes in the adult stage and 12 genes each in the egg and metacercaria stages were annotated. Highly abundant genes should be conserved based on the reasoning that they are major constituents of the transcriptome of the organism and are therefore likely to be functional, but the highly abundant genes found in C. sinensis had few homologs, indicating that this organism is developmentally quite different to other organisms for which gene abundance information is available. Genes expressed abundantly across all three stages coded for ubiquitin family proteins, elongation factor 1-alpha, fructose bisphosphate aldolase, which are all proteins involved in basal and energy metabolisms. Genes highly expressed in the adult compared to the metacercaria and egg encoded structural and reproduction-associated proteins (beta-tubulin, ferritin), detoxification proteins (glutathione S-transferase), transportation proteins (clonorporin 1, sodium/glucose co-transporter), energy production proteins (GAPDH, mitochondrial malate dehydrogenase), and enzymes (cysteine protease, PHGPx isoform 1). In particular, cysteine proteases have previously been shown to be the most abundantly expressed proteins in C. sinensis adults [19], [20], [24]. In the egg stage, the gene encoding acyl-CoA synthetase long chain family member 5 (ACSL5) was highly expressed among the known genes. ACSL5 is a member of the highly conserved ACSL family. In protozoan parasites, ACSL5 is thought to catalyze the conversion of long-chain fatty acids to CoA derivatives to enable parasite growth [46]. The majority of highly expressed CsAEs was unknown or hypothetical genes and therefore deserved further study.
Evolutionary and functionality conservation
To investigate the evolutionary and functional conservation of the transcriptome of C. sinensis, we estimated gene numbers and the degree of conservation among C. sinensis genes and the genomes of diverse eukaryotic organisms. We performed pair-wise sequence comparisons of all C. sinensis transcripts using BLASTX with an E-value cut-off from 1e−10 to 1e−200. The C. sinensis transcriptome shared 22.9% genes with Homo sapiens, 23.0% with Mus musculus, 20.5% with Drosophila melanogaster, 17.8% with Caenorhabditis elegans, and 26.6% Schistosoma japonicum at an E-value≤10−20. The CsAEs showed a moderate degree of sequence homology to the genes of the comparator organisms and a higher degree of sequence homology to S. japonicum genes ( Table 3 ). Genes highly conserved between the CsAEs and S. japonicum may be important for parasite survival. These genes are discussed in more detail in the next section. A small fraction of genes (37 genes; 0.3%) were highly conserved (E-value≤10−200) across all the animals compared in this study ( Table 4 ). These genes encode proteins such as actin, tubulin, translation elongation factor 1, valosin-containing protein, glycogen phosphorylase, and heat shock protein 70.
Table 3. Comparison of the transcriptomes of C. sinensis and selected eukaryotes.
| Unigene(%) | ||||||
| Homology | Homo sapiens | Mus musculus | Drosophila melanogaster | Caenorhabditis elegans | Schistosoma japonicum | All organisms+ |
| E-value≤1e−200 | 19(0.2%) | 18(0.2%) | 16(0.2%) | 18(0.2%) | 16(0.2%) | 37(0.3%) |
| E-value ≤1e−100 | 158(1.6%) | 159(1.6%) | 137(1.4%) | 114(1.3%) | 205(2.0%) | 346(3.0%) |
| E-value ≤1e−50 | 905(8.6%) | 906(8.7%) | 733(7.6%) | 564(6.2%) | 1,125(11.2%) | 1,674(14.7%) |
| E-value ≤1e−30 | 1,696(16.5%) | 1,687(16.4%) | 1,325(14.4%) | 1,078(12.3%) | 1,936(20.4%) | 2,850(26.0%) |
| E-value ≤1e−20 | 2,305(22.9%) | 2,302(23.0%) | 1,821(20.5%) | 1,468(17.8%) | 2,424(26.6%) | 3,654(33.9%) |
| E-value ≤1e−10 | 3,110(31.7%) | 3,092(31.8%) | 2,396(28.2%) | 1,946(24.9%) | 2,910(33.3%) | 4,643(43.6%) |
| Little homology * | 3,477(35.4%) | 3,412(35.2%) | 2,669(31.4%) | 2,193(28.2%) | 3,028(35.0%) | 5,269(49.1%) |
+: EST data were searched against the entire protein database by BLASTX.
*More than 25% homology with at least 30 amino acid residues deduced from CsAEs.
%the percent ratio of matched clusters of total 12,830 CsAEs to the protein database of model eukaryotes.
Table 4. Ultraconserved contigs of C. sinensis.
| Accession No. | Descriptions | H. sapiens | M. musculus | D. melanogaster | C. elegans | S. japonicum | All |
| CAO79607.1 | Beta-tubulin | ○ | ○ | ○ | ○ | ○ | ○ |
| AAQ16109.1 | Elongation factor1-alpha | ○ | ○ | ○ | ○ | ○ | ○ |
| ABS52704.1 | Heat shock protein 70 | ○ | ○ | ○ | ○ | - | ○ |
| NP_001086877.1 | Translation elongation factor 2 | ○ | ○ | ○ | ○ | - | ○ |
| ABS81352.1 | Phospho glucose isomerase | ○ | ○ | ○ | ○ | - | ○ |
| AAH83344.1 | Tubulin, alpha1A | ○ | ○ | ○ | ○ | ○ | ○ |
| CAQ13492.1 | Propionyl CoenzymeA carboxylase, beta polypeptide | ○ | ○ | - | ○ | - | ○ |
| AAW27581.1 | SJCHGC09453 protein | ○ | ○ | ○ | ○ | ○ | ○ |
| AAI61792.1 | Unknown | ○ | ○ | ○ | ○ | - | ○ |
| AAS93901.1 | Glycogen phosphorylase | ○ | ○ | ○ | ○ | ○ | ○ |
| AAM69406.1 | Heat shock protein HSP60 | ○ | ○ | ○ | ○ | ○ | ○ |
| BAB84579.1 | Actin2 | ○ | ○ | ○ | ○ | ○ | ○ |
| AAW27659.1 | SJCHGC00820 protein | ○ | ○ | ○ | ○ | ○ | ○ |
| XP_001118968.1 | Glucan(1,4-alpha-), branching enzyme1 | ○ | ○ | ○ | ○ | - | ○ |
| XP_001107041.1 | Oxoglutarate dehydrogenase-like isoform2 | ○ | ○ | ○ | ○ | - | ○ |
| XP_001089681.1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit | ○ | ○ | ○ | ○ | - | ○ |
| XP_001627515.1 | Predicted protein | - | ○ | ○ | - | - | ○ |
| AAW27320.1 | SJCHGC06322 protein | - | - | - | - | ○ | ○ |
| AAW27782.1 | SJCHGC00653 protein | - | - | - | - | ○ | ○ |
| AAW26140.1 | SJCHGC02536 protein | - | - | - | - | ○ | ○ |
| AAX27366.2 | SJCHGC05847 protein | - | - | - | - | ○ | ○ |
| AAW27345.1 | SJCHGC09272 protein | - | - | - | - | ○ | ○ |
| AAW26056.1 | SJCHGC05577 protein | - | - | - | - | ○ | - |
| AAW27129.1 | SJCHGC06305 protein | - | - | - | - | ○ | - |
| XP_001177707.1 | Gag-polpoly protein | - | - | - | - | - | ○ |
| XP_001628202.1 | Predicted protein | - | - | - | - | - | ○ |
| XP_001952362.1 | Zinc finger protein | - | - | - | - | - | ○ |
| ABI26619.1 | Enolase | - | - | - | - | - | ○ |
*Cut-off value <1e−200.
Clonorchis sinensis and parasitism
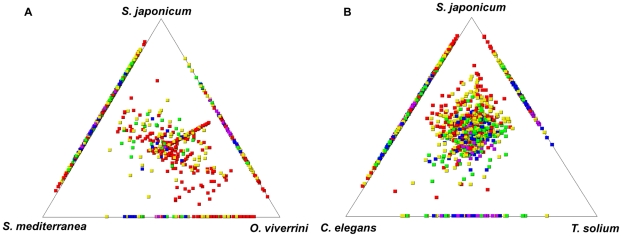
SimiTri graphically displays the relative similarity of one organism to others using bulk datasets from the respective organisms. The degree of similarity among helminth sequences was determined using two-dimensional plots [40]. We used SimiTri to analyze the global relative similarity of CsAEs to other parasitic or free-living trematodes, cestode and nematode ( Figure 2 ). C. sinensis (12,830 CsAEs) was more similar to the two parasitic flukes, S. japonicum and O. viverrini, than to the free-living helminths Schmidtea mediterranea (73,650 ESTs) and C. elegans (474,350 ESTs). When compared to both Opisthorchis viverrini (4,194 ESTs) and S. japonicum (99,069 ESTs), the closest neighbor of C. sinensis was O. viverrini, consistent with the general taxonomic classification of these trematodes and the recent molecular phylogeny of Digenean trematodes based on morphological characters and the sequence of the nuclear ribosomal small subunit (18S) [47].
Figure 2. Global relative similarity between C. sinensis and other species analyzed at the whole transcriptome scale.
. Each C. sinensis contigs and singlets were searched against the whole transcriptome using TBLASTX score (a cut-off of ≥50). Similarity comparison of parasitic organisms with a free-living flatworm (A) or with free-living nematode (B). Square tiles indicate genes, with the squares colored by their highest TBLASTX score to each of the databases: red ≥300; yellow ≥200; green ≥150, blue ≥100 and purple <100.
Based on the SimiTri analyses results, 23 CsAEs were considered to contain parasitism-related genes of C. sinensis (Table S2). The genes encoded by these 23 CsAEs showed higher sequence similarity to the parasitic platyhelminths, O. viverrini, S. japonicum, and Taenia solium than to free-living ones, S. mediterranea and C. elegans (Figures 2A and B). Of these, 12 CsAEs had an unknown function, whereas 11 CsAEs had cell communication, ion transport, metabolic processes, nucleotide and protein binding, or oxidoreduction functional annotations.
Membrane proteins, channels and transporters
Channels
In the adult C. sinensis cDNA library, EST encoding K+-channel was abundant but that of Ca+2-channel was rare ( Table 5 ). No Na+ -channel EST was found in any of the three developmental stages. From an evolutionary point of view, K+-channels are ancient, occurring in all three domains of life, and are abundant in invertebrate animals. K+-channels are ubiquitous and maintain cell homeostasis in organisms. Ca+2-channels emerged later in evolutionary time. In more basal animals, Ca+2-channels are activated in response to action potentials of nerve systems and provoke slow actions and movements. Na+-channels are more elaborate and are often multimeric, and are generally rare in invertebrates compared to higher vertebrate animals [48]. The frequency distribution of these cation channels is consistent with phylogenetic placement of C. sinensis in the tree of life. Specific structural motifs in the interacting domains of the beta subunit of Ca+2 channels render flukes PZQ susceptible. These specific structural motifs were found in the beta-subunits of C. sinensis adults, which explain why they are vulnerable to PZQ.
Table 5. Developmental expression of ion-channels and transporters in C. sinensis.
| No. of reads | ||||
| Category | Description | Adult | Metacercaria | Egg |
| K+-channel | Potassium channel protein | 10 | 4 | 2 |
| Ca+2-channel | High voltage-gated Ca+2-channel, batasubunit,CavB | 2 | 2 | 1 |
| High voltage-activated Ca+2-channel, CavA | 0 | 1 | 0 | |
| Na+-channel | Amiloide-sensitive sodium channel-related protein | 1 | 0 | 0 |
| Cl−-channel | Chloride channel protein 7 | 7 | 4 | 1 |
| Sodium Transporter | Solute carrier family 17 (sodium phosphate) | 2 | 0 | 0 |
| Solute carrier family 5 (sodium/glucose cotransporter) | 77 | 0 | 0 | |
| Sodium/sialic acid cotransporter, putative | 2 | 0 | 0 | |
| Sodium/bile acid cotransporter | 3 | 14 | 0 | |
| Potassium-dependent sodium-calcium exchanger | 1 | 0 | 0 | |
| Sodium/hydrogen exchanger 7, 9 | 1 | 0 | 0 | |
| Sodium/myo-inositol cotransporter | 1 | 0 | 0 | |
| Sodium/proton exchanger 3 | 1 | 0 | 0 | |
| Na+/K+-ATPase | Na+/K+-transporting ATPase beta | 4 | 0 | 0 |
| Na+/K+-transporting ATPase beta-2 chain | 1 | 0 | 0 | |
| Glucose transporter | Glucose transporter | 47 | 0 | 20 |
| Sodium/glucose co-transporter | 77 | 0 | 0 | |
| Amino acid transporters | Amino acid transporter | 22 | 2 | 9 |
| Zinc transporter | Zinc transporter | 15 | 5 | 4 |
| Phosphate transporter | Glycerol-3-phosphate transporter | 2 | 2 | 3 |
| Phosphate transporter | 2 | 0 | 1 | |
| Fatty acid transporter | Fatty acid transporter, member 1 | 16 | 18 | 2 |
Transporters
A large number of glucose transporters were found in both the adult and egg stages ( Table 5 ). Glucose/sodium co-transporters and Na+/K+-transporting ATPases were abundant only in adult C. sinensis, not in the metacercaria. This fluke species consumes large amounts of glucose to generate energy and metabolic intermediates for physiologic regulation. In adult C. sinensis, glucose appears to be imported actively from the environment through glucose/sodium co-transporters using ATP as an energy source. Glucose molecules may move passively through the glucose transporters between cells in fluke tissues. Anion channel proteins including chloride channels were 4-fold more frequent than cationic channels, which could compensate for the large amount of Na+ ions co-imported with exogenous glucose. These anionic channels may be suitable targets for vaccine or drug development.
CsAEs coding for bile acid beta-glucosidase and a sodium-bile acid cotransporter, a component of the bile acid transportation pathway, were present in the C. sinensis EST pool ( Table 5 ). Bile acid beta-glucosidase converts bile acid to a soluble conjugated form to facilitate its secretion. The sodium-bile acid cotransporter, which imports bile salts with Na+-dependency, was abundantly expressed in the metacercaria stage. This type of transporter is responsible for the influx of conjugated bile acids into hepatocytes, ileal enterocytes, and cholangiocytes in mammals [49], [50]. The presence of these transporters in C. sinensis suggests that C. sinensis thrives in bile juice by utilizing bile acid and its derivatives for normal physiologic pathways. C. sinensis is expected to have a bile acid exporting system comprising bile salt export pumps, organic solute transporters, and/or multidrug resistance protein 2 to maintain cellular homeostasis [49], [50].
Neurotransmitters & receptors
CsAEs encoding proteins involved in neurotransmission such as serotonin receptors, tryptophan hydroxylase, aromatic amino acid decarboxylase, glutamate receptors, glutaminase, GABA receptor-associated proteins, acetylcholine esterase, and DOPA-decarboxylase were found as rare species (Table S3). The presence of these neurotransmission-related proteins implies that C. sinensis has a web of serotoninergic, glutaminergic, and cholinergic neurons, which is consistent with the previous observation that the nervous systems of trematodes are highly conserved [51].
Proteases and protease inhibitors
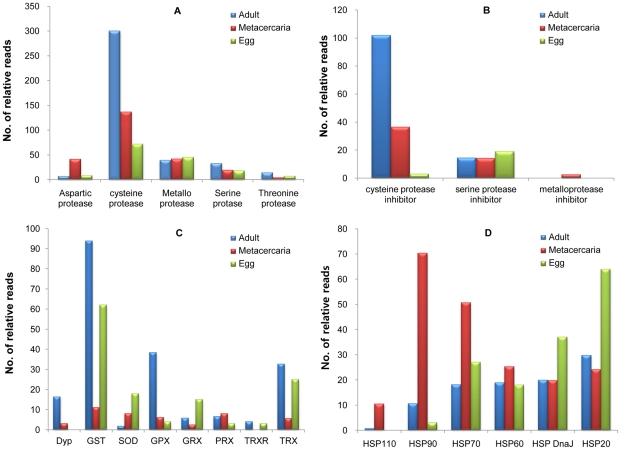
Proteases of parasitic origin are known to be important virulence factors based on genomic and proteomic analysis of several major global helminth species [52], [53], [54]. Secretory proteases of parasites are ubiquitous enzymes that have been implicated in several diverse physiological and adaptive mechanisms, such as tissue penetration, larval migration, immunoevasion, digestion, and excystation [55]. Because these proteases are indispensable for parasite viability and growth, they have been suggested as potential targets for vaccines or chemotherapeutic agents [56], [57]. We classified the CsAE proteases into four functional groups based on the catalytic type, namely serine, threonine, aspartate, and metallo- or cysteine proteases (Figure 3A). Cysteine proteases showed the highest expression (68.8%) levels among the four types of proteases during all three developmental stages. Cysteine proteases of C. sinensis are developmentally controlled and essential for survival because they are involved in processes such as nutrient uptake, tissue invasion, and evasion from host immune attacks [58]. Of the C. sinensis cysteine proteases, the cathepsin F-like isoenzyme (CsCF-6) was expressed across all developmental stages, and we observed that transcript levels of this protein increased according to the developmental stage of the parasite [59]. Metalloproteases were detected in all three developmental stages of C. sinensis ( Figure 3A ). Metalloproteases are crucial proteases for invasion and immune evasion of flukes in addition to the general roles they play in catabolic reactions and protein processing [57].
Figure 3. Developmental expression of proteases and protease inhibitors, antioxidant enzymes, and heat shock proteins in C. sinensis.
Relative reads refer to the number of calculated reads in proportion to the read size for each developmental stage (adult: metacercaria : egg = 2.76∶1.62∶1). (A) Proteases, (B) protease inhibitors, (C) antioxidant enzymes, (D) stress response proteins. Dyp, dye-decolorizing peroxidase; GST, glutathione-s-transferase; SOD, superoxide dismutase; GPX, glutathione peroxidase; GRX, glutaredoxin; PRX, Peroxiredoxin; TRXR, thioredoxin reductase; TRX, thioredoxin; HSP, heat hock protein.
Parasites utilize protease inhibitors to survive in their hosts; protease inhibitors can prevent damage by mature proteases prior to their secretion from the parasite and protect them against the digestive proteases of the hosts [60]. Diverse proteins function as protease inhibitors, but they have a common biochemical mechanism and are characterized by rapid evolution of their sequences [61]. In our study, cysteine protease inhibitor expression was highest in the adult stage among all three stages (Figure 3B). Cysteine protease inhibitors (cystatins) regulate cysteine proteases and modulate host immune responses [60]. Cystatins of C. elegans have been shown to inhibit cathepsin B while filarial cystatins have been shown to inhibit the proliferation of murine and human T-cells. Because C. sinensis cysteine proteases are expressed most abundantly in the intestinal epithelium for uptake of nutrients [59], cystatins could be expressed in the intestinal epithelium to fine-tune the intracellular activities of cysteine proteases. Expression of serine protease inhibitors remained stable in all stages with slightly greater expression levels observed in the egg stage ( Figure 3B ). This finding is consistent with a previous study that demonstrated that serine protease inhibitors were present mainly in the eggs of C. sinensis [62]. Transcripts encoding metalloprotease inhibitors were found only in the metacercaria library and inhibitors of aspartic and threonine proteases were not identified in any of the developmental stages.
Antioxidant enzymes
Several antioxidant enzymes constituting the oxidoreduction system were encoded by the CsAEs, with the expression levels of the various enzymes varying according to developmental stage ( Figure 3C ). Antioxidant enzymes catalyze reactions that neutralize endogenous and exogenous reactive oxygen species (ROS) that are produced either by aerobic cellular metabolism or by host immune responses [63]. Regulation of the expression levels of these enzymes during each development stage is therefore important to cope with host-produced ROS [64]. In the C. sinensis transcriptome, glutathione-S-transferases (GSTs) were the most highly expressed antioxidant enzymes, especially in the adult and egg stages. In Fasciola hepatica, GSTs are expressed at much lower levels in juvenile worms than in adult worms living in the bile duct, implying that adult worms require more protection against host immune responses [63]. In C. sinensis, two GST isoenzymes have been identified: a 26 kDa GST and a 28 kDa GST [65]. Glutathione peroxidase (GPx) and thioredoxins (TRX) were expressed differentially at high levels in the adult stage (Figure 3C). C. sinensis GPx has been reported to be specifically localized in the vitellocytes of vitelline glands and in the premature eggs [66]. GPx defends against ROS and repairs ROS-induced damage in trematodes that do not have catalases [53]. TRX was highly expressed (Figure 3). In Schistosoma mansoni, TRX is secreted from eggs and plays a crucial role in protecting eggs from host-induced ROS production [63]. All antioxidant enzymes were expressed at low levels in the metacercaria stage (Figure 3), which can be explained by the fact that this is a dormant state with a depressed metabolism that is protected by a cyst wall from exogenous oxidative stresses.
Stress responsive genes
Heat shock proteins (HSPs) function as molecular chaperones and play an important role in the stress response to a variety of biological stresses such as heat shock, hypoxia, mechanical stimuli, lack of glucose, and UV exposure by assisting in the refolding of denatured proteins into active forms or targeting them for degradation [67]. We identified six HSPs from the CsAEs: HSP110, HSP90, HSP70, HSP60, HSP DnaJ, and HSP20. Most HSPs were expressed at higher levels in the metacercaria and egg stages than in adult worms. The higher molecular weight HSPs such as HSP110, HSP90, HSP70 and HSP60 were more highly expressed in the metacercaria stage, while the lower molecular weight HSPs, HSP DnaJ and HSP20, were expressed at higher levels in the egg stage. In C. sinensis adults, HSPs were expressed at low levels relative to the metacercaria and egg stages ( Figure 3D ). In the life cycle of C. sinensis, the metacercariae experience a large thermal change when they move from the environment (ambient temperature) to the stomach of the mammalian host (37°C). Furthermore, as the metacercariae pass down and excyst in the duodenum, they face the osmotic stress of intestinal secretions, and then when they migrate into the bile duct, they are exposed to bile juices. The higher molecular weight C. sinensis HSPs are likely to responsd to these thermally- and environmentally-induced stresses [68], [69]. The C. sinensis eggs that ovulate in the bile duct are carried down the intestine and passed out in the feces of the mammalian host into the environment, thereby experiencing cold shock. In the C. sinensis eggs, the low molecular weight HSPs may function to protect the eggs against cold thermal shock [70], while the high molecular weight HSPs might contribute to recovery from the cold shock [71].
Cell proliferation and cholangiocarcinoma-related genes
The C. sinensis transcriptome had several contigs encoding proteins associated with cell proliferation and apoptosis such as granulin, epidermal growth factor (EGF), tumor growth factor (TGF) interacting protein, and inhibitors and regulators of apoptosis. Among these proteins, granulin, encoded for by 67 reads, was most abundantly expressed with a 8.6-fold higher expression level in adults than metacercariae. Similarly, the EGF gene was also expressed at 2.5-fold higher levels in the adult stage than the metacercaria stage (Table 6). Transcriptomic datasets of C. sinensis and O. viverrini were previously analyzed for proteins common to carcinogenesis and a large number of the amino acid sequences of these trematodes were inferred to have homologs to genes involved in human cancer development [25]. The C. sinensis genes associated with apoptosis, cell proliferation, and cancer development encoded laminins, c-Jun N-terminal kinase, catenins, cyclin-dependent kinases, histone deacetylases, MFS transporters, serine/threonine kinases, and transcription factors ( Table 6 ). C. sinensis infection provokes both acute and chronic pathological changes such as proliferation of the bile duct epithelium and periductal fibrosis that is disseminated over the biliary tree from proximal to remote biliary capillaries [5]. The mitogen-like proteins secreted or excreted from C. sinensis are likely to be provocative agents that cause biliary epithelial alterations, as has been documented for the granulin-like growth factor of O. viverrini [72], [73]. Proliferating cholangiocytes could be vulnerable to DNA damage from endogenous and exogenous carcinogens, bioreactive free radicals, and nitrosocompounds. The apoptosis inhibitor and regulatory proteins identified in the transcriptome of C. sinensis in this study (Table 6) may prevent the death of DNA-damaged cells and possibly facilitate their transformation into cancerous cells [73]. Epidemiologically, the incidence of cholangiocarcinoma is significantly higher in clonorchiasis endemic areas than in non-endemic areas [74]. Experimentally, infection with C. sinensis and the ingestion of dimethylnitrosamine by Syrian golden hamsters resulted in cholangiocarcinoma [9], [75].
Table 6. Expression of genes associated with apoptosis, cell proliferation, and cancer development in C. sinensis.
| Category | Description | No. of reads | ||
| Adult | Metacercaria | Egg | ||
| Apoptosis | Inhibitor of apoptosis protein | 9 | 8 | 0 |
| Apoptosis-linked gene 2 protein | 1 | 3 | 9 | |
| Cell-cycle and apoptosis regulatory protein 1 | 14 | 8 | 0 | |
| Cell proliferation | Granulin | 7 | 4 | 2 |
| Granulin precursor (Proepithelin) (PEPI) | 36 | 1 | 17 | |
| EGF | 1 | 2 | 0 | |
| EGF/Laminin | 9 | 2 | 13 | |
| Multiple EGF-like-domains 6 | 0 | 2 | 0 | |
| TGF-beta receptor interacting protein 1 | 2 | 3 | 0 | |
| Cancer development * | c-Jun N-terminal kinase | 23 | 11 | 0 |
| Catenin (cadherin-associated protein) | 5 | 15 | 8 | |
| Axin 1 | 0 | 3 | 0 | |
| Cell division control protein 42 | 2 | 2 | 3 | |
| Cyclin-dependent kinase | 11 | 4 | 3 | |
| Death-associated protein kinase | 2 | 0 | 0 | |
| DNA mismatch repair protein | 3 | 2 | 0 | |
| DNA repair protein | 10 | 0 | 0 | |
| Growth factor receptor-binding protein | 1 | 1 | 4 | |
| Histone deacetylase | 12 | 9 | 9 | |
| Integrin beta | 12 | 3 | 0 | |
| Laminin | 8 | 4 | 13 | |
| MFS transporter, SP family, solute carrier family | 77 | 0 | 0 | |
| Mitogen-activated protein kinase kinase | 6 | 0 | 2 | |
| RING-box protein 1 | 9 | 0 | 0 | |
| Serine/threonine-protein kinase | 32 | 29 | 23 | |
| Transcription elongation factor | 8 | 1 | 2 | |
| Transcription factor | 27 | 20 | 15 | |
| Tropomyosin | 30 | 38 | 12 | |
*Refer to Reference No. 25.
Drug target candidates
Praziquantel is currently used to treat clonorchiasis. However, the efficacy of praziquantel against clonorchiasis has been reported to be poor in northern Vietnam [76]. Tribendimidine has recently emerged as a promising alternative to praziquantel for the treatment of human opisthorchiasis [77]. Transcriptomic datasets facilitate the search for new drug targets by mining them with bioinformatic tools that employ statistical and network analyses, parasite-specific physico-metabolic pathways and developmental regulation can be found. Membrane proteins including channels, transporters, and permeases, which play important roles in host-parasite interactions, were some of the first novel targets identified using transcriptome data [78], [79]. A total of 435 CsAEs that had more than two transmembrane domains were screened using the TMHMM algorithm [36], and the localization of the predicted proteins was determined using PSORTb based on homology to proteins of known localization [35]. CsAEs homologous to proteins of the free-living flatworm S. mediterranea, or of humans were selected using a cut-off E-value≤1e−10 using BLASTX. Five genes not present in vertebrates were identified as putative drug targets using the screening strategy described above (Table S4). Tetraspanins, four-transmembrane-domain proteins, are present on the outer tegument of trematodes and function as receptors for host molecules [80]. Tetraspanins are a recognized vaccine target for S. mansoni [78]. ADP ribosylation-like factor-6 (ARL6) interacting protein, which interacts with the ARL6 protein [81], is involved in hematopoietic maturation processes such as protein transport, membrane trafficking, and cell signaling [82]. Myelin proteolipid protein is a transmembrane protein that has been suggested to serve as a structural component of myelin, contributing to both the stability and compact lamellar structure of myelin [83].
Diagnostic antigen candidates
Serodiagnostic methods are used to screen for patients infected with C. sinensis and as supportive diagnosis tools in individual patients. Antigen proteins have been identified from the excretory-secretory products of C. sinensis and have been purified from crude extracts [84]. As antigenic preparations, crude extracts of C. sinensis show high sensitivity but low specificity toward the sera of clonorchiasis patients. In contrast, some of the recombinant antigenic proteins used for screening have high specificity but low sensitivity [84]. Potent serodiagnostic antigens for clonorchiasis are therefore still lacking. As the first step to find antigen candidates, the C. sinensis transcriptome data was filtered for secretory signal peptides using the SignalP 3.0 server and a neural network/hidden Markov model [34], while proteins secreted into the extracellular space were searched for using PSORTb [35]. After removing proteins with transmembrane domains using TMHMM, 411 CsAEs were designated ‘secretory’. To determine which of these proteins is potentially antigenic [36], the secretory candidates were filtered using the ABCpred server [85] with default parameters, and 43 CsAEs with two or more B-cell epitopes and a score of 0.82 or higher were obtained. By removing proteins homologous to mammalian and nuclear proteins, a total of 19 CsAEs were identified as putative antigen candidates. Examples of these candidates include a male sterility protein, cathepsin L-like cysteine proteinase A, protein disulfide isomerase-related protein P5 precursor, cryptosporidial mucin, and TGF-beta receptor interacting protein 1 ( Table 7 ). Recombinant antigenic proteins encoded by the selected CsAEs could be synthesized in vitro using a high-throughput cell-free system [86], and their antigenicity for the serodiagnosis of clonorchiasis could be determined using various methods such as a protein chip [87].
Table 7. Putative serodiagnostic antigens.
| EST ID | Description |
| CL1051Contig1 | Male sterility protein |
| CSA22042 | Cathepsin L-like cysteine proteinase A |
| CL1502Contig1 | Protein disulfide isomerase-related protein P5 precursor |
| CL2119Contig1 | LAG1 (longevity assurance homolog) |
| CL2671Contig1 | Flagelliform silk protein |
| CL11Contig2 | Cryptopsoridial mucin, large thr stretch, signal peptide sequence |
| CSM15703 | TGF-beta receptor interacting protein 1 |
| CSA19952 | DJ-1_PfpI |
| CL3175Contig1 | Eukaryotic translation initiation factor 4A2 isoform 1 |
| CL1573Contig1 | Predicted protein |
| CL3180Contig1 | Peptidyl-prolyl cis-trans isomerase B precursor (PPIase B) (Cyclophilin B) |
| CL6354Contig1 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| CL6614Contig1 | Oxysterol binding protein |
| CL5658Contig1 | Vesicular mannose-binding lectin |
| CL222Contig1 | Glycoprotein X precursor |
| CSA21481 | Stromal interaction molecule 2 |
| CSA01410 | Calreticulin precursor (SM4 protein) |
| CL31Contig2 | Melibiase family protein |
| CSA17420 | Group 14 allergen protein |
Supporting Information
Contigs and singlets of the assembled C. sinensis EST pool according to developmental stage.
(TIFF)
The annotation of CsAEs using Interpro and BLASTX. Whole 12,830 CsAEs were searched for homologs in the NCBI NR database and the InterPro database, and the retrieved homologs were manually curated. A total of 7,132 CsAEs were annotated with homologs, but the remaining 5,698 (orange) found to have no homolog. Of the annotateds, 5,387 CsAEs (violet) matched homologs in both databases, 719 ones (green) in the InterPro, and 1,026 ones (blue) in the BLASTX.
(TIFF)
The 30 most abundantly expressed genes in the adult, metacercaria, and egg stages of C. sinensis.
(DOC)
Putative parasitism-related genes of C. sinensis.
(DOC)
CsAEs of neuro-receptors and neurotransmitter producing enzymes according to C. sinensis developmental stage.
(DOC)
Putative drug targets of C. sinensis
(DOC)
Acknowledgments
We thank Dr. Shunyu Li (Chung-Ang University) and Dr. Sung-Hwa Chae (GnCBIO Co., LTD.) for help with the laboratory experiments.
Footnotes
The authors have declared that no competing interests exist.
This study was funded by Korea National Institutes of Health grants 2008-E54003-00 and 2009-E54004-00 to Sung-Jong Hong and 2006-E54005-00 and 2007-E54004-00 to Hong-Seog Park. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rim HJ. Clonorchiasis: an update. J Helminthol. 2005;79:269–281. doi: 10.1079/joh2005300. [DOI] [PubMed] [Google Scholar]
- 2.Wang KX, Zhang RB, Cui YB, Tian Y, Cai R, et al. Clinical and epidemiological features of patients with clonorchiasis. World J Gastroenterol. 2004;10:446–448. doi: 10.3748/wjg.v10.i3.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu SH, Kawanaka M, Li XM, Xu LQ, Lan CG, et al. Epidemiological investigation on Clonorchis sinensis in human population in an area of South China. Jpn J Infect Dis. 2003;56:168–171. [PubMed] [Google Scholar]
- 4.Keiser J, Utzinger J. Emerging foodborne trematodiasis. Emerg Infect Dis. 2005;11:1507–1514. doi: 10.3201/eid1110.050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rim HJ. The current pathobiology and chemotherapy of clonorchiasis. Korean J Parasitol. 1986;24(Suppl):1–141. doi: 10.3347/kjp.1986.24.suppl.1. [DOI] [PubMed] [Google Scholar]
- 6.Lee JH, Rim HJ, Sell S. Heterogeneity of the “oval-cell” response in the hamster liver during cholangiocarcinogenesis following Clonorchis sinensis infection and dimethylnitrosamine treatment. J Hepatol. 1997;26:1313–1323. doi: 10.1016/s0168-8278(97)80467-9. [DOI] [PubMed] [Google Scholar]
- 7.Watanapa P, Watanapa WB. Liver fluke-associated cholangiocarcinoma. Br J Surg. 2002;89:962–970. doi: 10.1046/j.1365-2168.2002.02143.x. [DOI] [PubMed] [Google Scholar]
- 8.Shin HR, Oh JK, Lim MK, Shin A, Kong HJ, et al. Descriptive epidemiology of cholangiocarcinoma and clonorchiasis in Korea. J Korean Med Sci. 2010;25:1011–1016. doi: 10.3346/jkms.2010.25.7.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JH, Yang HM, Bak UB, Rim HJ. Promoting role of Clonorchis sinensis infection on induction of cholangiocarcinoma during two-step carcinogenesis. Korean J Parasitol. 1994;32:13–18. doi: 10.3347/kjp.1994.32.1.13. [DOI] [PubMed] [Google Scholar]
- 10.Yoon BI, Jung SY, Hur K, Lee JH, Joo KH, et al. Differentiation of hamster liver oval cell following Clonorchis sinensis infection. J Vet Med Sci. 2000;62:1303–1310. doi: 10.1292/jvms.62.1303. [DOI] [PubMed] [Google Scholar]
- 11.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, et al. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 12.Smout MJ, Laha T, Mulvenna J, Sripa B, Suttiprapa S, et al. A granulin-like growth factor secreted by the carcinogenic liver fluke, Opisthorchis viverrini, promotes proliferation of host cells. PLoS Pathog. 2009;5:e1000611. doi: 10.1371/journal.ppat.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ninlawan K, O'Hara SP, Splinter PL, Yongvanit P, Kaewkes S, et al. Opisthorchis viverrini excretory/secretory products induce toll-like receptor 4 upregulation and production of interleukin 6 and 8 in cholangiocyte. Parasitol Int. 2010;59:616–621. doi: 10.1016/j.parint.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaewpitoon N, Kaewpitoon SJ, Pengsaa P, Sripa B. Opisthorchis viverrini: the carcinogenic human liver fluke. World J Gastroenterol. 2008;14:666–674. doi: 10.3748/wjg.14.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vries E, Corton C, Harris B, Cornelissen AW, Berriman M. Expressed sequence tag (EST) analysis of the erythrocytic stages of Babesia bovis. Vet Parasitol. 2006;138:61–74. doi: 10.1016/j.vetpar.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 16.Verjovski-Almeida S, DeMarco R, Martins EA, Guimaraes PE, Ojopi EP, et al. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nat Genet. 2003;35:148–157. doi: 10.1038/ng1237. [DOI] [PubMed] [Google Scholar]
- 17.Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, et al. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nat Genet. 2003;35:139–147. doi: 10.1038/ng1236. [DOI] [PubMed] [Google Scholar]
- 18.Laha T, Pinlaor P, Mulvenna J, Sripa B, Sripa M, et al. Gene discovery for the carcinogenic human liver fluke, Opisthorchis viverrini. BMC Genomics. 2007;8:189. doi: 10.1186/1471-2164-8-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JS, Lee J, Park SJ, Yong TS. Analysis of the genes expressed in Clonorchis sinensis adults using the expressed sequence tag approach. Parasitol Res. 2003;91:283–289. doi: 10.1007/s00436-003-0962-y. [DOI] [PubMed] [Google Scholar]
- 20.Cho PY, Lee MJ, Kim TI, Kang SY, Hong SJ. Expressed sequence tag analysis of adult Clonorchis sinensis, the Chinese liver fluke. Parasitol Res. 2006;99:602–608. doi: 10.1007/s00436-006-0204-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Lu J, Hu W, Wang SY, Cui SJ, et al. New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. PLoS Pathog. 2006;2:e29. doi: 10.1371/journal.ppat.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho PY, Kim TI, Whang SM, Hong SJ. Gene expression profile of Clonorchis sinensis metacercariae. Parasitol Res. 2008;102:277–282. doi: 10.1007/s00436-007-0759-5. [DOI] [PubMed] [Google Scholar]
- 23.Young ND, Hall RS, Jex AR, Cantacessi C, Gasser RB. Elucidating the transcriptome of Fasciola hepatica - a key to fundamental and biotechnological discoveries for a neglected parasite. Biotechnol Adv. 2010;28:222–231. doi: 10.1016/j.biotechadv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Young ND, Campbell BE, Hall RS, Jex AR, Cantacessi C, et al. Unlocking the transcriptomes of two carcinogenic parasites, Clonorchis sinensis and Opisthorchis viverrini. PLoS Negl Trop Dis. 2010;4:e719. doi: 10.1371/journal.pntd.0000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young ND, Jex AR, Cantacessi C, Campbell BE, Laha T, et al. Progress on the transcriptomics of carcinogenic liver flukes of humans–unique biological and biotechnological prospects. Biotechnol Adv. 2010;28:859–870. doi: 10.1016/j.biotechadv.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelley JM, Field CE, Craven MB, Bocskai D, Kim UJ, et al. High throughput direct end sequencing of BAC clones. Nucleic Acids Res. 1999;27:1539–1546. doi: 10.1093/nar/27.6.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 29.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertea G, Huang X, Liang F, Antonescu V, Sultana R, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 32.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, et al. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min XJ, Butler G, Storms R, Tsang A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 2005;33:W677–680. doi: 10.1093/nar/gki394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 35.Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, et al. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 36.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 37.Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 38.Romualdi C, Bortoluzzi S, D'Alessi F, Danieli GA. IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. 2003;12:159–162. doi: 10.1152/physiolgenomics.00096.2002. [DOI] [PubMed] [Google Scholar]
- 39.Barker G, Batley J, H OS, Edwards KJ, Edwards D. Redundancy based detection of sequence polymorphisms in expressed sequence tag data using AutoSNP. Bioinformatics. 2003;19:421–422. doi: 10.1093/bioinformatics/btf881. [DOI] [PubMed] [Google Scholar]
- 40.Parkinson J, Blaxter M. SimiTri–visualizing similarity relationships for groups of sequences. Bioinformatics. 2003;19:390–395. doi: 10.1093/bioinformatics/btf870. [DOI] [PubMed] [Google Scholar]
- 41.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jongeneel CV. Searching the expressed sequence tag (EST) databases: panning for genes. Brief Bioinform. 2000;1:76–92. doi: 10.1093/bib/1.1.76. [DOI] [PubMed] [Google Scholar]
- 44.Simoes M, Bahia D, Zerlotini A, Torres K, Artiguenave F, et al. Single nucleotide polymorphisms identification in expressed genes of Schistosoma mansoni. Mol Biochem Parasitol. 2007;154:134–140. doi: 10.1016/j.molbiopara.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S, Sha Z, Sonstegard TS, Liu H, Xu P, et al. Quality assessment parameters for EST-derived SNPs from catfish. BMC Genomics. 2008;9:450. doi: 10.1186/1471-2164-9-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matesanz F, Tellez MM, Alcina A. The Plasmodium falciparum fatty acyl-CoA synthetase family (PfACS) and differential stage-specific expression in infected erythrocytes. Mol Biochem Parasitol. 2003;126:109–112. doi: 10.1016/s0166-6851(02)00242-6. [DOI] [PubMed] [Google Scholar]
- 47.Olson PD, Cribb TH, Tkach VV, Bray RA, Littlewood DT. Phylogeny and classification of the Digenea (Platyhelminthes: Trematoda). Int J Parasitol. 2003;33:733–755. doi: 10.1016/s0020-7519(03)00049-3. [DOI] [PubMed] [Google Scholar]
- 48.Hille B. Sunderland: Sinauer Associates, Inc.; 2001. Ion channels of excitable membranes. pp. 693–722. [Google Scholar]
- 49.Alrefai WA, Gill RK. Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharm Res. 2007;24:1803–1823. doi: 10.1007/s11095-007-9289-1. [DOI] [PubMed] [Google Scholar]
- 50.Xia X, Francis H, Glaser S, Alpini G, LeSage G. Bile acid interactions with cholangiocytes. World J Gastroenterol. 2006;12:3553–3563. doi: 10.3748/wjg.v12.i22.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sebelova S, Stewart MT, Mousley A, Fried B, Marks NJ, et al. The musculature and associated innervation of adult and intramolluscan stages of Echinostoma caproni (Trematoda) visualised by confocal microscopy. Parasitol Res. 2004;93:196–206. doi: 10.1007/s00436-004-1120-x. [DOI] [PubMed] [Google Scholar]
- 52.Robinson MW, Dalton JP, Donnelly S. Helminth pathogen cathepsin proteases: it's a family affair. Trends Biochem Sci. 2008;33:601–608. doi: 10.1016/j.tibs.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Robinson MW, Tort JF, Lowther J, Donnelly SM, Wong E, et al. Proteomics and phylogenetic analysis of the cathepsin L protease family of the helminth pathogen Fasciola hepatica: expansion of a repertoire of virulence-associated factors. Mol Cell Proteomics. 2008;7:1111–1123. doi: 10.1074/mcp.M700560-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Stack CM, Caffrey CR, Donnelly SM, Seshaadri A, Lowther J, et al. Structural and functional relationships in the virulence-associated cathepsin L proteases of the parasitic liver fluke, Fasciola hepatica. J Biol Chem. 2008;283:9896–9908. doi: 10.1074/jbc.M708521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lun HM, Mak CH, Ko RC. Characterization and cloning of metallo-proteinase in the excretory/secretory products of the infective-stage larva of Trichinella spiralis. Parasitol Res. 2003;90:27–37. doi: 10.1007/s00436-002-0815-0. [DOI] [PubMed] [Google Scholar]
- 56.Dalton JP, Neill SO, Stack C, Collins P, Walshe A, et al. Fasciola hepatica cathepsin L-like proteases: biology, function, and potential in the development of first generation liver fluke vaccines. Int J Parasitol. 2003;33:1173–1181. doi: 10.1016/s0020-7519(03)00171-1. [DOI] [PubMed] [Google Scholar]
- 57.Dvorak J, Mashiyama ST, Braschi S, Sajid M, Knudsen GM, et al. Differential use of protease families for invasion by schistosome cercariae. Biochimie. 2008;90:345–358. doi: 10.1016/j.biochi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 58.Song CY, Rege AA. Cysteine proteinase activity in various developmental stages of Clonorchis sinensis: a comparative analysis. Comp Biochem Physiol B. 1991;99:137–140. doi: 10.1016/0305-0491(91)90018-9. [DOI] [PubMed] [Google Scholar]
- 59.Na BK, Kang JM, Sohn WM. CsCF-6, a novel cathepsin F-like cysteine protease for nutrient uptake of Clonorchis sinensis. Int J Parasitol. 2008;38:493–502. doi: 10.1016/j.ijpara.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Knox DP. Proteinase inhibitors and helminth parasite infection. Parasite Immunol. 2007;29:57–71. doi: 10.1111/j.1365-3024.2006.00913.x. [DOI] [PubMed] [Google Scholar]
- 61.Rawlings ND, Tolle DP, Barrett AJ. Evolutionary families of peptidase inhibitors. Biochem J. 2004;378:705–716. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang JM, Sohn WM, Ju JW, Kim TS, Na BK. Identification and characterization of a serine protease inhibitor of Clonorchis sinensis. Acta Trop. 2010;116:134–140. doi: 10.1016/j.actatropica.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Chiumiento L, Bruschi F. Enzymatic antioxidant systems in helminth parasites. Parasitol Res. 2009;105:593–603. doi: 10.1007/s00436-009-1483-0. [DOI] [PubMed] [Google Scholar]
- 64.Dzik JM. Molecules released by helminth parasites involved in host colonization. Acta Biochim Pol. 2006;53:33–64. [PubMed] [Google Scholar]
- 65.Hong SJ, Lee JY, Lee DH, Sohn WM, Cho SY. Molecular cloning and characterization of a mu-class glutathione S-transferase from Clonorchis sinensis. Mol Biochem Parasitol. 2001;115:69–75. doi: 10.1016/s0166-6851(01)00270-5. [DOI] [PubMed] [Google Scholar]
- 66.Cai GB, Bae YA, Kim SH, Sohn WM, Lee YS, et al. Vitellocyte-specific expression of phospholipid hydroperoxide glutathione peroxidases in Clonorchis sinensis. Int J Parasitol. 2008;38:1613–1623. doi: 10.1016/j.ijpara.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 67.Nollen EA, Morimoto RI. Chaperoning signaling pathways: molecular chaperones as stress-sensing ‘heat shock’ proteins. J Cell Sci. 2002;115:2809–2816. doi: 10.1242/jcs.115.14.2809. [DOI] [PubMed] [Google Scholar]
- 68.Devaney E. Thermoregulation in the life cycle of nematodes. Int J Parasitol. 2006;36:641–649. doi: 10.1016/j.ijpara.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Schlesinger MJ. Heat shock proteins: the search for functions. J Cell Biol. 1986;103:321–325. doi: 10.1083/jcb.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez-Matas MA, Nunez P, Soto A, Allona I, Casado R, et al. Protein cryoprotective activity of a cytosolic small heat shock protein that accumulates constitutively in chestnut stems and is up-regulated by low and high temperatures. Plant Physiol. 2004;134:1708–1717. doi: 10.1104/pp.103.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yocum GD, Joplin KH, Denlinger DL. Upregulation of a 23 kDa small heat shock protein transcript during pupal diapause in the flesh fly, Sarcophaga, crassipalpis. Insect Biochem Mol Biol. 1998;28:677–682. doi: 10.1016/s0965-1748(98)00046-0. [DOI] [PubMed] [Google Scholar]
- 72.Kim YJ, Choi MH, Hong ST, Bae YM. Proliferative effects of excretory/secretory products from Clonorchis sinensis on the human epithelial cell line HEK293 via regulation of the transcription factor E2F1. Parasitol Res. 2008;102:411–417. doi: 10.1007/s00436-007-0778-2. [DOI] [PubMed] [Google Scholar]
- 73.Sripa B, Kaewkes S, Sithithaworn P, Mairiang E, Laha T, et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007;4:e201. doi: 10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shin HR, Lee CU, Park HJ, Seol SY, Chung JM, et al. Hepatitis B and C virus, Clonorchis sinensis for the risk of liver cancer: a case-control study in Pusan, Korea. Int J Epidemiol. 1996;25:933–940. doi: 10.1093/ije/25.5.933. [DOI] [PubMed] [Google Scholar]
- 75.Lee JH, Rim HJ, Bak UB. Effect of Clonorchis sinensis infection and dimethylnitrosamine administration on the induction of cholangiocarcinoma in Syrian golden hamsters. Korean J Parasitol. 1993;31:21–30. doi: 10.3347/kjp.1993.31.1.21. [DOI] [PubMed] [Google Scholar]
- 76.Tinga N, De N, Vien HV, Chau L, Toan ND, et al. Little effect of praziquantel or artemisinin on clonorchiasis in Northern Vietnam. A pilot study. Trop Med Int Health. 1999;4:814–818. doi: 10.1046/j.1365-3156.1999.00499.x. [DOI] [PubMed] [Google Scholar]
- 77.Keiser J, Utzinger J, Xiao SH, Odermatt P, Tesana S. Opisthorchis viverrini: efficacy and tegumental alterations following administration of tribendimidine in vivo and in vitro. Parasitol Res. 2008;102:771–776. doi: 10.1007/s00436-007-0838-7. [DOI] [PubMed] [Google Scholar]
- 78.Loukas A, Tran M, Pearson MS. Schistosome membrane proteins as vaccines. Int J Parasitol. 2007;37:257–263. doi: 10.1016/j.ijpara.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 79.Han ZG, Brindley PJ, Wang SY, Chen Z. Schistosoma genomics: new perspectives on schistosome biology and host-parasite interaction. Annu Rev Genomics Hum Genet. 2009;10:211–240. doi: 10.1146/annurev-genom-082908-150036. [DOI] [PubMed] [Google Scholar]
- 80.Tran MH, Pearson MS, Bethony JM, Smyth DJ, Jones MK, et al. Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat Med. 2006;12:835–840. doi: 10.1038/nm1430. [DOI] [PubMed] [Google Scholar]
- 81.Ingley E, Williams JH, Walker CE, Tsai S, Colley S, et al. A novel ADP-ribosylation like factor (ARL-6), interacts with the protein-conducting channel SEC61beta subunit. FEBS Lett. 1999;459:69–74. doi: 10.1016/s0014-5793(99)01188-6. [DOI] [PubMed] [Google Scholar]
- 82.Pettersson M, Bessonova M, Gu HF, Groop LC, Jonsson JI. Characterization, chromosomal localization, and expression during hematopoietic differentiation of the gene encoding Arl6ip, ADP-ribosylation-like factor-6 interacting protein (ARL6). Genomics. 2000;68:351–354. doi: 10.1006/geno.2000.6278. [DOI] [PubMed] [Google Scholar]
- 83.Gudz TI, Schneider TE, Haas TA, Macklin WB. Myelin proteolipid protein forms a complex with integrins and may participate in integrin receptor signaling in oligodendrocytes. J Neurosci. 2002;22:7398–7407. doi: 10.1523/JNEUROSCI.22-17-07398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim TI, Na BK, Hong SJ. Functional genes and proteins of Clonorchis sinensis. Korean J Parasitol. 2009;47(Suppl):S59–68. doi: 10.3347/kjp.2009.47.S.S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen J, Liu H, Yang J, Chou KC. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33:423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- 86.Tsuboi T, Takeo S, Arumugam TU, Otsuki H, Torii M. The wheat germ cell-free protein synthesis system: a key tool for novel malaria vaccine candidate discovery. Acta Trop. 2010;114:171–176. doi: 10.1016/j.actatropica.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 87.Khan F, He M, Taussig MJ. Double-hexahistidine tag with high-affinity binding for protein immobilization, purification, and detection on ni-nitrilotriacetic acid surfaces. Anal Chem. 2006;78:3072–3079. doi: 10.1021/ac060184l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contigs and singlets of the assembled C. sinensis EST pool according to developmental stage.
(TIFF)
The annotation of CsAEs using Interpro and BLASTX. Whole 12,830 CsAEs were searched for homologs in the NCBI NR database and the InterPro database, and the retrieved homologs were manually curated. A total of 7,132 CsAEs were annotated with homologs, but the remaining 5,698 (orange) found to have no homolog. Of the annotateds, 5,387 CsAEs (violet) matched homologs in both databases, 719 ones (green) in the InterPro, and 1,026 ones (blue) in the BLASTX.
(TIFF)
The 30 most abundantly expressed genes in the adult, metacercaria, and egg stages of C. sinensis.
(DOC)
Putative parasitism-related genes of C. sinensis.
(DOC)
CsAEs of neuro-receptors and neurotransmitter producing enzymes according to C. sinensis developmental stage.
(DOC)
Putative drug targets of C. sinensis
(DOC)