Abstract
Termination signals induce rapid and irreversible dissociation of the nascent transcript from RNA polymerase. Terminators at the end of genes prevent unintended transcription into the downstream genes, whereas terminators in the upstream regulatory leader regions adjust expression of the structural genes in response to metabolic and environmental signals. Premature termination within an operon leads to potentially deleterious defects in the expression of the downstream genes, but also provides an important surveillance mechanism. This Review discusses the actions of bacterial and phage antiterminators that allow RNA polymerase to override a terminator when the circumstances demand it.
Transcription is the first, and probably the most highly regulated, step in gene expression. Cellular multisubunit RNA polymerases (RNAPs) initiate RNA synthesis at a promoter, extend the nascent RNA chain for many steps and then release the completed message at a terminator1. The distance between the promoter and the terminator can be up to 105 bp in bacteria, but the transcribing enzyme must traverse it in just one attempt. RNAP initiates RNA synthesis de novo from single nucleoside 5′-triphosphates (NTPs) and, although it may sometimes use short 2–4-nucleotide RNAs as primers2, longer prematurely released RNAs are irreversibly lost.
At most positions along the DNA template, the transcription elongation complex (FIG. 1) is very stable and can move against a large applied force3, rapidly adding 1 nucleotide and moving 1 nucleotide forward at each step. However, certain nucleic acid signals and auxiliary factors may slow RNAP down (at a pause site), induce it to move backwards a few steps (at an arrest site) or trigger its dissociation from RNA and DNA (at a terminator). Some of these signals serve as checkpoints of gene expression; for example, a pause may mediate recruitment of a regulatory factor to the RNAP4,5 or provide sufficient time for the ribosome to initiate translation6. Other signals are simply roadblocks that the enzyme must bypass to complete synthesis of an RNA chain; for example, sequences that favour backtracking and arrest7 or proteins that are bound to DNA8 hinder RNAP movement. Successful execution of the gene expression programme requires that RNAP properly responds to the genuine regulatory signals and avoids falling into spurious traps along the way.
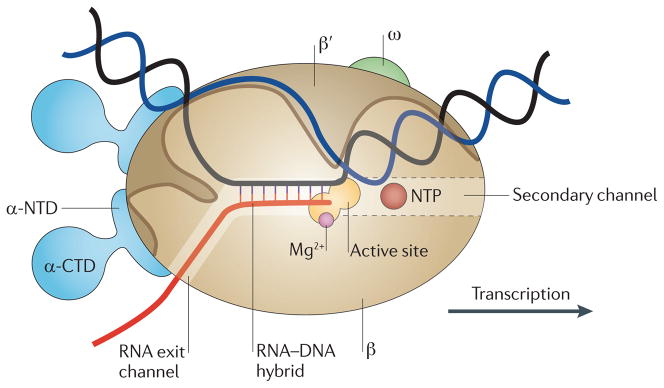
Figure 1. Schematic model of the elongation complex.
Core RNA polymerase (RNAP) (in bacteria, a complex composed of an α-dimer, a β-subunit, a β′-subunit and an ω-subunit) is bound to the DNA duplex composed of the template strand (black) and the non-template strand (blue), and the nascent RNA (red). The α-amino-terminal domains (α-NTDs) serve as a scaffold for complex assembly; the α-carboxy-terminal domains (α-CTDs) and ω-subunit play regulatory roles during initiation. The β- and β′-subunits jointly form the active site and make all the contacts to the nucleic acids. The substrate nucleoside 5′-triphosphate (NTP) (bound to a second Mg2+ ion) is thought to enter through the secondary channel. 12–14 bp of the DNA are melted in the transcription bubble. The non-template DNA strand is exposed on the surface, where it may interact with regulatory proteins. The nascent RNA is annealed to the template strand to form 8–9 bp of the RNA–DNA hybrid, which is the key determinant of elongation complex stability7,98,99. The upstream RNA is extruded through the RNA exit channel formed between the β-flap and β′-clamp.
In this Review, we describe bacterial antitermination mechanisms that suppress the action of terminators and termination factors to increase the expression of downstream genes. Some antitermination factors allow bypass of a single terminator in response to a regulatory signal, whereas others act on RNAP to increase its processivity. The second mechanism could be particularly important in higher eukaryotes, in which it can take hours for RNAP to transcribe genes, as they are tightly packaged into nucleosomes and can be very long (for example, the human dystrophin gene is 2.4 × 106 bp). Given that the structural and functional organization of all cellular RNAPs is remarkably conserved9, lessons learned from bacteria have been, and will probably be in the future, applicable to higher organisms.
Bacterial termination signals
Termination signals have a dramatic effect on the elongation complex, which normally has a half-life of days, but falls apart within seconds when reaching a terminator. Terminators serve as punctuation marks that demarcate gene boundaries and as targets for regulation. They can be grouped into two classes: intrinsic and factor-dependent terminators (FIG. 2).
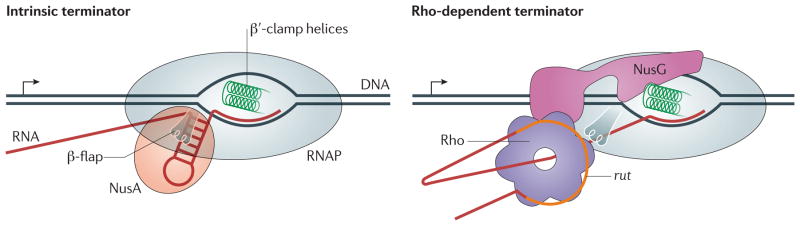
Figure 2. Bacterial termination signals.
Intrinsic terminators are composed of a stable RNA hairpin that can extend to within 8 nucleotides of the 3′ end of the RNA and disrupt the upstream edge of the RNA–DNA hybrid. Transcription elongation protein NusA interacts with the nascent RNA near the exit channel and can stimulate termination107. At Rho-dependent terminators, a Rho utilization (rut) element (orange) encoded in the nascent RNA binds to Rho. The initial Rho–rut interactions trigger formation of a stable complex in which the Rho hexamer encircles the RNA and translocates towards RNA polymerase (RNAP). Contacts with rut may persist115 until Rho reaches RNAP at the actual site of RNA release, which may be located far downstream; however, recent data suggest that Rho–rut contacts are lost earlier 116,117. The carboxy-terminal domain of NusG binds to Rho and strongly stimulates its activity in vivo and in vitro67,68.
At intrinsic sites, a nucleic acid signal triggers elongation complex dissociation. Recognition of these signals by the elongation complex does not require any factors but can be enhanced by accessory proteins, such as the general transcription elongation protein NusA10. A canonical intrinsic terminator that has been characterized in great detail in Escherichia coli is an RNA signal composed of a GC-rich RNA hairpin followed by a run of U residues. Termination occurs in two steps: RNAP pausing within the U track11, followed by RNA release. In contrast to the direct role of RNA hairpins that induce pausing by interactions with the β-flap domain of RNAP12, the role of the termination hairpin appears to be indirect as it can be replaced by oligonucleotides that pair to the nascent RNA to mimic the hairpin13–16. Intrinsic terminator signals can be easily identified by computational approaches and generate ~80% of the RNA ends in E. coli17. However, Thermus thermophilus RNAP responds poorly to canonical E. coli terminators in vitro18, and many bacterial and archaeal genomes lack intrinsic terminator signals, implying the existence of a different type of signal or dependence on a termination factor in these species19.
Termination at factor-dependent signals depends on the action of a regulatory protein, such as Rho20. Rho-dependent termination generates the 3′ ends of ~20% of messages, including both mRNAs and stable small RNAs, and may regulate widespread antisense transcription17. Rho also carries out several quality control tasks within genes: it mediates polarity21, guards E. coli against expression of horizontally acquired DNA22 and helps to resolve deleterious R-loops23. Rho has been proposed to remove the slow RNAPs that escaped modification to an antitermination state from the rrn operons24 and, most recently, to guard the chromosome against double-stranded DNA breaks by removing obstructing elongation complexes from the path of an advancing replisome25. In every case, Rho acts on a naked nascent transcript that is not engaged in translation or bound to RNA-binding proteins.
Rho-dependent termination signals are complex and cannot be easily predicted by sequence analysis. The Rho utilization (rut) site is a pyrimidine-rich RNA element that is over 30-nucleotides long which can be separated from the site of its action by hundreds of nucleotides26. Rho is an ATP-dependent translocase that binds to unstructured and ribosome-free RNAs and uses the energy that is liberated from ATP hydrolysis to move along the transcript until it catches up with the elongation complex27. Rho can then use its translocase activity to push the RNAP forward or pull the nascent RNA out, or use its helicase activity to unwind the RNA–DNA hybrid. Even though recent studies suggest that Rho can associate with RNAP throughout all the steps of transcription28,29, it drives RNA release only at certain sites17. The positions at which Rho induces RNA release frequently correspond to the pause sites where RNAP halts in the absence of Rho30. Whether Rho contacts RNAP at all times or only at a terminator site, it must be recruited to — and translocate along — the RNA to induce its release from the elongation complex.
Types of antitermination mechanisms
Given the dramatic outcome of termination, mechanisms that control it would be expected to have similarly dramatic effects on gene expression. Proteins, small molecules, RNAs and even temperature can modulate the efficiency of termination, thereby affecting the expression of downstream genes. Antiterminators can have a passive or an active effect on RNAP. In the first case, the action of the termination signal itself is compromised (for example, by the antiterminator preventing the formation of the RNA hairpin or the action of Rho), but RNAP is unaltered. For example, factors that inhibit Rho binding to RNA or translocating along the RNA (such as YaeO31 and the phage protein polarity suppression protein (Psu)32, respectively) will inhibit termination. In the second case, the action of a bound protein or RNA promotes RNAP bypassing otherwise functional termination signals. In this scenario, also called processive antitermination33, RNAP reads through many consecutive terminators.
Overriding a single terminator
The majority of known antitermination mechanisms are passive (TABLE 1) and are specific for intrinsic terminators, in part because these signals are simple and induce termination at a defined position. Exclusion of Rho binding has also been reported34, but this is a unique case. The transcribed leader regions of many operons fold into at least two mutually exclusive RNA structures: a terminator and an antiterminator (FIG. 3a). Switching between different conformations of the leader, and thus the expression of the downstream structural genes, is controlled by diverse regulators that include accessory proteins, small molecules, uncharged tRNAs and translating ribosomes. Importantly, once the RNA structure is formed, it cannot be remodelled within the timescale of transcription. Thus, each effector must act while the regulatory region is being transcribed. In this Review, we briefly describe a few examples; several excellent reviews offer a comprehensive overview of these mechanisms35–38.
Table 1.
Antitermination regulators in bacteria and phages
| Antiterminator | DNA and/or RNA requirements | Sites of action | Cofactors and mechanisms | Known or predicted elongation complex interaction sites |
|---|---|---|---|---|
| Processive antiterminators encoded by bacteria | ||||
| RfaH | ops sequence | Horizontally acquired operons | No cofactors | β′-clamp helices, β-gate loop and the non-template strand of the transcription bubble |
| S4 | nut and boxA | rrn operons | NusA, NusB, S10 and NusG | RNA and the RNA exit channel |
| Processive antiterminators encoded by phages | ||||
| Phage λ protein N | nut and boxA | Phage λ tL and tR | Can function alone; NusA, NusB, S10 and NusG provide processivity | RNA and the RNA exit channel |
| Phage λ protein Q | qut | Phage λ tR′ | Can function alone; but NusA can stimulate activity | β-flap and the RNA exit channel |
| Phage HK022 put RNAs | None | Phage HK022 tL and tR | No cofactors | β′-zinc-finger and the β′-clamp |
| Phage Xp10 p7 | None | Phage Xp10 operons | No cofactors; direct binding to Xanthomonas oryzae RNAP inhibits RNA release | β′-subunit amino terminus and the upstream edge of the transcription bubble |
| Passive, site-specific cellular antiterminators encoded by bacteria | ||||
| YaeO | None | Binds directly to Rho, blocking Rho–RNA interactions | No cofactors | None |
| HutP | Intergenic RNA downstream of hutP | Bacillus subtilus hut operon | His binding is necessary to allow RNA binding | None |
| GlcT | Leader sequence of pstG | pstG | Only non-phosphorylated GlcT can bind the leader RNA | None |
| LicT | RAT element in the licS leader | Terminator sequence upstream of licS | β-glucan-binding is necessary to allow RNA binding | None |
| BglG | Leader sequence of the bgl operon | bgl operon | BglF-mediated phosphorylation dictates dimerization and RNA binding | None |
| Riboswitches and T-box systems | Leader RNA | Many and varied | Small molecules, cations, proteins, metabolites and tRNAs | None |
| Attenuation through ribosome positioning | Leader RNA | Typical of amino acid biosynthesis operons | Positioning of the stalled ribosome blocks terminator formation | None |
| Rho inhibition through ribosome positioning | Leader RNA | Typical of amino acid biosynthesis operons | Stalled ribosome occludes rut or hinders Rho–NusG interactions | None |
| Csp proteins | RNA hairpins | Global cellular activities | Direct Csp–RNA interactions destabilize RNA structures | None |
| Passive, site-specific antiterminators encoded by phages | ||||
| Psu | None | Binds directly to Rho and reduces ATP hydrolysis and translocation | No cofactors | None |
bgl, β-glucoside utilization; Csp, cold-shock protein; hut, histone utilization; HutP, hut operon positive regulatory protein; GlcT, PtsGHI operon antiterminator; licS, a secreted β-endoglucanase; LicT, transcription antiterminator protein LicT; N, phage λ protein N; nut, N utilization; ops, operon polarity suppressor; Psu, polarity suppression protein; put, polymerase utilization; pstG, glucose permease; Q, phage λ protein Q; qut, Q utilization site; RAT, ribonucleic antiterminator; rut, Rho utilization site; RNAP, RNA polymerase; S4, ribosomal protein S4; S10, 30S ribosomal protein S10 (also known as NusE); tL, terminator of the left operon; tR, terminator of the right operon; YaeO, inhibitor of Rho.
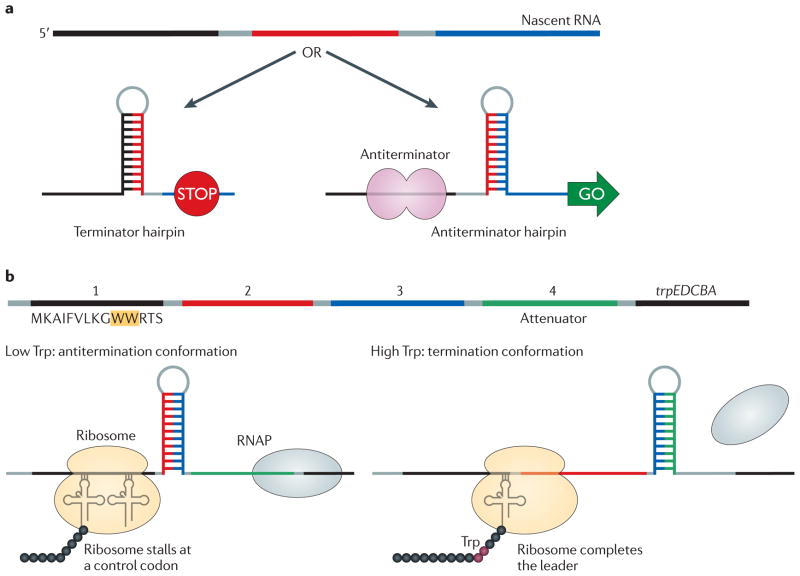
Figure 3. Differential folding of a nascent RNA.
a | The leader regions of many operons encode RNA elements that can base pair with different segments of the same mRNA. In a simple scenario, an upstream terminator will form and transcription will stop unless the formation of the terminator hairpin is prevented — for example, by an RNA-binding antiterminator protein. In this case, an antiterminator hairpin will form instead. The number and complexity of alternative structures and the regulatory mechanisms that control the fate of the nascent RNA vary greatly among operons, and new regulators remain to be identified. b | Attenuation in the Escherichia coli trp operon. The leader region upstream of the structural trp genes encodes a 14-residue leader peptide (region 1) and several RNA elements that can form alternative secondary structures. Region 3 can base pair with either region 2 (to form an antiterminator hairpin) or region 4 (to form a terminator hairpin). When Trp levels are low, the ribosome stalls at one of two Trp codons (at positions 10 and 11 within the leader peptide), the antiterminator hairpin forms and Trp biosynthesis genes are expressed, leading to an increase in the concentration of Trp. When Trp levels are high, the ribosome advances into region 2 and blocks the antiterminator. The terminator structure forms instead, and RNA polymerase (RNAP) dissociates upstream of the trpE gene.
Antitermination by an RNA-bound protein
Many proteins bind to RNA and disfavour formation of RNA hairpins, either directly (by preferentially binding to single-stranded RNA) or indirectly (by stabilizing a competitive alternative structure). In most cases, the RNA binding is sequence specific, and the antitermination activity regulates expression of only a few genes. An exception is the global response to low temperature mediated by cold shock proteins, which are RNA chaperones that are overproduced after a temperature downshift and that bind and stabilize single-stranded RNAs39. Binding of site-specific antiterminators to RNA is usually controlled by conformational changes in the antiterminator upon ligand binding or as a result of covalent modification such as phosphorylation. The structural basis of the underlying molecular mechanisms has been described for only a few RNA-binding regulators. Among these are antiterminator proteins His utilization (hut) operon positive regulatory protein (HutP)40, PtsGHI operon antiterminator GlcT41 and transcription antiterminator LicT42.
HutP regulates the hut operon in Bacillus subtilis in response to L-His. A HutP hexamer binds to an untranslated RNA region that separates the hutP gene from the downstream genes encoding enzymes which degrade His. HutP binds RNA only when activated by His, which induces significant structural rearrangements of the residues involved in RNA binding40. HutP binds to six NAG triplets, three just upstream of and three within the terminator structure. Binding of HutP disrupts the terminator and allows expression of the hut genes43. Several homologous antitermination systems regulate the expression of sugar-metabolizing operons in B. subtilis44. Each of these systems is composed of a sensory sugar permease, an antiterminator protein and a regulatory RNA region that folds into at least two mutually exclusive structures. GlcT is a dimeric antiterminator that binds to the leader of the ptsG gene, which encodes a glucose permease. GlcT binds to and stabilizes an antiterminator hairpin, thereby preventing the formation of an overlapping terminator and allowing transcription into the ptsG gene. This regulatory system couples the availability of the inducer, glucose, to the phosphorylation state of the antiterminator: only the non-phosphorylated GlcT can form a dimer and bind RNA41. In the absence of glucose, the permease transfers a phosphate to a His residue on GlcT, thereby inactivating the antiterminator. If glucose is present, the phosphate groups are transferred from PtsG to the sugar instead. In LicT, an antiterminator homologous to GlcT that controls transport and metabolism of β-glucosides, the sensor domain is connected to the RNA-binding domain by a linker that undergoes a helix–coil transition upon inducer binding to allow LicT to bind its RNA target42. The related antiterminator, BglG, is regulated both by reversible phosphorylation and by sequestration into an inactive complex45. Regulators that contain ANTAR RNA-binding domains46 may use similar mechanisms to block terminator hairpin formation47,48.
Antitermination by a stalled ribosome
In most Gram-negative bacteria, the ribosome controls expression of amino acid biosynthesis operons in response to the availability of the cognate amino acid by sensing the level of charged tRNA49. In the E. coli trp operon, a small leader peptide contains two tandem Trp codons (FIG. 3b). When the levels of Trp and charged tRNATrp are high, the ribosome completes the synthesis of the leader peptide, the terminator forms and RNAP is released. Conversely, at low levels of charged tRNATrp the ribosome stalls within the leader peptide, which allows the antiterminator hairpin to form; the RNAP then proceeds into the Trp biosynthesis genes. Ribosome stalling is also used to control expression of the E. coli tna operon, which allows bacteria to use Trp as a nitrogen or carbon source. The structural tryptophanase genes are preceded by a leader region that encodes a short peptide, the tryptophanase leader peptide (TnaC). In the absence of Trp, Rho terminates transcription downstream of the tnaC stop codon50. When Trp is present, it binds to the ribosome and inhibits RF2-mediated cleavage of the nascent leader peptide, stalling the ribosome that is translating tnaC mRNA51. The stalled ribosome occupies the overlapping rut sequences, thus preventing Rho binding and increasing transcription of the tna operon50.
Antitermination by uncharged tRNAs
In most Gram-positive bacteria, tRNA charging is sensed directly — independently of the ribosome52 — through interactions between the tRNA and the leader transcript, which also encodes a terminator and an alternative antiterminator53. The antiterminator includes a conserved 14-nucleotide sequence called the T-box (see REF. 54 for a recent review). Expression of genes that are regulated by a T-box (including genes that encode proteins involved in amino acid metabolism and the aminoacyl tRNA synthetases) is induced by stabilization of an antiterminator in the leader RNA by the cognate uncharged tRNA. The specificity of this response depends on a specifier codon in the leader that pairs with the anticodon of the corresponding tRNA53. Interactions between a T-box leader mRNA (some of which fold into very complex structures) and a tRNA are independent of accessory proteins, involve several parts of the tRNA, are accompanied by structural changes in both partners and are kinetically controlled54. Direct sensing of tRNA charging may also be used to co-regulate the translational capacity of the cell in response to stress55.
Antitermination by a small molecule
Riboswitches are mRNA leader regions that undergo structural rearrangements56 in response to changes in cellular ion concentrations57 or upon binding of a small molecule such as flavin mononucleotide, a purine, lysine, S-adenosyl-L-methionine and thiamin pyrophosphate (recently reviewed in REFS 35, 37, 58). These ligands bind to their RNA targets with high affinity and selectivity in the absence of accessory proteins59–61. Riboswitches encode various regulatory elements: binding of an effector favours one structural configuration of the riboswitch over another, thereby altering expression of the downstream genes. Most known riboswitches stabilize an ‘off’ state; however, the leader of the B. subtilis gene ydhL (which encodes an adenine efflux pump) folds into a very stable terminator structure that is disrupted upon binding to adenine62, leading to derepression of the gene. As the mechanism of switching is conceptually the same whether a terminator or an antiterminator is favoured, other riboswitches that activate expression are likely to exist.
Antitermination by translation
Transcription and translation are coupled in bacteria and archaea63,64, allowing for their coordinated regulation. Concurrent translation plays a key part in uninterrupted RNA synthesis27,65; if a message is not translated (for example, when a premature nonsense codon is inserted), it will be terminated by the action of Rho, unless the RNAP is modified by an antiterminator (see below). In one model, translating ribosomes occlude the nascent RNA, blocking the binding of Rho to the rut element. In another model, the ribosome competes with Rho for binding to NusG66. Contacts between Rho and NusG, which play a key part in Rho-mediated termination67,68, cannot be established if NusG forms a complex with the 30S ribosomal protein S10 (also known as NusE), as recently observed by NMR69. Interactions between NusG and S10 are thought to tether the trailing ribosome to the elongation complex to establish a functional connection between the two complexes69, which controls the rate of transcription in response to the cellular translational capacity70. Thus, a tethered ribosome may function as an antiterminator by preventing Rho binding to mRNA, inhibiting Rho-mediated RNA release by sequestering the Rho-interaction surface on NusG, and blocking formation of a terminator hairpin. The trailing ribosome may also push RNAP forward, inhibiting pausing and preventing transcription arrest70.
Bypassing multiple consecutive terminators
The signal-specific mechanisms described above balance gene expression of the target operon in response to a regulatory signal, such as the concentration of a metabolite. By contrast, processive antitermination mechanisms increase the probability that RNAP reaches the end of an operon containing several intragenic termination signals. Phage and bacterial antiterminators differ in several respects: phage regulators typically modify RNAP to allow read-through of all terminators, whereas the bacterial regulators ribosomal protein S4 and RfaH do not have strong effects at intrinsic sites5,71. Some bacterial antiterminators may have evolved to decrease the efficiency of intrinsic terminators, but inhibition of the activity of Rho is probably their main target. The essential ribosomal RNA operons are not translated and would be an easy target for Rho in the absence of the S4-containing rrn antitermination complex. Many other key genes in bacteria (such as the cell wall biosynthesis operons, which are the targets of RfaH) have been acquired through horizontal transfer and are translated inefficiently because of their codon bias; transcription of these genes would be terminated by Rho22 unless an anti-Rho mechanism were in place. In addition, cellular antiterminators should permit RNAP release at the end of an operon, and most of the intergenic terminators are intrinsic17. By contrast, phage antiterminators are likely to have a more complex set of tasks to execute. Numerous hairpin-dependent and Rho-dependent terminators exist in the genome of phageλ, and some of these signals are conserved in other lambdoid phages, suggesting that they may play important roles during the phage life cycle. Many proposals have been offered to explain why phage λ retained its terminators and antiterminators, from stabilization of the lysogenic state by the phage λ protein N to a rapid restart of late gene transcription by phage λ protein Q72.
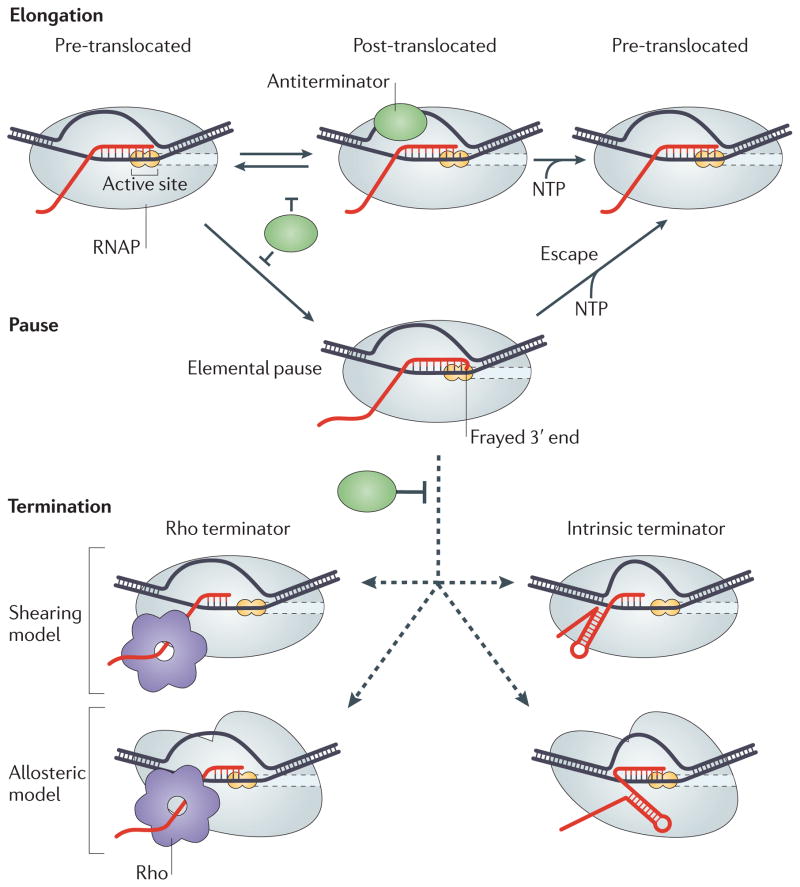
How can a regulator prevent termination at multiple sites? The current model of transcript elongation control posits that RNAP undergoes structural rearrangements at certain sites in response to signals that are encoded in DNA and RNA (FIG. 4). These rearrangements lead to the formation of an ‘elemental pause’ state from which long-lived pause, arrest and termination intermediates arise73. The elemental pause state forms from a pre-translocated elongation complex and is characterized by a frayed 3′ end of the nascent RNA74 that precludes rapid nucleotide addition. There are three mechanisms by which an antiterminator can act on RNA: it can favour the post-translocated state of the elongation complex; it can anchor the 3′ end in the active site; and it can block Rho access to the RNA or prevent formation of the terminator hairpin. The first two mechanisms will necessarily affect both intrinsic and factor-dependent terminators, as well as RNAP pausing and arrest, whereas the third mechanism can be signal specific. We briefly describe the known antitermination mechanisms (FIG. 5) below and discuss E. coli RfaH as an example of an antiterminator that uses several strategies to maximize its effects.
Figure 4. A model for termination and antitermination.
During rapid elongation (top row), the active site of RNA polymerase (RNAP) is optimized for catalysis. After the enzyme has added a nucleotide to the nascent RNA, the enzyme is in the ‘pre-translocated state’ where the 3′ nucleotide of the RNA occupies the downstream half of the active site. The enzyme then translocates along the DNA to form the ‘post-translocated state’, in which the 3′ OH group of the nascent RNA is positioned in the upstream half of the active site, and the incoming nucleoside 5′-triphosphate (NTP) can readily bind to the vacant downstream half-site of the active site and is subsequently incorporated into the RNA. When the RNAP reaches a pause site, the 3′ OH group remains in the pre-translocated configuration, inhibiting binding of the NTP substrate, and the active site undergoes a rearrangement to yield the elemental pause intermediate in which the 3′ OH may be frayed74,109. From this intermediate, RNAP can either escape upon addition of another nucleotide or isomerize into a termination complex. Two mechanisms for formation of the termination complex are currently debated. In the first (shearing or hyper-translocation) model, the RNA 3′ end is lost from the active site when the nascent RNA is pulled upstream by Rho or an RNA hairpin or when the RNAP is pushed forward14,15,104. In the second (allosteric) model, Rho or terminator hairpin formation induces dramatic changes within the complex (indicated by an altered shape of the enzyme) but the 3′ end of the RNA is retained in the active site28,101. An antiterminator protein can directly block formation of the termination complex, prevent isomerization into the elemental pause or stabilize the elongation complex against dissociation.
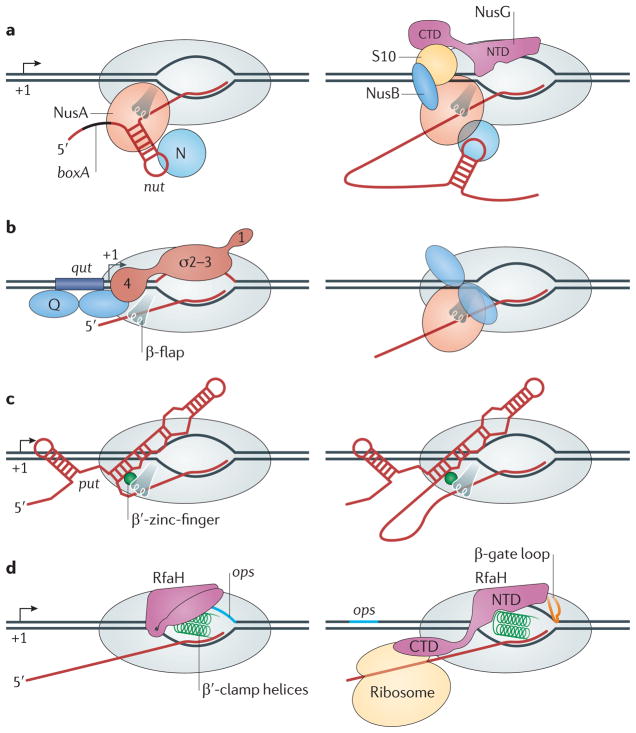
Figure 5. Processive antitermination mechanisms.
In each panel, only the RNA polymerase (RNAP) elements implicated in antitermination are shown. a | Antitermination by phage λ protein N requires assembly of a large ribonucleoprotein complex on two RNA elements, boxA and the N utilization (nut) RNA hairpin. N can directly bind the nut hairpin on its own and it allows RNAP to read through a single terminator118 (left). However, establishing the long-lived termination-resistant modification of RNAP also requires several host Nus proteins (NusA, NusB, 30S ribosomal protein S10 (also known as NusE) and NusG) to stabilize the antiterminator complex through a network of interactions (right)76. b | Phage λ protein Q recruitment to RNAP requires the Q utilization (qut) DNA element, which overlaps the promoter and directly binds to Q, and a promoter-proximal pause, which is induced by interactions of region 2 of initiation factor-σ with a –10-like promoter element in the transcribed DNA81; σ-factor region 4 also interacts with Q (REF. 80). After recruitment, Q turns into an RNAP subunit and modifies the enzyme into a processive state. NusA can stimulate Q activity. c | Antitermination by polymerase utilization (put) RNA is independent of accessory proteins but does require stable binding of put RNA to the enzyme119. d | The amino-terminal domain (NTD) of RfaH binds to the non-template DNA in the elongation complex paused at the operon polarity suppressor (ops) site (left), triggering domain dissociation, which in turn unmasks the site that binds to the β′-clamp helices92. RfaH remains bound to the elongation complex through the NTD contacts with the β′-clamp helices, β-gate loop and non-template DNA (right). CTD, carboxy-terminal domain.
Phage λ protein N
N was the first reported example of an antiterminator. It allows the transition between the early and middle stages of phage λ transcription by suppressing several Rho-dependent terminators20. N is recruited to the elongation complex through interactions with an N utilization (nut) RNA hairpin (FIG. 5a) and can act alone in the vicinity of the nut site75; however, its action at a distance requires the assembly of a multipartite complex that includes the bacterial NusA, NusB, S10 and NusG proteins76. N suppresses pausing and termination at both intrinsic and Rho-dependent sites, in part by stabilizing the elongation complex, thereby preventing RNAP dissociation77. At intrinsic termination sites, N, in conjunction with NusA, is proposed to bind to the 5′ portion of the terminator hairpin and preclude its formation78. It is not known whether the N-containing antiterminator complex actively inhibits Rho by blocking its access to RNA79 or simply speeds up the RNAP, allowing it to escape from the advancing Rho.
Phage λ protein Q
Q mediates antitermination in the phage λ late operon. During recruitment, a Q dimer interacts with a specific double-stranded DNA site (the Q utilization (qut) site) just upstream of the transcription start site of the late promoter, PR′, and with region 4 of initiation factor-σ (REF. 80) in an elongation complex paused near the late promoter4 (FIG. 5b). After recruitment, Q travels with RNAP over long distances and modifies the enzyme into a termination-resistant form; Q activity is enhanced by NusA in vitro, but it is unclear whether other cellular factors are involved81. The model for Q-mediated antitermination14 suggests that Q acts by inhibiting pausing and, thus, increasing the kinetic barrier to termination and interfering with the formation of a terminator hairpin. Acting jointly with NusA, Q protects the nascent RNA from the action of Rho and termination-inducing oligonucleotides, which anneal to the nascent RNA to mimic the hairpin16, implying that Q and NusA form a shield around the transcript. The recent identification of the β-flap of RNAP as a target of Q is consistent with this direct exclusion mechanism and suggests an explanation for the Q-mediated stabilization of the elongation complex82. When it acts by itself, Q is likely to prevent termination through its anti-pausing activity16.
Antitermination in the rRNA operons
The untranslated rrn operons are especially vulnerable to termination by Rho owing to their length and lack of translation. Several mechanisms of antitermination prevent the effect of Rho in these operons. First, ribosomal RNA transcripts fold into elaborate secondary structures that can mask rut sites. Second, ribosomal proteins interact with the nascent rRNA co-transcriptionally83, shielding the transcript from Rho. Third, an rrn antitermination complex modifies the RNAP into a Rho-resistant state84. This complex is composed of the same nucleic acid and protein determinants as the phage λ N complex85, except that N is substituted with S4, which directly binds to RNAP and can act as an antiterminator on its own71. It is unknown whether Rho activity is inhibited directly (by steric occlusion) or kinetically (the synthesis rate of rRNA is much higher than that of coding genes) and whether S4 is the key antiterminator in this complex.
Phage HK022 put RNAs
The transcripts of the genes polymerase utilization L (putL) and putR fold into two RNA stem-loop structures (FIG. 5c). One arm of each put RNA binds directly to RNAP throughout elongation; put-modified enzymes transcribe at faster rates than enzymes that are not bound by put RNAs and read through terminators located thousands of base pairs downstream86. The activity of the put transcripts can be reconstituted in vitro in the absence of accessory factors and is thought to be mediated by a direct interaction with the β′-zinc-finger87. The put RNAs inhibit RNAP backtracking near the site of recruitment by blocking the re-entry of the transcript into the RNA exit channel88, but the mechanism of long-range antitermination modification and the involvement of any cellular proteins remain unknown. A recently described cis-acting RNA element (eps-associated RNA) that activates expression of exopolysaccharide genes in B. subtilis89 may use a similar mechanism to modify RNAP into the antitermination state.
RfaH
RfaH is a specialized paralogue of NusG that increases the expression of distal genes in several operons in E. coli and related bacteria90. Unlike NusG, which is essential in wild-type E. coli68 and is associated with RNAP during transcription of most of the E. coli str. K12 substr. MG1655 genes29, RfaH is dispensable for growth of commensal E. coli and targets only those operons that have a 12-nucleotide operon polarity suppressor (ops) element in their untranslated leader regions91. The ops element (FIG. 5d) mediates sequence-specific binding of RfaH to the non-template DNA5 and may induce elongation complex isomerization into the distinct state that is necessary for RfaH recruitment. After recruitment, RfaH remains bound to RNAP throughout the transcription of an entire operon in vivo91 and reduces pausing and termination in vitro91,92.
E. coli RfaH and NusG consist of two domains92,93. The amino-terminal domains are very similar; they interact with the β′-clamp helices and mediate the anti-pausing effects of RfaH and NusG in vitro. By contrast, the carboxy-terminal domains of these two proteins have very different structures (an α-helical hairpin in RfaH and a β-barrel in NusG) and confer protein-specific functions. In RfaH, the C-terminal domain masks the RNAP-binding site on the N-terminal domain unless the ops site is present in the transcribed DNA, thereby restricting RfaH action to ops-containing operons92. In NusG, the C-terminal domain interacts with Rho to increase Rho-dependent termination93,94 and with the ribosomal protein S10 to couple transcription to translation69,70.
Recent studies reveal that NusG and RfaH have opposite regulatory roles in the cell. NusG acts in concert with Rho to inhibit the expression of foreign genes22, whereas RfaH inhibits Rho action and, thus, activates expression of laterally acquired genes91. RfaH reduces Rho-mediated polarity by several independent mechanisms.
First, RfaH tightly binds to the β′-clamp helices and competes against a large excess of NusG91 or σ-factor95 in vitro. NusG is also absent from RfaH-controlled operons in vivo91. Thus, RfaH excludes NusG from the elongation complex, thereby inhibiting Rho-mediated RNA release. Second, RfaH modifies RNAP into a pause-resistant state, thereby inhibiting Rho, which preferentially targets paused RNAPs30. This effect is composed of two components. RfaH induces forward translocation of RNAP96, which is likely to be because it favours the DNA strand reannealing at the upstream portion of the transcription bubble, and RfaH may also inhibit isomerization into the elemental pause state (FIG. 4) by controlling the β′-clamp movements through its contacts with the β′-clamp helices96. Finally, RfaH may limit Rho access to poorly translated ops-containing operons by recruiting ribosomes. We argue that the stable in vivo association of RfaH with the elongation complex91 requires sequestration of the RfaH C-terminal domain, probably by the ribosome. Such an interaction has been detected directly97, but it remains unknown whether, similarly to the NusG–elongation complex interaction69, it has a regulatory role.
Summary and perspectives
Two classes of regulators help RNAP to read through terminators. The larger class comprises proteins, RNAs and small molecules that act on a terminator. These regulators control transcription in response to, for example, metabolites, cellular translation capacity and environmental conditions. Their mechanisms are incredibly diverse but share a common theme: they all affect the elongation complex indirectly. By contrast, regulators from the second class modify RNAP into a processive, pause- and terminator-resistant state.
Although antiterminator proteins have been studied for more than 40 years33, the molecular mechanism of antitermination has been slow to emerge. First, the structures of antiterminators and their binding sites on the elongation complex remained unknown. Second, the detailed molecular mechanism of termination remains unclear, in part owing to the transient nature of termination intermediates that are difficult to characterize even in solution, let alone capture in a crystal.
The RNA–DNA hybrid is the key element that determines the high stability of the elongation complex7,98,99. Three models can explain how Rho or a terminator hairpin destabilizes the hybrid, bringing about the dissociation of the elongation complex (see REFS 66, 100 for detailed analyses). In the allosteric model, structural changes in the active-site cleft that are induced by an RNA hairpin12,101 or Rho28 weaken interactions with the hybrid and destabilize the elongation complex. In the forward translocation model, RNAP is pushed forward without adding nucleotides to the RNA, thereby shortening the hybrid using the energy of hairpin formation, or ATP hydrolysis by Rho or Mfd (also known as TRCF)15,102, or the energy of Trp RNA-binding attenuation protein (TRAP) binding103. In the hybrid-shearing model, the RNA is extracted from the stationary RNAP in the absence of translocation11,104, and the elongation complex dissociates. Some biochemical data argue in favour of the allosteric model of intrinsic termination101, whereas other biochemical and single-molecule analyses support either the forward translocation or the shearing mechanism, depending on the signal15,104–106. A combination of approaches and analysis of diverse termination signals will be required to identify the features that dictate the preferred termination pathway at each site.
During termination, RNAP interactions with the nascent RNA are altered, and pausing is thought to precede RNA release. An antiterminator can shield the RNA near the exit channel or inhibit pausing, thereby kinetically favouring the elongation pathway. The exit channel is formed between two mobile domains: the β-flap and the β′-clamp (which includes the clamp helices and several flexible loops, the lid, the zipper and the zinc-finger (FIG. 6)). An antiterminator that stabilizes the RNA in the exit channel through contacts with either the β-flap or β′-clamp will resist the action of Rho or a nascent hairpin. Indeed, many antiterminators appear to target the exit channel: Q binds to the β-flap82, N binds to the nut hairpin that probably binds to NusA and the β-flap12,78,107, put RNA is likely to interact with the β′-zinc-finger88, and phage Xp10 p7 protein binds to the N terminus of the β′-subunit that is disordered in all current structures but may lie near the RNA exit108. N appears to inhibit formation of the terminator hairpin78. Q, acting in concert with NusA, protects ~10 additional nucleotides of the emerging transcript from nuclease digestion and resists the action of Rho and hairpin-mimicking oligonucleotides16.
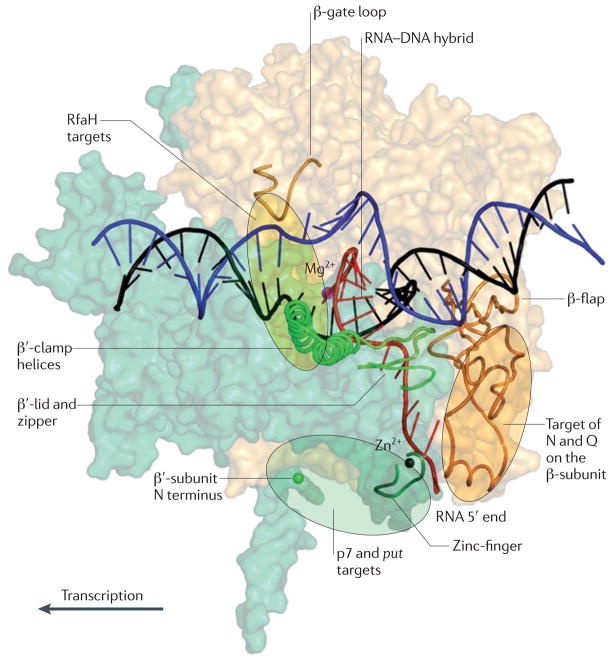
Figure 6. Targets for processive antiterminators.
The β-subunit (light yellow) and β′-subunit (light green) of RNA polymerase (RNAP) are shown in surface representation, with the key elements shown as cartoons. The non-template DNA strand is shown in blue and the template DNA strand in black. The active-site Mg2+ ion (magenta) and Zn2+ ion (black) in the zinc-finger motif are shown. The amino terminus of the β′-subunit is marked with a green sphere. The target sites for the antitermination factors are shown as transparent ovals. RfaH has two targets, the β′-clamp helices and the β-gate loop. N, phage λ protein N; put, polymerase utilization; Q, phage λ protein Q.
Anti-pausing activity is also common among antiterminators but its detailed mechanism remains unknown. Antiterminators may target the RNAP clamp73 because the closed state of the clamp is thought to be required for processive elongation. At a pause site, the clamp may partially open to allow for the fraying of the 3′ nucleotide out of the active site74,109. At a terminator, the clamp could open even further to permit release of the DNA; the allosteric models of termination invoke large movements of the clamp28,101. By stabilizing the clamp, a regulator could favour the active, catalytically competent state of the elongation complex. Our recent studies suggest that RfaH reduces pausing by this mechanism. RfaH binds simultaneously to the β′-clamp helices and β-gate loop110, the two elements located on the opposite sides of the main channel (FIG. 6), and bridges the gap across the downstream DNA. The continuous clamp formed by the β′-clamp helices, RfaH and β-gate loop ensures that the clamp remains locked even when the RNAP encounters a pause signal. Other, structurally unrelated proteins may inhibit pausing and termination by locking the clamp through binding to the same or different determinants on RNAP.
The clamp locking mechanism may be ancient and ubiquitous. In all domains of life, RNAP must overcome numerous barriers, such as sequences that induce pausing, DNA-bound regulators or DNA-packaging proteins (for example, nucleoid-associated proteins in bacteria and crenarchaeota, and nucleosomes in eukaryotes and euryarchaeota). The structural organization of the multi-subunit RNAPs is highly conserved, suggesting that control of RNAP processivity is also universal. Indeed, RfaH belongs to the only known group of ubiquitous transcription elongation factors111, and the archaeal Spt5 protein has been reported to increase elongation through contacts to the clamp helices112 and to bridge the main channel113,114.
Recent structural studies of RNAP and its complexes, genome-wide localization of transcription factors, single-molecule analyses and conventional solution studies have led to tremendous advances in our understanding of the mechanism and regulation of RNA chain elongation. To understand termination and its control by accessory factors, we must now probe the structure and dynamics of termination intermediates in real time.
Acknowledgments
We thank N. Ruiz and the anonymous referees for their help in improving the manuscript. This work was supported by the US National Institutes of Health grants GM67153 (to I.A.) and F32-GM073336 (to T.J.S.).
Glossary
- Pause site
A signal that causes the elongation complex to stall temporarily.
- Arrest site
A signal at which the elongation complex comes to a complete halt but does not dissociate.
- Polarity
A quality control mechanism in which Rho terminates the transcription of mRNAs that are not translated.
- R-loops
DNA loops behind RNA polymerase that are created when the nascent RNA invades the DNA duplex and pairs with the template DNA strand.
- rrn operon
The operons that contain ribosomal RNA and tRNA genes.
- Leader regions
Regions of DNA that separate promoters from structural genes. Leaders play diverse regulatory roles as riboswitches, attenuators and targets for auxiliary factors.
- RF2
Release factor 2. A release factor that mediates hydrolysis of the peptidyl-tRNA ester bond at UAA and UGA stop codons.
- Zinc-finger
A small protein motif in which the structure is stabilized by a bound Zn2+ ion
- Transcription bubble
A 12–14-nucleotide region in which the template and the non-template DNA strands are separated by the RNA polymerase
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Irina Artsimovitch’s homepage:http://microbiology.osu.edu/faculty/artsimovitch-irina
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Mooney RA, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grachev MA, et al. Oligonucleotides complementary to a promoter over the region −8 + 2 as transcription primers for E. coli RNA polymerase. Nucleic Acids Res. 1984;12:8509–8524. doi: 10.1093/nar/12.22.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin H, et al. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 4.Ring BZ, Yarnell WS, Roberts JW. Function of E. coli RNA polymerase σ factor σ 70 in promoter-proximal pausing. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 5.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 6.Landick R, Carey J, Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci USA. 1985;82:4663–4667. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 8.Toulme F, Mosrin-Huaman C, Artsimovitch I, Rahmouni AR. Transcriptional pausing in vivo: a nascent RNA hairpin restricts lateral movements of RNA polymerase in both forward and reverse directions. J Mol Biol. 2005;351:39–51. doi: 10.1016/j.jmb.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 9.Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt MC, Chamberlin MJ. NusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol. 1987;195:809–818. doi: 10.1016/0022-2836(87)90486-4. [DOI] [PubMed] [Google Scholar]
- 11.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 12.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 13.Artsimovitch I, Landick R. Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release. Genes Dev. 1998;12:3110–3122. doi: 10.1101/gad.12.19.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 15.Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 16.Shankar S, Hatoum A, Roberts JW. A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mol Cell. 2007;27:914–927. doi: 10.1016/j.molcel.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters JM, et al. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sevostyanova A, Artsimovitch I. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 2010;38:7432–7445. doi: 10.1093/nar/gkq623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santangelo TJ, Cubonova L, Skinner KM, Reeve JN. Archaeal intrinsic transcription termination in vivo. J Bacteriol. 2009;191:7102–7108. doi: 10.1128/JB.00982-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- 21.Richardson JP, Grimley C, Lowery C. Transcription termination factor Rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci USA. 1975;72:1725–1728. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardinale CJ, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harinarayanan R, Gowrishankar J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J Mol Biol. 2003;332:31–46. doi: 10.1016/s0022-2836(03)00753-8. [DOI] [PubMed] [Google Scholar]
- 24.Klumpp S, Hwa T. Stochasticity and traffic jams in the transcription of ribosomal RNA: intriguing role of termination and antitermination. Proc Natl Acad Sci USA. 2008;105:18159–18164. doi: 10.1073/pnas.0806084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Washburn RS, Gottesman ME. Transcription termination maintains chromosome integrity. Proc Natl Acad Sci USA. 2011;108:792–797. doi: 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson J, Greenblatt J. In: Escherichia coli and Salmonella. Neidhardt FC, et al., editors. Vol. 1. ASM Press; Washington DC: 1996. pp. 822–848. [Google Scholar]
- 27.Ciampi MS. Rho-dependent terminators and transcription termination. Microbiology. 2006;152:2515–2528. doi: 10.1099/mic.0.28982-0. [DOI] [PubMed] [Google Scholar]
- 28.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mooney RA, et al. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan WD, Bear DG, von Hippel PH. Specificity of release by Escherichia coli transcription termination factor Rho of nascent mRNA transcripts initiated at the λ PR. J Biol Chem. 1984;259:8664–8671. [PubMed] [Google Scholar]
- 31.Gutierrez P, et al. Solution structure of YaeO, a Rho-specific inhibitor of transcription termination. J Biol Chem. 2007;282:23348–23353. doi: 10.1074/jbc.M702010200. [DOI] [PubMed] [Google Scholar]
- 32.Pani B, et al. Mechanism of inhibition of Rho-dependent transcription termination by bacteriophage P4 protein Psu. J Biol Chem. 2006;281:26491–26500. doi: 10.1074/jbc.M603982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisberg RA, Gottesman ME. Processive antitermination. J Bacteriol. 1999;181:359–367. doi: 10.1128/jb.181.2.359-367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166:217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkin TM. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Smith AM, Fuchs RT, Grundy FJ, Henkin TM. Riboswitch RNAs: regulation of gene expression by direct monitoring of a physiological signal. RNA Biol. 2010;7:104–110. doi: 10.4161/rna.7.1.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amster-Choder O. The bgl sensory system: a transmembrane signaling pathway controlling transcriptional antitermination. Curr Opin Microbiol. 2005;8:127–134. doi: 10.1016/j.mib.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Phadtare S, Severinov K. RNA remodeling and gene regulation by cold shock proteins. RNA Biol. 2010;7:788–795. doi: 10.4161/rna.7.6.13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumarevel T, Mizuno H, Kumar PK. Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand. Nature. 2005;434:183–191. doi: 10.1038/nature03355. [DOI] [PubMed] [Google Scholar]
- 41.Schmalisch MH, Bachem S, Stulke J. Control of the Bacillus subtilis antiterminator protein GlcT by phosphorylation. Elucidation of the phosphorylation chain leading to inactivation of GlcT. J Biol Chem. 2003;278:51108–51115. doi: 10.1074/jbc.M309972200. [DOI] [PubMed] [Google Scholar]
- 42.Demene H, et al. Structural mechanism of signal transduction between the RNA-binding domain and the phosphotransferase system regulation domain of the LicT antiterminator. J Biol Chem. 2008;283:30838–30849. doi: 10.1074/jbc.M805955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gopinath SC, et al. Insights into anti-termination regulation of the hut operon in Bacillus subtilis: importance of the dual RNA-binding surfaces of HutP. Nucleic Acids Res. 2008;36:3463–3473. doi: 10.1093/nar/gkn199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Tilbeurgh H, Declerck N. Structural insights into the regulation of bacterial signalling proteins containing PRDs. Curr Opin Struct Biol. 2001;11:685–693. doi: 10.1016/s0959-440x(01)00267-6. [DOI] [PubMed] [Google Scholar]
- 45.Raveh H, Lopian L, Nussbaum-Shochat A, Wright A, Amster-Choder O. Modulation of transcription antitermination in the bgl operon of Escherichia coli by the PTS. Proc Natl Acad Sci USA. 2009;106:13523–13528. doi: 10.1073/pnas.0902559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fox KA, et al. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci USA. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chai W, Stewart V. RNA sequence requirements for NasR-mediated, nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader. J Mol Biol. 1999;292:203–216. doi: 10.1006/jmbi.1999.3084. [DOI] [PubMed] [Google Scholar]
- 49.Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 50.Gong F, Yanofsky C. Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J Biol Chem. 2002;277:17095–17100. doi: 10.1074/jbc.M201213200. [DOI] [PubMed] [Google Scholar]
- 51.Gong M, Cruz-Vera LR, Yanofsky C. Ribosome recycling factor and release factor 3 action promotes TnaC-peptidyl-tRNA dropoff and relieves ribosome stalling during tryptophan induction of tna operon expression in Escherichia coli. J Bacteriol. 2007;189:3147–3155. doi: 10.1128/JB.01868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grundy FJ, Winkler WC, Henkin T. M tRNA-mediated transcription antitermination in vitro: codon–anticodon pairing independent of the ribosome. Proc Natl Acad Sci USA. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grundy FJ, Henkin T. M tRNA as a positive regulator of transcription antitermination in B subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 54.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murray KD, Bremer H. Control of SpoT-dependent ppGpp synthesis and degradation in Escherichia coli. J Mol Biol. 1996;259:41–57. doi: 10.1006/jmbi.1996.0300. [DOI] [PubMed] [Google Scholar]
- 56.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park SY, Cromie MJ, Lee EJ, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 59.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 62.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 63.French SL, Santangelo TJ, Beyer AL, Reeve JN. Transcription and translation are coupled in Archaea. Mol Biol Evol. 2007;24:893–895. doi: 10.1093/molbev/msm007. [DOI] [PubMed] [Google Scholar]
- 64.Miller OL, Jr, Hamkalo BA, Thomas CA., Jr Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- 65.Santangelo TJ, et al. Polarity in archaeal operon transcription in Thermococcus kodakaraensis. J Bacteriol. 2008;190:2244–2248. doi: 10.1128/JB.01811-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol. 2011 Mar 23; doi: 10.1016/j. jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burns CM, Richardson JP. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc Natl Acad Sci USA. 1995;92:4738–4742. doi: 10.1073/pnas.92.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sullivan S, Gottesman M. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- 69.Burmann BM, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 70.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torres M, Condon C, Balada JM, Squires C, Squires CL. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 2001;20:3811–3820. doi: 10.1093/emboj/20.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campbell A. Comparative molecular biology of lambdoid phages. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 73.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 75.Barik S, Ghosh B, Whalen W, Lazinski D, Das A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987;50:885–899. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- 76.Mogridge J, Mah T, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 1995;9:2831–2845. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 77.Rees WA, Weitzel SE, Das A, von Hippel PH. Regulation of the elongation-termination decision at intrinsic terminators by antitermination protein N of phage λ. J Mol Biol. 1997;273:797–813. doi: 10.1006/jmbi.1997.1327. [DOI] [PubMed] [Google Scholar]
- 78.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 79.Vieu E, Rahmouni AR. Dual role of boxB RNA motif in the mechanisms of termination/antitermination at the lambda tR1 terminator revealed in vivo. J Mol Biol. 2004;339:1077–1087. doi: 10.1016/j.jmb.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 80.Nickels BE, Roberts CW, Sun HI, Roberts JW, Hochschild A. The σ70 subunit of RNA polymerase is contacted by the λ Q antiterminator during early elongation. Mol Cell. 2002;10:611–622. doi: 10.1016/s1097-2765(02)00648-2. [DOI] [PubMed] [Google Scholar]
- 81.Roberts JW, et al. Antitermination by bacteriophage λ Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- 82.Deighan P, Diez CM, Leibman M, Hochschild A, Nickels BE. The bacteriophage λ Q antiterminator protein contacts the β-flap domain of RNA polymerase. Proc Natl Acad Sci USA. 2008;105:15305–15310. doi: 10.1073/pnas.0805757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greive SJ, Lins AF, von Hippel PH. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J Biol Chem. 2005;280:36397–36408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- 86.Sen R, King RA, Weisberg RA. Modification of the properties of elongating RNA polymerase by persistent association with nascent antiterminator RNA. Mol Cell. 2001;7:993–1001. doi: 10.1016/s1097-2765(01)00243-x. [DOI] [PubMed] [Google Scholar]
- 87.King RA, Markov D, Sen R, Severinov K, Weisberg RA. A conserved zinc binding domain in the largest subunit of DNA-dependent RNA polymerase modulates intrinsic transcription termination and antitermination but does not stabilize the elongation complex. J Mol Biol. 2004;342:1143–1154. doi: 10.1016/j.jmb.2004.07.072. [DOI] [PubMed] [Google Scholar]
- 88.Komissarova N, et al. Inhibition of a transcriptional pause by RNA anchoring to RNA polymerase. Mol Cell. 2008;31:683–694. doi: 10.1016/j.molcel.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Irnov I, Winkler WC. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol Microbiol. 2010;76:559–575. doi: 10.1111/j.1365-2958.2010.07131.x. [DOI] [PubMed] [Google Scholar]
- 90.Bailey MJ, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 91.Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. EMBO J. 2009;28:112–122. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Belogurov GA, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chalissery J, et al. Interaction surface of the transcription terminator Rho required to form a complex with the C-terminal domain of the antiterminator NusG. J Mol Biol. 2011;405:49–64. doi: 10.1016/j.jmb.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 95.Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I. The elongation factor RfaH and the initiation factor σ bind to the same site on the transcription elongation complex. Proc Natl Acad Sci USA. 2008;105:865–870. doi: 10.1073/pnas.0708432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–5705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bailey MJ, Hughes C, Koronakis V. In vitro recruitment of the RfaH regulatory protein into a specialised transcription complex, directed by the nucleic acid ops element. Mol Gen Genet. 2000;262:1052–1059. doi: 10.1007/pl00008648. [DOI] [PubMed] [Google Scholar]
- 98.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 99.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 100.Nudler E. RNA polymerase active center: the molecular engine of transcription. Ann Rev Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 102.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Potter KD, Merlino NM, Jacobs T, Gollnick P. TRAP binding to the Bacillus subtilis trp leader region RNA causes efficient transcription termination at a weak intrinsic terminator. Nucleic Acids Res. 2011;39:2092–2102. doi: 10.1093/nar/gkq965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 105.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Datta K, von Hippel PH. Direct spectroscopic study of reconstituted transcription complexes reveals that intrinsic termination is driven primarily by thermodynamic destabilization of the nucleic acid framework. J Biol Chem. 2008;283:3537–3549. doi: 10.1074/jbc.M707998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Biol. 2010;401:708–725. doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yuzenkova Y, Zenkin N, Severinov K. Mapping of RNA polymerase residues that interact with bacteriophage Xp 10 transcription antitermination factor p7. J Mol Biol. 2008;375:29–35. doi: 10.1016/j.jmb.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sydow JF, et al. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 110.Belogurov GA, Sevostyanova A, Svetlov V, Artsimovitch I. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol Microbiol. 2010;76:286–301. doi: 10.1111/j.1365-2958.2010.07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nature Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 112.Hirtreiter A, et al. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Klein BJ, et al. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci USA. 2011;108:546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011 Mar 8; doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Steinmetz EJ, Platt T. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc Natl Acad Sci USA. 1994;91:1401–1405. doi: 10.1073/pnas.91.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rabhi M, et al. Mutagenesis-based evidence for an asymmetric configuration of the ring-shaped transcription termination factor Rho. J Mol Biol. 2011;405:497–518. doi: 10.1016/j.jmb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 117.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rees W, Weitzel S, Yager T, Das A, von Hippel P. Bacteriophage λ N protein alone can induce transcription antitermination in vitro. Proc Natl Acad Sci USA. 1996;93:342–346. doi: 10.1073/pnas.93.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sloan S, Rutkai E, King RA, Velikodvorskaya T, Weisberg RA. Protection of antiterminator RNA by the transcript elongation complex. Mol Microbiol. 2007;63:1197–1208. doi: 10.1111/j.1365-2958.2006.05579.x. [DOI] [PubMed] [Google Scholar]