Abstract
Network reconstructions are a common denominator in systems biology. Bottom-up metabolic network reconstructions have developed over the past 10 years. These reconstructions represent structured knowledge-bases that abstract pertinent information on the biochemical transformations taking place within specific target organisms. The conversion of a reconstruction into a mathematical format facilitates myriad computational biological studies including evaluation of network content, hypothesis testing and generation, analysis of phenotypic characteristics, and metabolic engineering. To date, genome-scale metabolic reconstructions for more than 30 organisms have been published and this number is expected to increase rapidly. However, these reconstructions differ in quality and coverage that may minimize their predictive potential and use as knowledge-bases. Here, we present a comprehensive protocol describing each step necessary to build a high-quality genome-scale metabolic reconstruction as well as common trials and tribulations. Therefore, this protocol provides a helpful manual for all stages of the reconstruction process.
INTRODUCTION
Metabolic network reconstructions have become an indispensable tool for studying the systems biology of metabolism1–7. The number of organisms for which metabolic reconstructions have been created is increasing at a pace similar to whole genome sequencing. However, the quality of metabolic reconstructions differs considerably, which is partially caused by varying amounts of available data for the target organisms, but also partially by a missing standard operating procedure that describes the reconstruction process in detail. This protocol details a procedure by which a quality-controlled quality-assured (QC/QA) reconstruction can be built to ensure high quality and comparability between reconstructions. In particular, the protocol points out data that are necessary for the reconstruction process and that should accompany reconstructions. Moreover, standard tests are presented, which are necessary to verify functionality and applicability of reconstruction-derived metabolic models. Finally, this protocol presents strategies to debug non- or malfunctioning models. While the reconstruction process has been reviewed conceptually by numerous groups8–11 and a good general overview of the necessary data and steps is available, no detailed description of the reconstruction, debugging, and iterative validation process has been published. This protocol seeks to make this process explicit and generally available.
The presented protocol describes the procedure necessary to reconstruct metabolic networks intended to be used for computational modeling, including the constraint-based reconstruction and analysis (COBRA) approach11, 12. These network reconstructions, and in silico models, are created in a bottom-up fashion based on genomic and bibliomic data, and thus represent a biochemical, genetic, and genomic (BiGG) knowledge-base for the target organism9. These BiGG reconstructions can be converted into mathematical models and their systems and physiological properties can be determined. For example, they can be used to simulate maximal growth of a cell in a given environmental condition using flux balance analysis (FBA)13, 14. In contrast, the generation of networks derived from top-down approaches (high-throughput data based interference of component interactions) is not discussed here, as they do not generally result in functional, mathematical models.
The metabolic reconstruction process described herein is usually very labor- and time intensive, spanning from six months for well-studied, medium genome sized bacteria, to two years (and six people) for the metabolic reconstruction of human metabolism15. Often, the reconstruction process is iterative, as demonstrated by the metabolic network of Escherichia coli, whose reconstruction has been expanded and refined over the last 19 years7. As the number of reconstructed organisms increases, the need to find automated, or at least semi-automated, ways to reconstruct metabolic networks straight from the genome annotation is growing. Despite growing experience and knowledge, to date, we are still not able to completely automatically reconstruct high-quality metabolic networks that can be used as predictive models. Recent reviews highlight current problems with genome annotations and databases, which make automated reconstructions challenging and thus, require manual evaluation8, 9. Organism-specific features such as substrate and cofactor utilization of enzymes, intracellular pH, and reaction directionality remain problematic, and thus, requiring manual evaluation. However, some organism-specific databases and approaches exist, which can be used for automation. We describe here the manual reconstruction process in detail.
A limited number of software tools and packages are available (freely and commercially), which aim to assist and facilitate the reconstruction process (Table 1). The protocol presented can, in principle, be combined with those reconstruction tools. For generality, we present the entire procedure using a spreadsheet, namely Excel workbook (Microsoft Inc), and a numeric computation and visualization software package, namely Matlab (Mathwork Inc). Free spreadsheets (e.g., Open office and Google Docs) could be used instead of the listed spreadsheet. Alternatively, MySQL databases may be used, as they are very helpful to structure and track data. Matlab was also used to encode the COBRA Toolbox, which is a suite of COBRA functions commonly used for simulation16. This Toolbox was extended to facilitate the reconstruction, debugging, and manual curation process described herein.
Table 1.
Data sources frequently used for metabolic reconstructions.
| Name | Link | Comment |
|---|---|---|
| Genome Databases | ||
| Comprehensive Microbial Resource (CMR) | http://cmr.jcvi.org/cgi-bin/CMR/CmrHomePage.cgi. The | |
| Genomes OnLine Database (GOLD) | http://www.genomesonline.org/ | |
| TIGR | http://www.tigr.org/db.shtml | |
| NCBI Entrez Gene | http://www.ncbi.nlm.nih.gov/sites/entrez | |
| SEED database32 | theseed.uchicago.edu/FIG/index.cgi | Comparative genomics tool. |
| Biochemical Databases | ||
| KEGG41 | www.genome.jp/kegg/ | |
| BRENDA42 | www.brenda-enzymes.info/ | |
| Transport DB89 | http://www.membranetransport.org/ | |
| PubChem86 | http://pubchem.ncbi.nlm.nih.gov/ | |
| Transport Classification Database (TCDB) | http://www.tcdb.org/ | TCDB is a curated database of factual information from over 10,000 published references. |
| pKa Plugin | http://www.chemaxon.com/product/pka.html | Free for academic users |
| pKa DB | http://www.acdlabs.com/products/phys_chem_lab/pka/ | Commercial software package to determine acid-base ionization/dissociation constant, pKa. |
| Organism-specific databases | ||
| Ecocyc43 | http://ecocyc.org/ | Escherichia coli database |
| PyloriGene37 | http://genolist.pasteur.fr/PyloriGene | Helicobacter pylori database |
| Gene Cards | www.genecards.org/ | Human gene database |
| Protein Localization databases | ||
| PSORT47 | http://www.psort.org/psortb/ | Support vector machine (SVM) based. |
| PA-SUB48 | http://www.cs.ualberta.ca/~bioinfo/PA/Sub/ | Proteome Analyst Specialized Subcellular Localization Server (SVM based). |
| Bio-numbers | ||
| CyberCell Database (CCDB)88 | http://redpoll.pharmacy.ualberta.ca/CCDB/cgi-bin/STAT_NEW.cgi | |
| B10NUMB3R5 | http://bionumbers.hms.harvard.edu/ | |
| Available reconstruction software packages | ||
| Simpheny | http://www.genomatica.com/technology/technologySuite.html | Commercial software |
| COBRA simulation environments | ||
| CellNetAnalyzer90/FluxAnalyzer91 | http://www.mpi-magdeburg.mpg.de/projects/cna/cna.html | Matlab is required |
| COBRA Toolbox16 | http://systemsbiology.ucsd.edu/Downloads/Cobra_Toolbox | Matlab is required |
| FluxExplorer92 | ||
| MetaFluxNet93, 94 | http://mbel.kaist.ac.kr/lab/mfn/ | Stand alone package |
The protocol describes in detail the process to generate metabolic reconstructions applicable for representatives of all domains of life. The process of reconstructing prokaryotic and eukaryotic metabolic networks is, in principle, identical, although eukaryote reconstructions are more challenging due to size of genomes, coverage of knowledge, and the multitude of cellular compartments. Specific properties and pitfalls are highlighted.
The described reconstruction and debugging process requires organism specific information. The minimum information includes the genome sequence, from which key metabolic functions can be obtained, and physiological data, such as growth conditions, which allow the comparison of model prediction to refine the network’s content. In general, the more information about physiology, biochemistry, and genetics is available for the target organism, the better the predictive capacity of the models. This property becomes obvious considering that the network evaluation and validation process relies on comparing predicted phenotypes (e.g., growth rate) with experimental observations. Additional cellular objectives (other than maximal growth rate) may be compared with experimental data but they are not detailed in this protocol15, 17–20.
Although this protocol presents the reconstruction process in terms of metabolic networks, the same approach can, and has been, applied for reconstructing signaling21, 22 and transcription/translation networks23. Regulatory networks have not been constructed in a fully stoichiometric manner yet, although a pseudo-stoichiometric approach has been proposed24, 25. The reconstruction process for these networks is not as well established as for metabolic networks, and is thus still subject to active research.
Lastly, myriad data sources are used during the reconstruction process rendering metabolic network reconstructions as knowledge-bases, which summarize and structure the available BiGG knowledge about the target organism. Frequently used organism-unspecific, and some of the organism-specific, resources are listed in Table 1. Note that the quality and wealth of organism-specific information will directly affect the quality and coverage of the metabolic reconstruction. Great resources are organism-specific books that have been published for a growing number of organisms26–29. In cases where organism-specific information is scarce, data from phylogenic neighbors may be of great help. It is important to ensure that, in cases where the reconstruction relies extensively on relative information, the overall behavior of the model matches the target organism. This assurance can be achieved by carefully comparing the predictions with experimental and physiological data, such as growth conditions, secretion products, and knock-out phenotypes.
The resulting knowledge-bases can be queried, used for mapping experimental data (e.g., gene expression, proteomic, fluxomic, and metabolomic data), and converted into a mathematical format to investigate metabolic capabilities and generate new biological hypotheses. The multitude of possible applications of BiGG knowledge-bases distinguishes them from other, automated efforts. By introducing standards in content and format with this protocol it will soon be possible to compare metabolic reconstructions between different organisms, which will further enhance our understanding of the evolutionary processes and may provide a complementary approach to comparative genomics.
GENERAL PROCEDURE
The metabolic network reconstruction process described herein consists of four major stages followed by its prospective use in stage 5 (Figure 1). The order of steps in the different stages is a recommendation and may be altered within each stage, and with some limitations between stages, as long as they are completed. The quality of the reconstruction is generally ensured by performing all steps.
Figure 1. Overview of the procedure to iteratively reconstruct metabolic networks.
In particular stages 2 to 4 are continuously iterated until model predictions are similar to the phenotypic characteristics of the target organism and/or all experimental data for comparison are exhausted.
Stage 1: Creating a draft reconstruction
Note that the creation of a draft reconstruction and the manual reconstruction refinement (next stage) may be combined for bacterial reconstructions with main emphasis on reconstruction refinement.
The first stage consists of the generation of a draft reconstruction based on the genome annotation of the target organism and biochemical databases. This draft reconstruction, or automated reconstruction, is thus a collection of genome encoded metabolic functions, some of which may be falsely included while other ones are missing (e.g., due to missing, wrong, or incomplete annotations). Software tools such as Pathway tools30 or metaSHARK31 can be used for the generation of the draft reconstruction but they do not replace the manual curation.
Genome annotation (Step 1)
Genomic information is important to unambiguously define the gene properties in respect to the organism’s genome as well as to allow data mapping (e.g., gene expression) in subsequent studies. Since the draft reconstruction, and to some extent the curated reconstruction, relies mainly on the genome annotation, it is important to download the most recent version available to ensure that updates and corrections since the genome’s original publication are accounted for. Thus, the quality and reliability of the genome annotation is crucial to the reconstruction quality. Note that the manual reconstruction refinement tries to identify those low confidence gene annotations by retrieving further, experimental evidence for the presence of the gene product and its metabolic function. The reconstruction assembly and refinement may also require re-annotation of genes but the procedure is not further discussed here. Please refer to available work and reviews32–36. Furthermore, in some cases, the genome-sequencing group created organism specific database (e.g., for Helicobacter pylori37 and E. coli38), which are very valuable during the reconstruction process. Table 1 lists some of the commonly used databases for annotations.
Candidate metabolic functions (Step 2)
To obtain the draft reconstruction, one can automatically retrieve metabolic genes from the genome annotation by using, for example, key words or gene ontology (GO) catergories39 (see Supplemental methods 1, Figure S1). Metabolic reactions catalyzed by the identified gene products can be connected with the draft reconstruction by using the enzyme commission (E.C.) numbers40 and biochemical reaction databases, e.g., KEGG41 and Brenda42. Note that this first stage aims to obtain a list of candidates that will not necessarily be complete or comprehensive. Many false-positives may be present in the list. For example, proteins involved in DNA methylation or rRNA modification also have E.C. numbers, but their functions are normally not considered in metabolic reconstructions. Another example involves kinases that may be involved in signal transfer reactions or annotated as ‘histidine kinase-like’ and thus, no specific function can be derived from this annotation. A more targeted query for metabolic annotations could be designed to reduce the number of false-positives but it does not replace manual curation.
Stage 2: Manual reconstruction refinement
In this stage, the entire draft reconstruction will be re-evaluated and refined. For each gene and reaction entry, two questions will be asked: 1) Should this entry be here? 2) Is there an entry missing to connect the entry with the remainder of the network?
The second stage of the reconstruction process concentrates on curation and refinement of the network content. We highlight in this protocol parts that need special attention. In particular, the metabolic functions and reactions collected in the draft reconstruction are individually evaluated against organism-specific literature (and expert opinion). This manual evaluation is important since 1) not all annotations have a high confidence score (e.g., low e-value), and 2) biochemical databases are mostly organism-unspecific, listing enzymes activities found in various organisms, not all of which may be present in the target organism (Figure 2). Including organism-unspecific reactions can affect the predictive behavior of the resulting models. Furthermore, information about biomass composition, maintenance parameters, and growth conditions are collected in this stage, which will provide a basis for the simulations in stage 3 and 4.
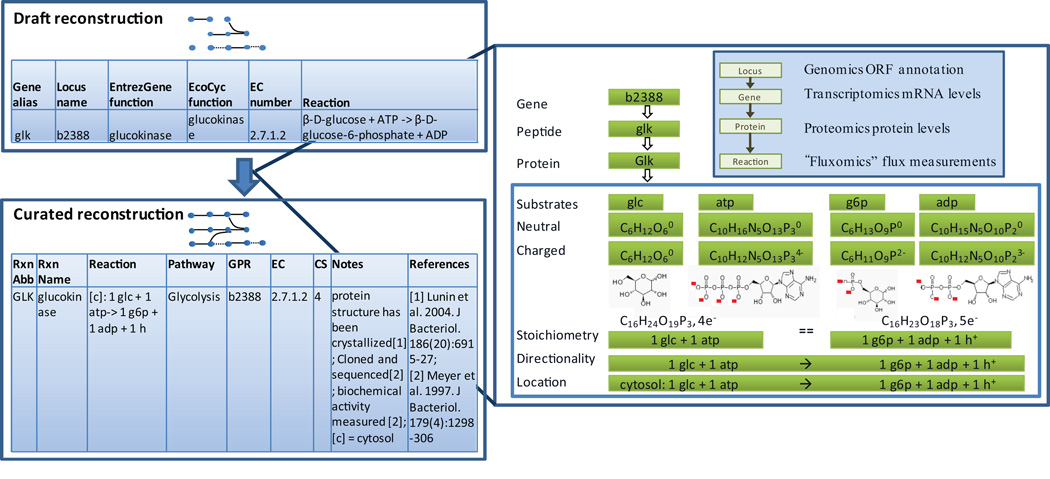
Figure 2. Refinement of reconstruction content.
The draft reconstruction is converted into a curated reconstruction by re-evaluation of the content. In particular, the metabolic reactions, obtained from biochemical databases or the literature, need to be tested for mass- and charge balancing. Many resources omit protons and water. Furthermore, adjusting metabolites to a particular pH may change their charged formulae and thus may require correction of the network reaction. For instance, the reaction catalyzed by the glucokinase which was obtained from KEGG86 is not mass- and charge-balanced when charged metabolite formula at pH 7.2 is considered. The right hand side (RHS) is missing an H and the charge is unbalanced. Adding a proton to the RHS balances both sides of the equation in terms of protons and electrons/charge. Abbreviations: glc – D-glucose, g6p – D-glucose-6-phosphate, atp – adenosine-triphosphate, adp – adenosine-diphosphate, H+ - proton. CS – confidence level.
Reconstruction assembly
It is generally recommended to refine and assemble the curated reconstruction in a pathway by pathway manner, starting from the canonical pathway. Peripheral pathways and reactions/gene products without clear pathway assignment are added in a later step. This approach has the advantage that reactions are evaluated within their metabolomic context and missing gene annotations can be readily identified, facilitating gap analysis and debugging in stage 4. However, this approach will also result in identification and/or additional information for reactions that are not in the pathway currently under investigation. One can choose to only include the main reaction(s) associated with the pathway that is currently considered. The remaining reactions may be noted somewhere so that they can be readily retrieved if necessary.
Verification of metabolic function (Step 6)
The draft reconstruction identified a set of metabolic genes and functions that are thought to be present in the target organism. Due to potential errors or incomplete in genome annotation, the presence of the annotated gene and its function should be supported using experimental data or literature.
Use of phylogenetically close organisms (Step 6)
If no organism-specific information can be found in the literature, information for phylogenetically close organisms can be used and should be marked as such. If enzyme-associated reactions are included purely based on gene annotation, they should receive with the lowest confidence score (Table 2). In the case of problems during subsequent simulations, these low confidence reactions can be easily identified.
Table 2.
Confidence score system that is currently employed for metabolic reconstructions.
| Evidence type | Confidence score | Examples |
|---|---|---|
| Biochemical data | 4 | Direct evidence for gene product function and biochemical reaction: Protein purification, biochemical assays, experimentally solved protein structures, and comparative gene-expression studies (e.g., Chhabra et al. 95). |
| Genetic data | 3 | Direct and indirect evidence for gene function: Knock–out characterization, knock-in characterization, and over-expression. |
| Physiological data | 2 | Indirect evidence for biochemical reactions based on physiological data: secretion products or defined medium components serve as evidence for transport and metabolic reactions. |
| Sequence data | 2 | Evidence for gene function: Genome annotation, SEED annotation32. |
| Modeling data | 1 | No evidence is available but reaction is required for modeling. The included function is a hypothesis and needs experimental verification. The reaction mechanism may be different from the included reaction(s). |
| Not evaluated | 0 |
Generic reaction terms (Step 6)
In some cases, it is appropriate to exclude certain reactions to be entered in the reconstruction. Reactions containing generic terms, such as protein, DNA, electron acceptor, etc. should not be included as they are not specific enough and normally serve in databases as space holders until more knowledge and biochemical evidence becomes available.
Substrate and cofactor usage (Step 6)
Substrate and cofactor specificity of enzymes may differ between organisms. Organism-unspecific databases, such as KEGG41 and Brenda42, list all possible transformations of an enzyme that have been identified in any organism. Additionally, Brenda lists organism-specific information along with relevant references and kinetic parameters. As a rule of thumb, one can assume that enzymes, which have only one reaction associated in, for example, KEGG41do not require organism refinement. However, enzymes that are associated with multiple reactions, with varying substrates and/or cofactors, require manual refinement. Information about substrate and cofactor utilization can be obtained from organism-specific biochemical studies and may also be listed in organism-specific databases (e.g., Ecocyc43). This part of the curation process can be very time consuming and laborious as it may be difficult to find the necessary information. Often, this requires intensive literature search. It is important to pay great attention as false inclusion of substrates or cofactors can greatly change the in silico behavior (i.e., predictive potential) of the reconstruction.
Charged formula for each metabolite (Step 7 and 8)
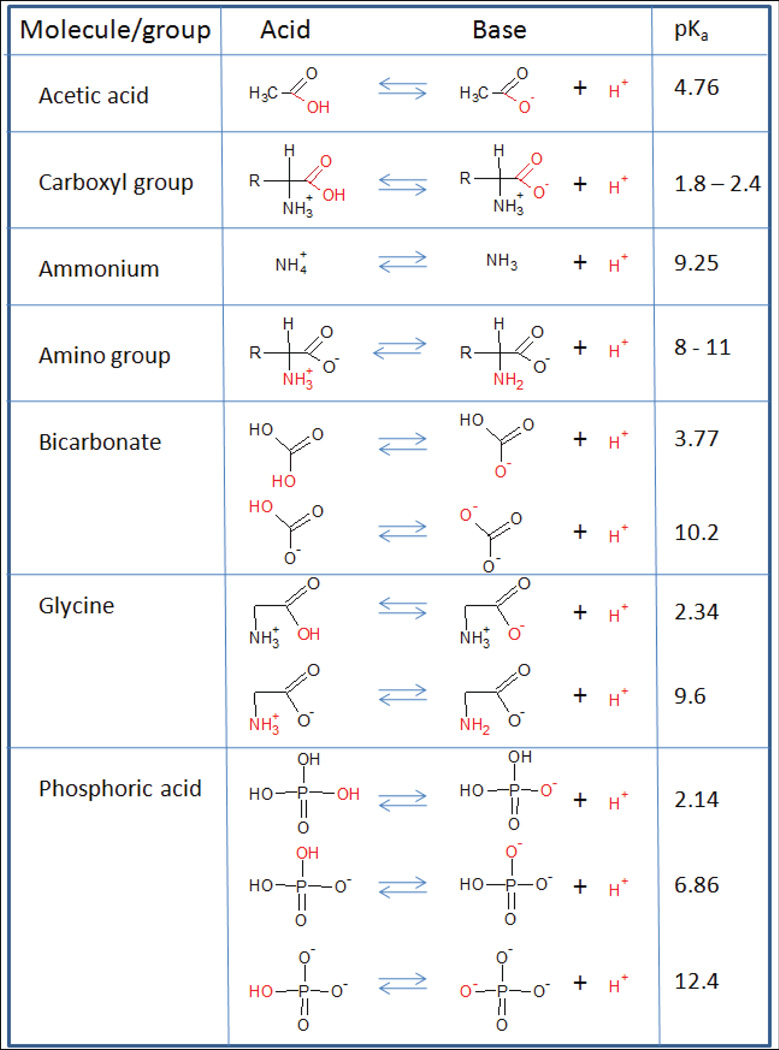
In databases, metabolites are generally listed with their uncharged formula. In contrast, in medium and in cells, many metabolites are protonated or deprotonated. The protonation state, and thus the charged formula, depends on the pH of interest. Often metabolic networks are reconstructed assuming an intracellular pH of 7.2. However, the intracellular pH of bacterial cells may vary depending on environmental conditions and bacteria. Also, the pH of organelles may be different, e.g., peroxisome and lysosome. The protonated formula is calculated based on the pKa value of the functional groups (Figure 3). Software packages, such as Pipeline Pilot and pKa DB, can predict pKa values for a given compound (Table 1). Figure 2 shows some examples of charged molecules and their pKa values.
Figure 3.
List of functional groups, their charge formula and the corresponding pKa.
Reaction stoichiometry (Step 9)
Once the charged formula is obtained for each metabolite, the reaction stoichiometry can be determined by counting the different elements on the left- and right-hand side of the reaction. Protons and water may need to be added to the reaction in this step as some databases and many biochemical textbooks omit these molecules. Therefore, every element and the charge need to balance on both sides of the reaction. This step is easy for many central metabolic reactions but may become challenging for more complex reactions. Note that unbalanced reactions may lead to synthesis of protons or energy (ATP) out of nothing (see also Figure 4 for examples).
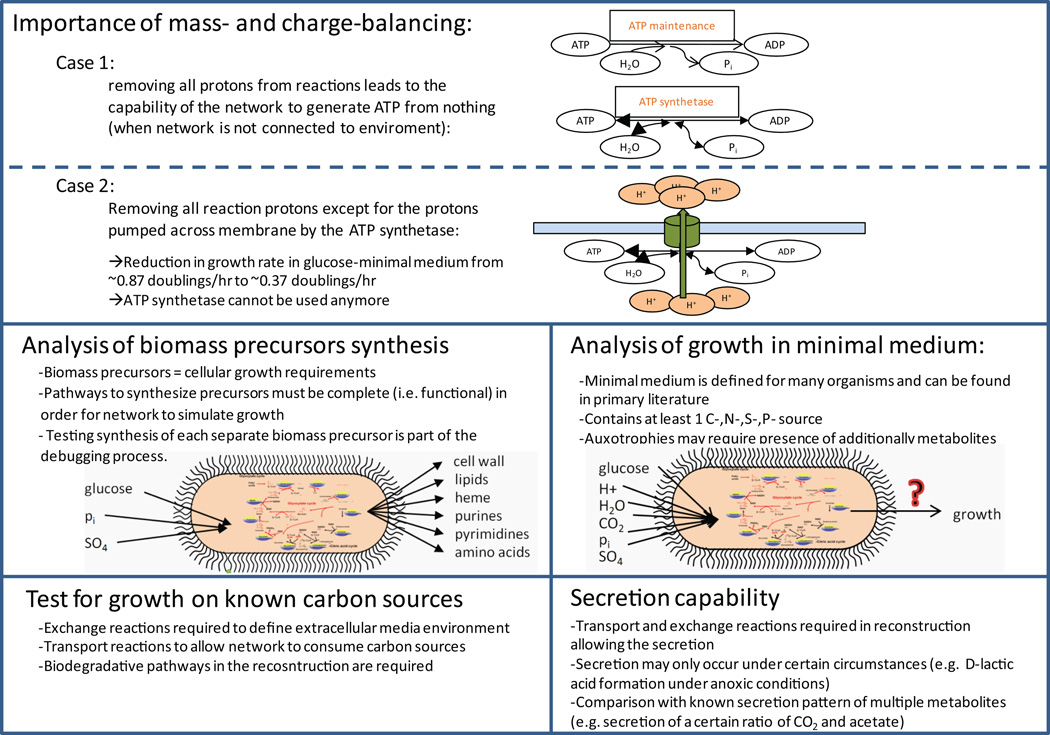
Figure 4. Examples of network evaluation.
The network evaluation and debugging stage (stage 4) includes various QC/QA tests, some of which are illustrated in this figure. For instance, mass-and charge-balancing of the network reaction is crucial to ensure similar properties of the model and the cell or organism. A standard test for most metabolic reconstructions is to verify that each biomass precursor, which makes up a new cell, can be produced by the model in different growth conditions (e.g., minimal medium, different carbon sources, etc.). Other QC/QA tests may include the capability to secrete certain metabolites given a particular growth condition. At its end, the models will have similar properties as the cell and error cases can be used to systematically refine the models and thus the reconstruction content.
Reaction Directionality (Step 10)
Biochemical data for the target organism are very important for determination of reaction directionality but may not be available. New approaches are available, such as the estimation of the standard Gibbs free energy of formation (ΔfGo) and of reaction (ΔrGo) in a biochemical system44, 45. The standard Gibbs free energy of formation (ΔfGo) and of reaction (ΔrGo) can be obtained for most KEGG41 reactions from Web GCM44. Another approach combines thermodynamic information with network topology and heuristic rules to assign reaction directionality46. Biochemical textbooks may also report reaction directionalities. Additionally one can use the following rules of thumb: 1) all reactions involving transfer of phosphate from ATP to an accepter molecule should be irreversible (with the exception of the ATP synthetase, which is known to occur in reverse direction); 2) reactions involving quinones are generally irreversible.
Note that assigning the wrong direction to a reaction may have significant impact on the model’s performance. In general, one should leave a reaction reversible if no information is available and the aforementioned rules of thumb do not apply. However, models with too many reversible reactions (too loose constraints) may have so called futile cycle, which overcome the proton gradient by freely exchanging metabolites and protons across compartments. Therefore, assigning the correct reversibility to transport reactions is especially important (see below).
Information for gene and reaction localization (Step 11)
This information may also be difficult to obtain. The compartments that have been considered in various metabolic reconstructions are listed in Supplemental methods 1, Table S1. Algorithms such as PSORT47 and PASUB48 can be used to predict the cellular localization of proteins based on nucleotide or amino acid sequences. A recently published protocol describes the use of internet-accessible tools to predict the subcellular location of eukaryotic and prokaryotic proteins49. High-throughput experimental approaches are available to locate individual proteins, including immunofluorescence50 and GFP tagging of individual proteins51. In the absence of appropriate data, proteins should be assumed to reside in the cytosol. Incorrect assignment of the location of a reaction can lead to additional gaps in the metabolic network and misrepresentation of the network properties, especially, if intracellular transport reactions need to be added for which no evidence is available either.
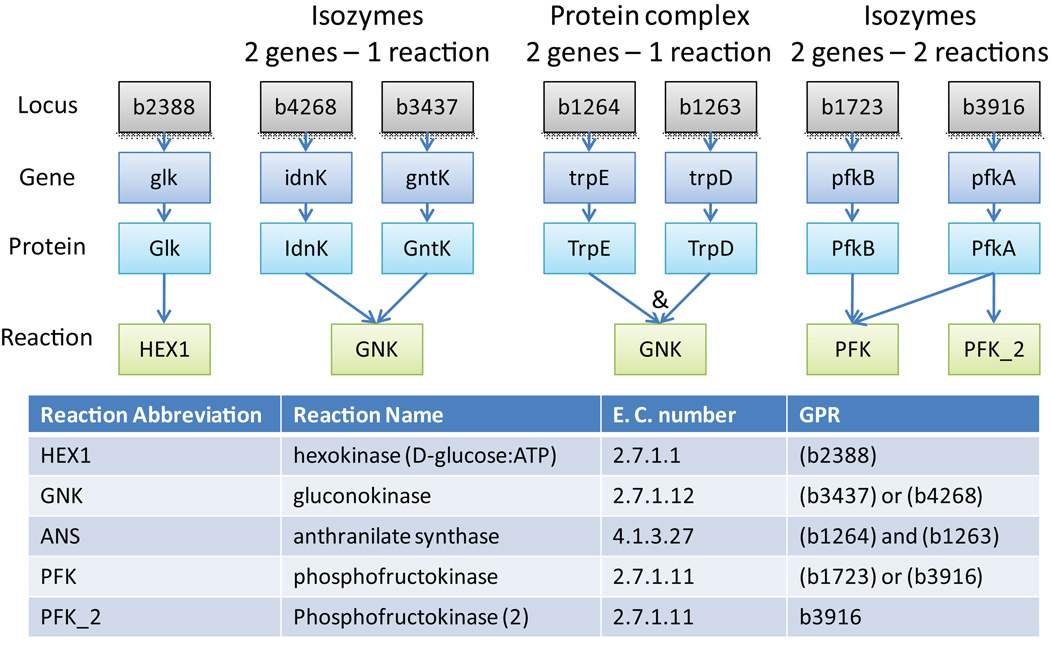
Gene-protein-reaction (GPR) association (Step 13)
The genome annotation often provides information about the GPR association, i.e., it indicates which gene has what function (Figure 5). The verification and refinement necessary in this step includes determining: i) if the functional protein is a heteromeric enzyme complex; ii) if the enzyme (complex) can carry out more than one reaction and iii) if more than one protein can carry out the same functions (i.e., isozymes exist). For the first case (i), the genome annotation often has refined information, e.g.: ‘protein X, catalytic subunit’ - which indicates that there is at least one more subunit needed for the function of the protein complex. Furthermore, KEGG41 lists subunits in some cases. Often, a more comprehensive database and/or literature search is required. Also, the protein complex composition may differ between organisms. The second case can also be identified from biochemical databases and/or literature. Multitasking of enzymes may also differ between organisms. Note that mistakes or mis-assignments in the GPR associations will change results of in silico gene deletion studies. However, discrepancies between in silico and in vivo results can be used to refine knowledge and reconstructions (see Step 79 and 80).
Figure 5. Gene-protein-reaction (GPR) associations.
Examples of GPR associations and their representation in Boolean format are shown for E. coli.
Linear pathways, such as fatty acid oxidation, have often been combined into few lumped reactions. The genes associated with these reactions are all required, with the exception of isozymes. Subsequently, the GPR association should reflect the requirement for all genes within the lumped reaction by using the Boolean rule AND.
Metabolite identifier (Step 14)
Metabolite identifiers are necessary to enable the use of reconstructions for high-throughput data mapping (e.g., metabolomic or fluxomic data) and for comparison of network content with other metabolic reconstructions. Therefore, metabolites and reactions need to be recognizable by other scientists and by software tools. Each metabolite should be associated with at least one of the following identifiers: ChEBI52, Kegg41, and PubChem53. In many cases, having one of the identifiers is sufficient to automatically obtain the other two identifiers. Furthermore, database-independent representations of metabolites such as SMILES54 and InCHI strings55, 56 are also helpful when associated with each metabolite. These representations represent the exact chemical structure of compounds. Additionally, collecting Molfiles (MDL file format, http://www.symyx.com/), which hold information about the atoms, bonds, connectivity and coordinates of a molecule, will be very useful, e.g., if you are using online software for pKa determination (see Step 10 for details).
Confidence scoring system (Step 15)
The confidence score provides a fast way of assessing the amount of information available for a metabolic function, pathway, or the entire reconstruction15, 57. Every network reaction is associated with a confidence score reflecting the information and evidence currently available. The confidence score ranges from 0 to 4, where 0 is the lowest and 4 is the highest evidence score (Table 2). Note that multiple information types result in a cumulative confidence score. For example, a confidence score of 4 may represent physiological and sequence evidence.
Spontaneous reactions (Step 19)
An excerpt of typical spontaneous reactions included in metabolic reconstructions is listed in Supplemental methods 1, Table S2. Note that only those spontaneous reactions should be added that have at least one metabolite connecting them to the rest of the reconstruction. This is to avoid too many dead-end metabolites caused by spontaneous reactions. In more recent reconstructions, spontaneous reactions have been associated with an artificial gene (s0001) and protein (S0001). By doing so, reaction and gene essentiality studies are easier to analyze. Furthermore, this artificial GPR association makes it easy to distinguish between spontaneous and orphan reactions, i.e., reactions without known gene.
Intracellular transport reactions (Step 22)
When multi-compartment networks are constructed, intracellular transport reactions need to be added for all metabolites that are supposed to “move” between compartments. Inner cellular transport systems are not very well studied and many of these are not annotated in the genome. Finding experimental data is often not easy. A general approach should be to minimize the number of intracellular transport reactions to the ones that really need to be there. If too many transport reactions are added in a reconstruction, they can cause cycles (futile cycles or Type III pathways). This is a common problem in reconstructions with multiple compartments. For the directionality of intracellular transport reactions, one should consider the nature of the pathway in the compartment. For instance, if the pathway is biosynthetic, it is very likely that i) the precursor(s) is only imported, ii) the product(s) of the pathway is only exported from the compartment, and iii) intermediates are not transported at all. Another issue is the transport mechanism. Many transport reactions are in symport or antiport with either protons, cations, or other metabolites. However, not much information is available for intracellular transporters, but the mechanism used in the model may affect the predictive potential. To minimize the error and increase consistency, one can adopt the intracellular transport mechanism from a corresponding transport reaction from extracellular/periplasmic space to cytoplasm when it is known (and is not an ABC transport reaction); Otherwise (facilitated) diffusion reaction may be assumed as mechanism. In any case, these reactions should receive a low confidence score (1 for modeling purpose) to enable easy identification (Table 2) as well as a note and references describing where the mechanism was taken from.
Identification of missing functions
The refinement stage of the reconstruction process is also an ideal point to identify missing functions in the draft reconstruction. Using KEGG41 maps, for example, one can analyze the metabolic “environment” of the reaction(s) under inspections. If the genome annotation of the target organism is present in KEGG41, one can highlight the genes on the map. This gives an estimate of the “connectivity” of the reaction with its metabolic surrounding (Supplemental methods 1, Figure S2). Missing reactions/functions may become evidence for which experimental/annotation evidence should be collected (see also gap analysis). Creating organism-specific maps, using specific drawing software, is of great use for identifying missing functions as well as for network evaluation and debugging.
Biomass composition (Step 24–33)
The biomass reaction accounts for all known biomass constituents and their fractional contributions to the overall cellular biomass (Table 3). The detailed biomass composition of the target organism needs to be experimentally determined for cells growing in log phase58–60. However, it may be not possible to obtain a detailed biomass composition for the target organism. In this case, one can estimate the relative fraction of the precursors from the genome (e.g., by using the Comprehensive Microbial Resource (CMR) database, Table 1). Note that we do not suggest taking the RNA composition from E. coli, rather than estimating it using organism-specific genome data. One reason is that the number of rRNA operons, which contains rRNA and tRNA molecules, can differ significantly between organisms. For instance, E. coli has 7 rRNA operons per genome61, while Mycoplasma capricolum has two62 and Halobacterium cutirubrum has only one rRNA operon63.
Table 3.
Chemical composition of cells. Here is listed the cellular content of E. coli taken from Neidhardt et al.64.
| Cellular Component | Cellular Content %(w/w) |
|---|---|
| Protein | 55% |
| RNA | 20.5% |
| DNA | 3.1% |
| Lipids | 9.1% |
| LPS | 3.4% |
| Peptidoglycan | 2.5% |
| Glycogen | 2.5% |
| Polyamines | 0.4% |
| Other | 3.5% |
| Total | 100.00% |
In comparison to other biomass precursors, it is slightly more difficult to determine the lipid composition of the cell. The contribution fatty acids and phospholipids needs to be determined from experiments and/or experimental data. Note that compounds, such as phospholipids, can consist of many different fatty acids (different chain length, saturated and unsaturated). Available data often reports the average composition of these compounds, listing the fraction of the fatty acids with different chain length and saturation status. Thus, the model compounds will not represent all possible combinations but only average compounds, consistent with the experimental data.
The composition of the biomass reaction plays an important role for in silico gene deletion experiments. If a biomass precursor is not accounted for in the biomass reactions, the synthesis reactions may not be required for growth (i.e., it is non-essential). Therefore, the associated genes may not be essential either. Subsequently, the presence or absence of metabolites in the biomass reaction may affect the in silico essentiality of reactions and their associated gene(s). In contrast, the fractional contribution of each precursor plays a minor role for gene and reaction essentiality studies. When one wishes to predict the optimal growth rate accurately, the fractional distribution of each compound plays an important role. The unit of the biomass reaction is 1/h since all biomass precursor fractions are converted to mmol/gDW. Therefore, the biomass reaction sums the mole fraction of each precursor necessary to produce 1 g dry weight of cells.
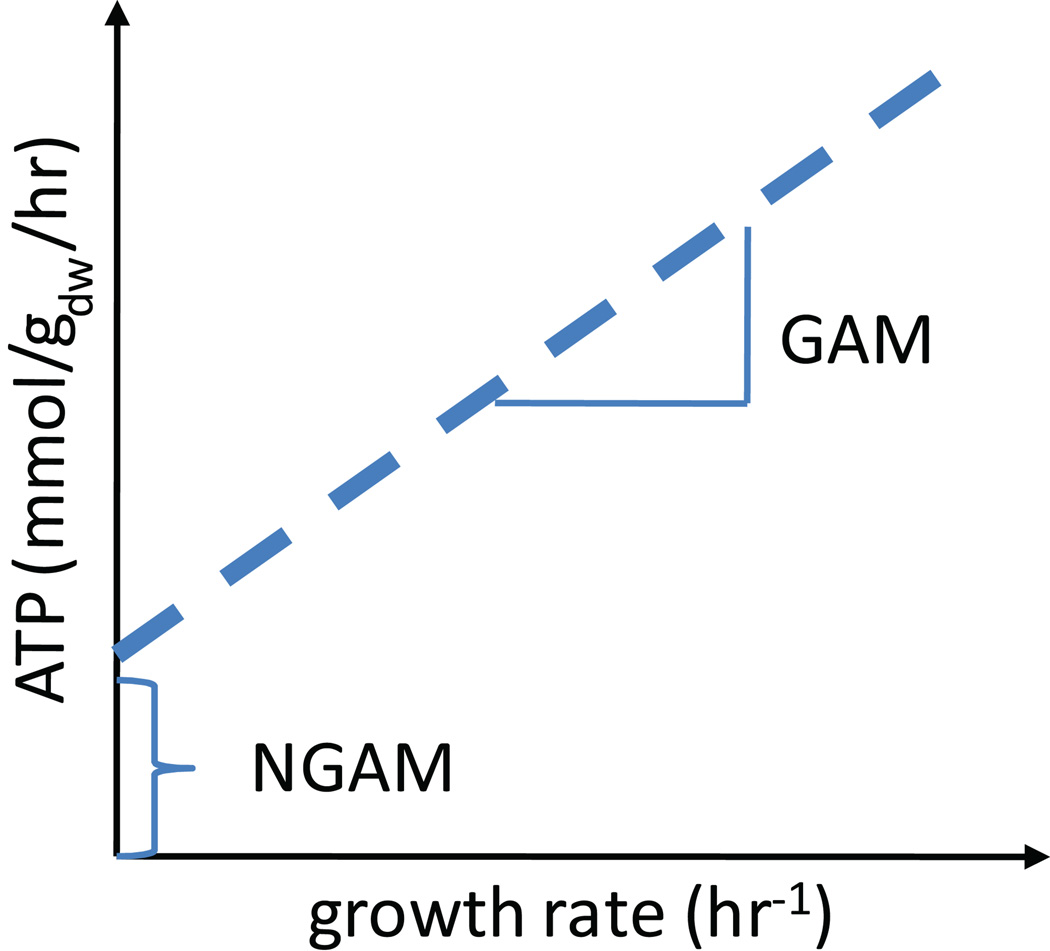
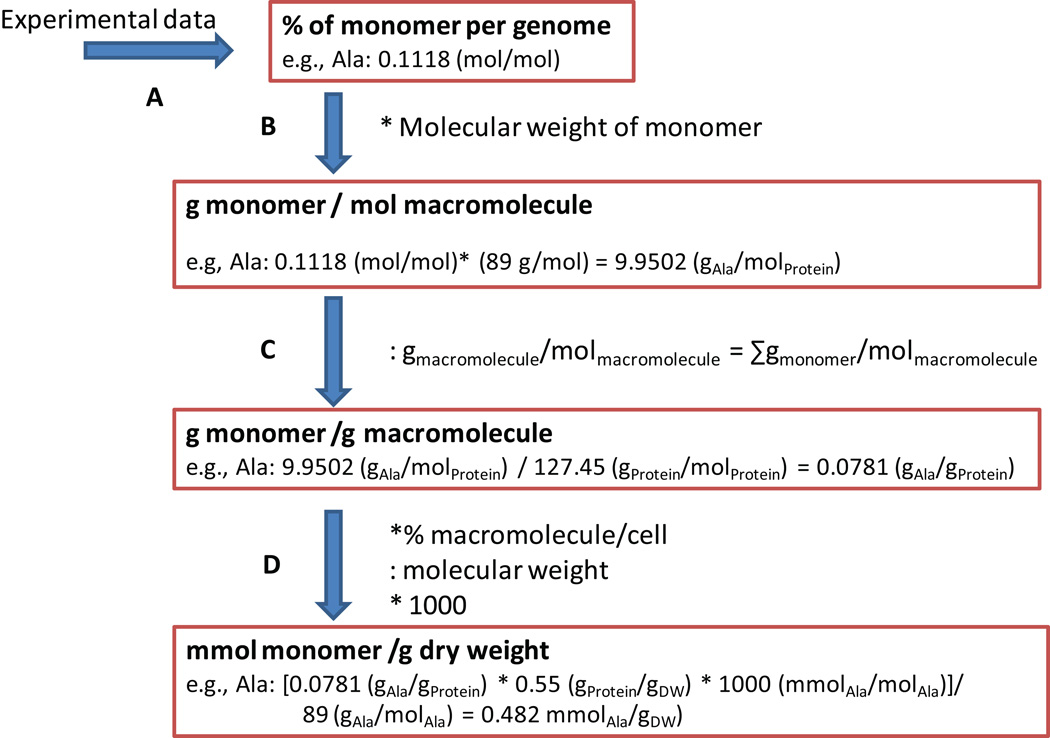
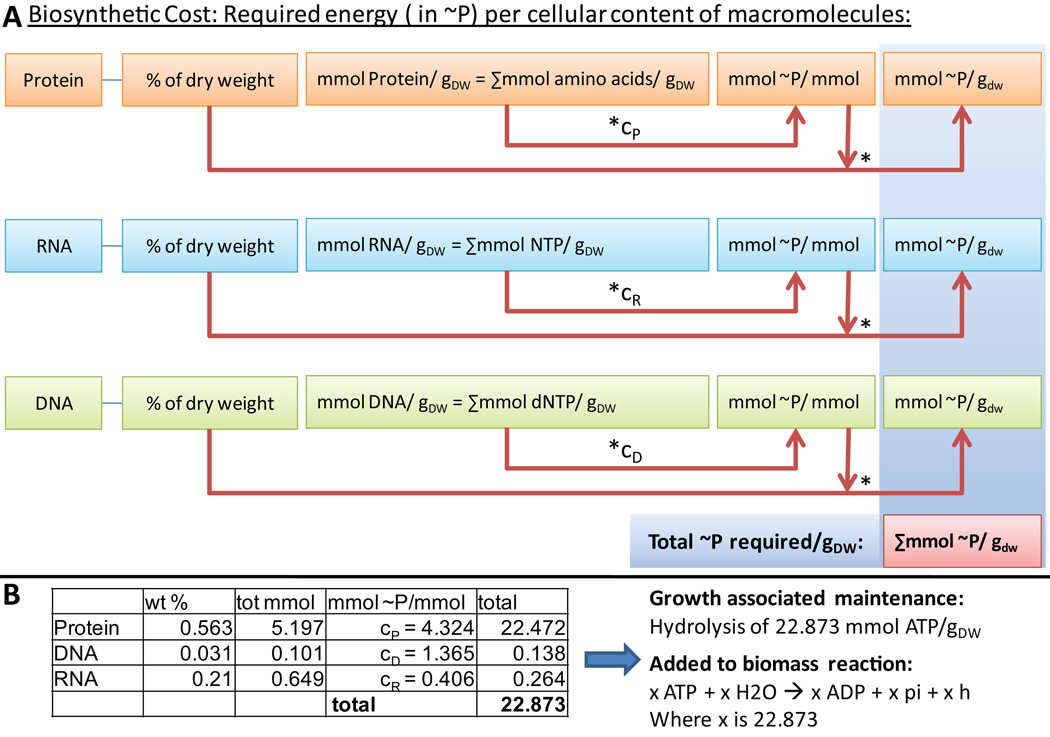
Growth associated ATP maintenance reaction (GAM) (Step 32)
The GAM accounts for the energy (in form of ATP) necessary to replicate a cell, including for macromolecular synthesis (e.g., proteins, DNA, and RNA). The GAM is best determined in chemostat growth experiments (see also Figure 6). Alternatively, if experimental data is not available the GAM can be estimated by determining the energy required for macromolecular synthesis. Therefore, the total amount of macromolecule (Protein, DNA, and RNA) is determined from databases or other resources. Neidhardt et al.64 lists the amount of phosphate bonds necessary to synthesize a macromolecule which is then multiplied with the total amount of phosphate bonds necessary. These phosphate bonds are accounted for by adding ATP hydrolysis to the biomass reaction (x ATP + x H2O → x ADP + x Pi + x H+, where x is the number of required phosphate bonds). Note that this estimate will be too low, as other growth-associated cellular processes also require ATP.
Figure 6. Growth associated maintentance (GAM) and non-growth associated maintenance (NGAM).
The best way to obtain accurate information regarding the GAM and NGAM is by plotting growth data obtained from chemostat growth experiments. GAM and NGAM can be directly read from the plot.
Non-growth associated ATP maintenance reaction (NGAM) (Step 34)
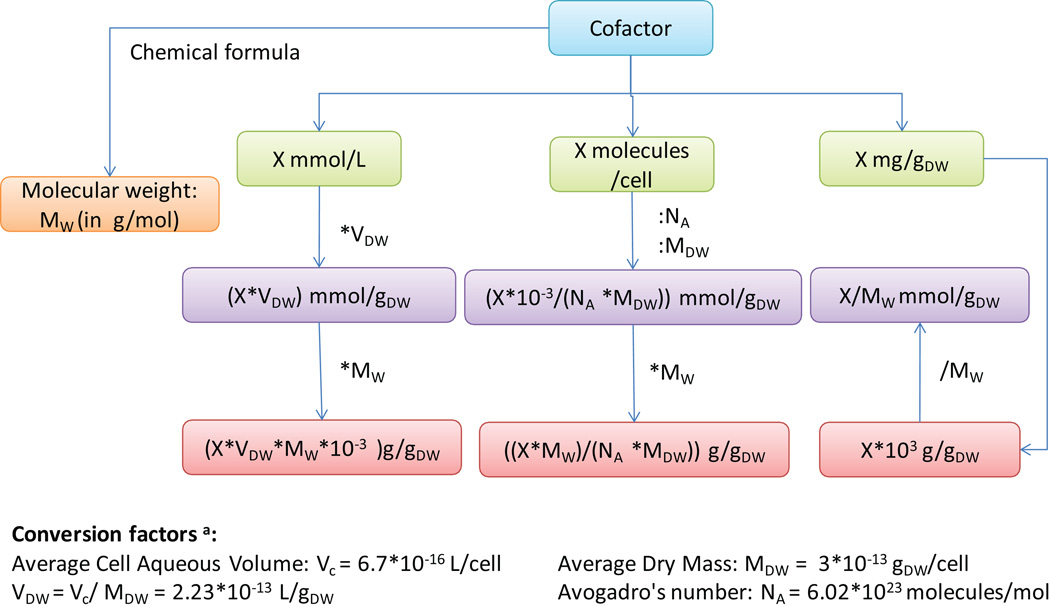
More recent reconstructions include an ATP hydrolysis reaction (1 ATP + 1 H2O → 1 ADP + 1 Pi + 1 H+), which represents non-growth associated ATP requirements of the cell to maintain, for example, Turgor pressure65. The value for the reaction rate can be estimated from growth experiments. For example, based on such measurements, the reaction flux rate was constrained to 8.39 mmol/gDW/h in the E. coli metabolic model65 (Figure 6).
Demand reaction (Step 35)
Demand reactions are unbalanced network reactions that allow the accumulation of a compound, which otherwise is not allowed in steady-state models due to mass-balancing requirement (i.e., in steady state the sum of influx equals the sum of efflux for each metabolite) (Figure 7). Most of the demand reactions will be added in the gap filling process (Steps 46 to 48). At this stage, demand functions should only be added for compounds that are known to be produce by the organism, e.g., certain cofactors, lipopolysaccharide, and antigens, but i) for which information is available about their fractional distribution to the biomass or ii) which may be only produced in some environmental conditions. By including a demand reaction for a particular metabolite one can turn otherwise blocked reactions (cannot carry flux) into active reactions (can carry flux). In general, most reconstructions contain only few demand reactions. However, during the debugging and network evaluation process (Stage 4) demand reactions may be temporarily added to the model to test or verify certain metabolic functions. They will be removed from the model before versioning.
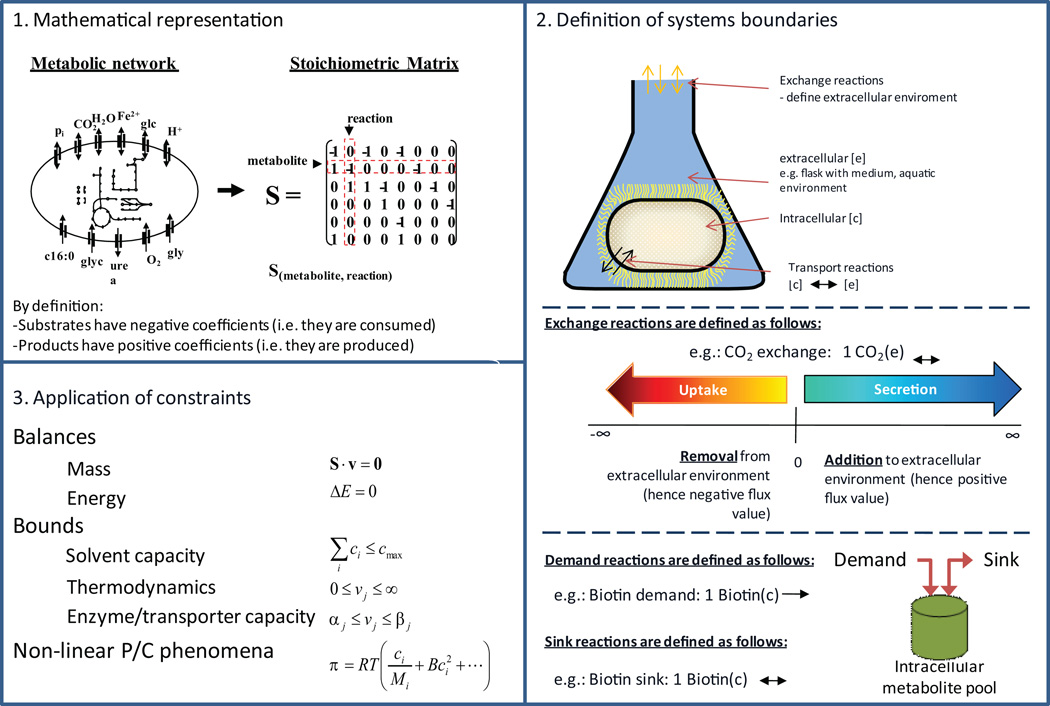
Figure 7. Conversion of reconstruction into a condition-specific model.
This conversion requires three main steps. 1. The first step involves the mathematical representation by a stoichiometric matrix, S, of the network reaction list. The columns of S correspond to the network reactions, while the rows represent the network metabolites. The substrates in a reaction are defined to have a negative coefficient, while products have a positive value. The metabolites participating in a reaction have non-zero entry in the S matrix. 2. Now that the reconstruction is in a computer-readable format, the systems boundaries need to be defined. In particular, this means that for all metabolites that can be consumed or secreted by the target cell a so-called exchange reaction needs to be added to the reconstruction. The exchange reactions can be employed in later simulation to define for example environmental conditions (e.g., carbon source). 3. As a last step, constraints will be added to the reconstruction, thus rendering it to a condition-specific model. Mass conservation is a basic physical law. All steady-states can be thus described by S.v = 0 where v is a vector of reaction fluxes. Adding further constraints such as thermodynamics (reaction directionality), enzyme capacity or regulation (i.e., presence or absence of an enzyme) to the model will lead to a smaller, more confined set of feasible steady-states flux solutions.
Sink reactions (Step 36)
Sink reactions are similar to demand reactions but are defined to be reversible and thus provide the network with metabolites (see Figure 7 for examples). These sink reactions are of great use for compounds that are produced by non-metabolic cellular processes but need to be metabolized. Adding too many sink reactions may enable the model to grow without any resources in the medium. Therefore, sink reactions have to be added with care. As for demand reactions, sink reactions are mostly used during the debugging process. They help to identify the origin of a problem (e.g., why a metabolite cannot be produced). These sink reactions are functionally replaced by filling the identified gap.
Growth medium requirements (Step 37)
Information about growth-enabling media is of great help in the following two stages. Thus, if possible, they should be collected prior to the conversion and debugging stage. The following information should be collected: 1) Which metabolites are present? 2) Are there any auxotrophies? 3) Define the composition of a base medium, e.g., water, protons, ions, etc. 4) Obtain information about rich medium composition. This data will be crucial for simulations and network evaluations. If uptake or secretion rates are available, they should also be documented and collected. While this step is easy for the experimentalist, researchers which cannot grow the target organism have to identify growth requirements from literature (or genome annotation). In some cases, research studies describe minimal, defined, or rich medium compositions. In other cases, the culturing conditions reported in some experimental study must be sufficient.
Stage 3: Conversion from reconstruction to mathematical model
In the third stage, the reconstruction is converted into a mathematical format and condition-specific models are defined. This stage can be mostly automated. Moreover, systems boundaries are defined, converting the general reconstruction into a condition-specific model. Note that the initial model may differ in scope and boundaries to the final model, which is obtained after multiple iterations of validation and refinement, and which is used to simulate phenotypic behavior in a prospective manner. Figure 7 illustrates the conversion of a reconstruction into mathematical format.
Simulation constraints (Step 42)
Using the functions in the COBRA Toolbox, it is very easy to change reaction constraints but it is sometimes difficult to keep track of all the changes. In fact, one of the most common reasons for errors in simulation is that reaction constraints are not correctly set (Table 4). Therefore, it is important to have an expectation of the results before running a simulation to avoid erroneous conclusions. It is recommended that the constraints are checked by copying the model reaction abbreviations as well as lower and upper bounds into a spreadsheet. For most models, this is the easiest way to see where problems are with the constraints. Similarly, copying calculated solution(s) into a spreadsheet is very helpful.
Table 4.
General error modes in metabolic networks.
| Error mode | Action |
|---|---|
| Wrong reaction constraints. | Check reaction constraints if they are applied correctly. |
| Missing transport reactions. | Add transport reactions. |
| Missing exchange reactions. | Add exchange reactions. |
| Cofactor cannot be consumed or produced. | Follow Figure 13. |
| Shuttling of compounds across compartment. | Adjust reversibility of transport reactions. |
Stage 4: Network evaluation = ‘Debugging mode’
The fourth stage in the reconstruction process consists of network verification, evaluation, and validation. Common error modes in metabolic reconstructions are listed in Table 4. The metabolic model created in the third step is tested, among other thing, for its ability to synthesize biomass precursors (such as amino acids, nucleotides triphosphates, and lipids). This evaluation generally leads to the identification of missing metabolic functions in the reconstruction, so called network gaps, which are added by repeating partially stage 2 and 3. This illustrates how the reconstruction process is an iterative procedure. An important issue is to decide when to stop the iterative process and call a reconstruction “finished”. This decision is normally based on the definition of the scope and purpose of the reconstruction.
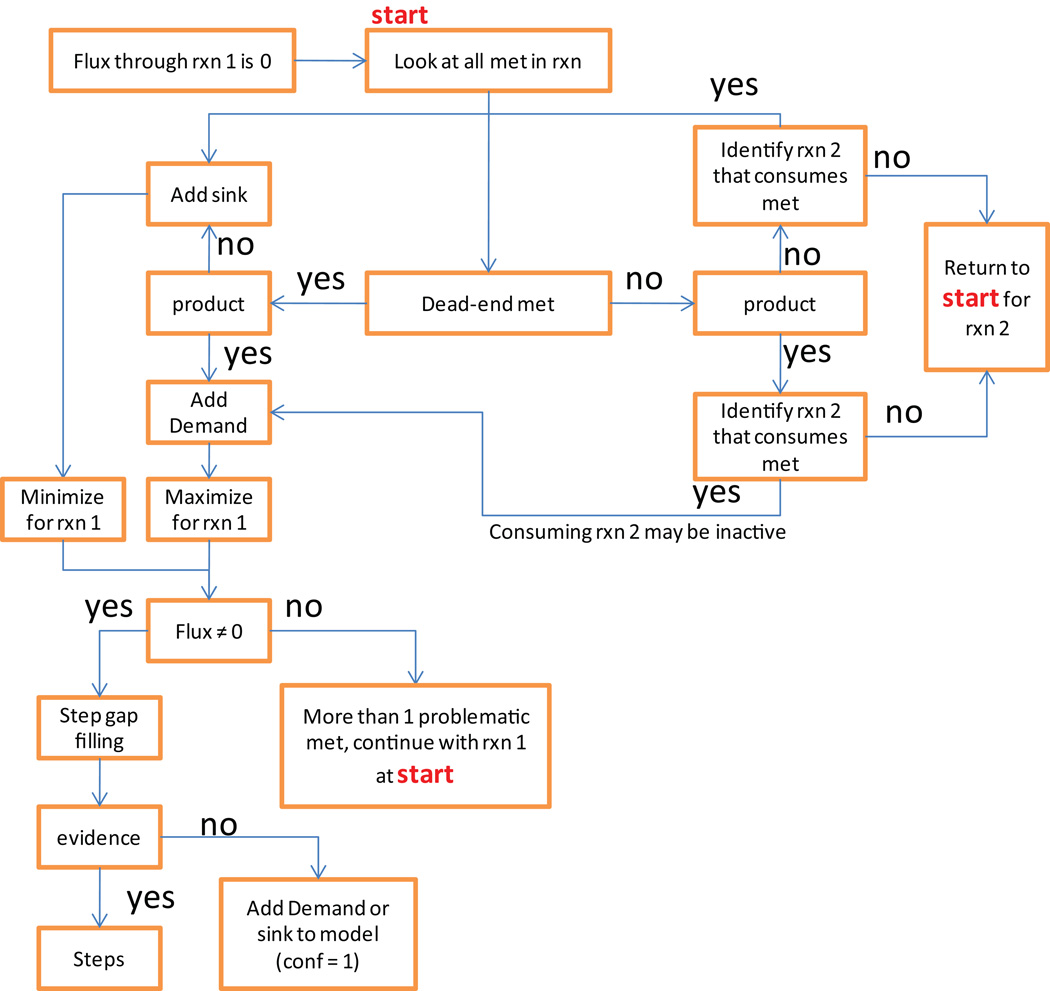
Metabolic dead-end (Step 45)
At this point, the first iteration of manual curated reconstruction is finished. It is expected that the network contain a significant number of gaps, i.e., missing reactions and functions. We recommend performing a first gap analysis at this stage of the reconstruction process as it will ease the subsequent computation and reduce the number of “bugs” in the model. Comparing dead-end metabolites identified in this step with the list generated in Stage 2 will accelerate the debugging process.
Candidate reactions for gap filling (Step 46 and 47)
This step will require an intensive literature search and may include re-annotation of a genome to find candidate genes and reactions to fill the gap (see Table 1 and supplemental methods 1, Table S3) for some example tools). KEGG41 maps, biochemical textbooks, or other available biochemical maps can be used to identify the metabolic ‘environment’ of the dead-end metabolite. If the genome annotation of the target organism is present in KEGG41, one can highlight the dead-end metabolite on the map (Supplemental methods, Figure S2). This context analysis may give an indication of which enzyme(s) may be able to produce or synthesize the dead-end metabolite and thus provide a good starting point for literature and/or genome search.
Gap-filling is a tricky business. In some cases, a gap should be filled to ensure that the model is functional, i.e., biomass precursor synthesis or a certain physiological function can be simulated. In other cases, filling a gap may enable the model to perform a function that the organism is not able to do (see Figure 8 for some examples). In general, if no information supports the existence of a particular gap reaction, the gap should only be filled if it is required for the model’s functionality. In such cases, the confidence score should be set to 1, which corresponds to “modeling purpose” only, and allows retrieving these low confidence reactions readily, if desired. Earlier, we highlighted that enzymes, which are listed in biochemical databases to catalyze multiple reactions should be included in the reconstruction with care and that it should be noted if evidence for all of the reactions could be found. Some of the identified dead-end metabolites will originate from such secondary reactions of these “multitasking” enzymes. Closing these gaps may affect the predictive potential of the reconstruction, therefore, only gaps should be filled which are required for network functionality (e.g., biomass precursor synthesis) or which have supporting data. Keep in mind that adding new reactions to the network may cause new gaps. Therefore, when adding reactions you should make sure that all metabolites are connected to the network.
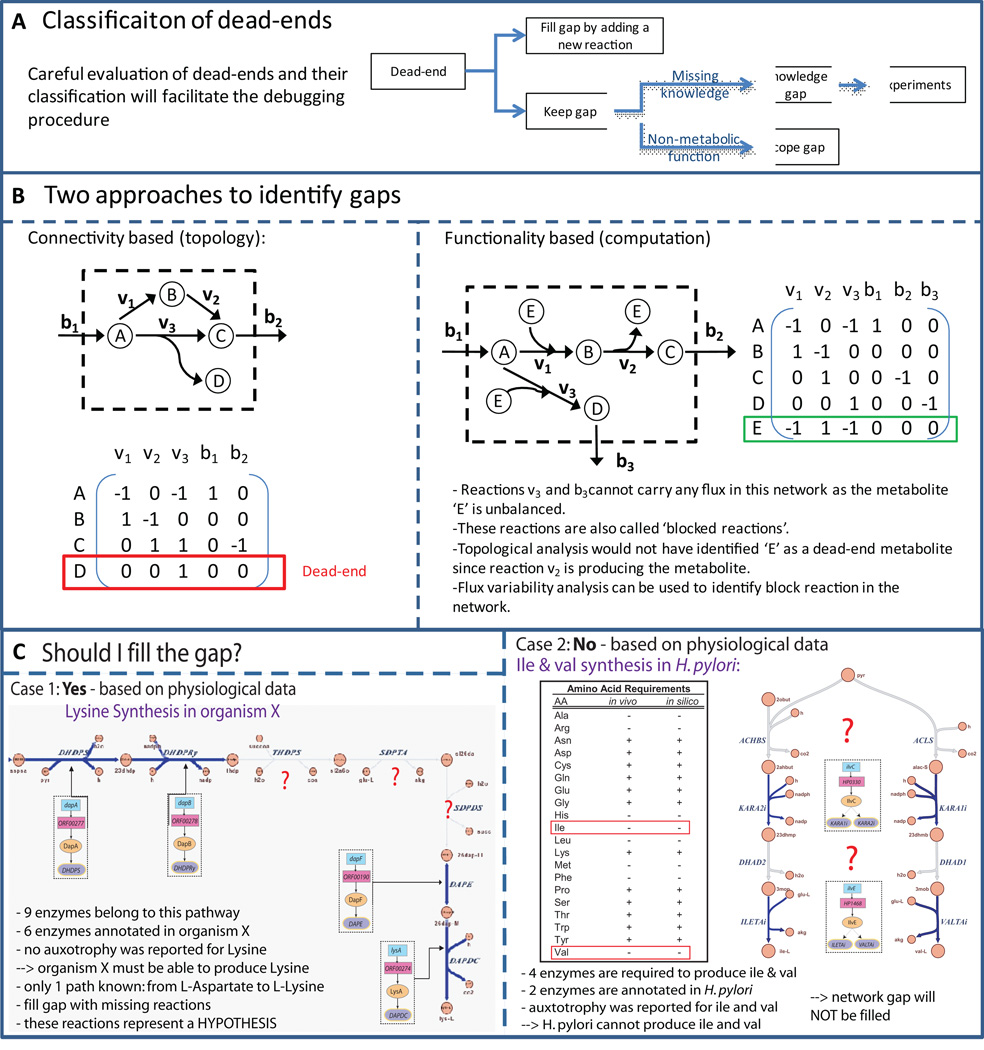
Figure 8. Gap analysis.
The gap analysis includes the identification and the tentative filling of network gaps. A. While many dead-end metabolites that create network gaps can be connected to the network by re-evaluating genomic and experimental data, some dead-end metabolites will remain in the refined, curated reconstruction. These dead-end metabolites can be grouped into two groups, depending on which type of reactions could connect them to the remaining network: knowledge gaps and scope gaps. While knowledge gaps represent missing biochemical knowledge for the target organism, the scope gaps include reactions and cellular processes, which are currently not accounted for in the metabolic reconstruction (e.g., DNA methylation). B. There are at least two approaches to identify gaps in the reconstruction. In the connectivity based approach, one can count the non-zero entries in each row of the S matrix and identify those metabolites, which are only produced or consumed. In the example, metabolite D is only produced by reaction v3 and the S matrix contains only one entry in the row corresponding to metabolite D. A second approach is based on model functionality: In this approach the models capability to carry flux through every network reaction is tested. This approach identifies blocked reactions, which are directly or indirectly associated with one or more dead-end metabolites. In the shown example, one would not identify metabolite E as a dead-end metabolite with the connectivity based approach as it is produced and consumed in the network. However, testing for flux through reactions containing E will show that reaction v3 and b3 cannot carry any flux in this model. C. Two sample cases are shown which address the question of filling a gap or not.
Stoichiometrically balanced cycles (SBCs) (Step 51–59)
SBC, or Type III extreme pathways66, are formed by internal network reactions and can carry fluxes despite closed exchange reactions (closed system). Examples for simple or more complex Type III pathways in metabolic networks can be found in67, 68. These SBCs are artifacts of metabolic reconstructions due to insufficient constraints (e.g., thermodynamic constraints and regulatory constraints). Recent efforts have concentrated on dealing with these SBCs67. Note that SBCs are not futile cycles. This protocol shows how to identify SBCs and highlights some possible approaches to eliminate them. However, no systematic, universally valid approach has been developed yet to eliminate SBCs. For practical purposes, in simulation one can use the ‘min norm’ option for the LP solver, which will minimize the sum of the squares of fluxes and thus, will return an optimal solution without netflux around SBCs.
The following steps will test if the model can or cannot grow. This means that we will test for qualitative behavior but not focus the correctness of predicted growth rates.
Biomass precursor production (Step 60–66)
The composition of the biomass reaction was determined in stage 2. It is best to test for model’s ability to produce each individual biomass component in standard medium condition (e.g., minimal medium M9 supplemented with D-Glucose) (Figure 4). This sequential approach will facilitate the debugging process and make it easier to find causes of error. It is very likely that these tests will lead to addition of further reactions by repeating steps listed in the second stage. Furthermore, this step may lead to the addition of reactions for which no experimental evidence and candidate genes can be identified. These reactions should be marked with the tag “modeling purposes” only (confidence score of 1). Be careful with such reactions as too many of them may change the overall properties of the network (in this or other simulation conditions). Moreover, the overall performance of the model in standard medium condition is determined and, in some cases, corrected. This step needs great care since there may be many possible ways of filling a gap.
Subsequently, the capability to produce biomass precursors needs to be tested in other growth media. Therefore, the correctness of the network content is evaluated in respect to all known growth conditions of the target organism. This includes all known carbon, nitrogen, sulfur, and phosphor sources. Physiological information is of great value to determine all growth conditions. For example, Gutnick et al.69 have tested about 600 compounds and have found that 100 can serve as carbon-or nitrogen source for Salmonella typhimurium. The model should be able to produce biomass in the majority of these instances. However, not all known conditions may be reproduced by the model – this is not a problem as it represents a starting point for experimental studies to identify missing metabolic functions. Nevertheless, great attention should be given to collecting and documenting those cases and thus enabling other researchers to pursue them.
By-product secretion (Step 70)
If such information is available, they can be used to further refine the model. The first question is if the model can produce the secretion product(s) given a substrate, while the subsequent question could be if a specific ratio of by-product secretion is correct. Classical biochemical studies often reported measured secretion products given a certain carbon source (e.g., Schroeder et al.70). This information is very helpful to evaluate the phenotypic traits of the model with those of the target organism.
Blocked reactions (Step 76–78)
Reactions that cannot carry any flux in any simulation conditions are called blocked reactions. These reactions are directly or indirectly associated with dead-end metabolites, which cannot be balanced and give rise to so-called blocked compounds71. It is good to be aware of those reactions, especially, if one expects different results in a simulation (e.g., false-negative analysis of single gene deletion). In the early phase of the debugging stage, the reconstruction can contain many blocked reactions that one might decide to fill if supporting information is available or if they are required for the overall function of the network. Targeted use of sink and demand reactions around a pathway of block reactions will facilitate the identification of the source problem. Other blocked reactions may remain if the terminal dead-end metabolite is beyond the scope of the metabolic reconstruction or no information and evidence for filling the gap is available. The easiest way to determine blocked reactions is by performing flux variability analysis72, 73.
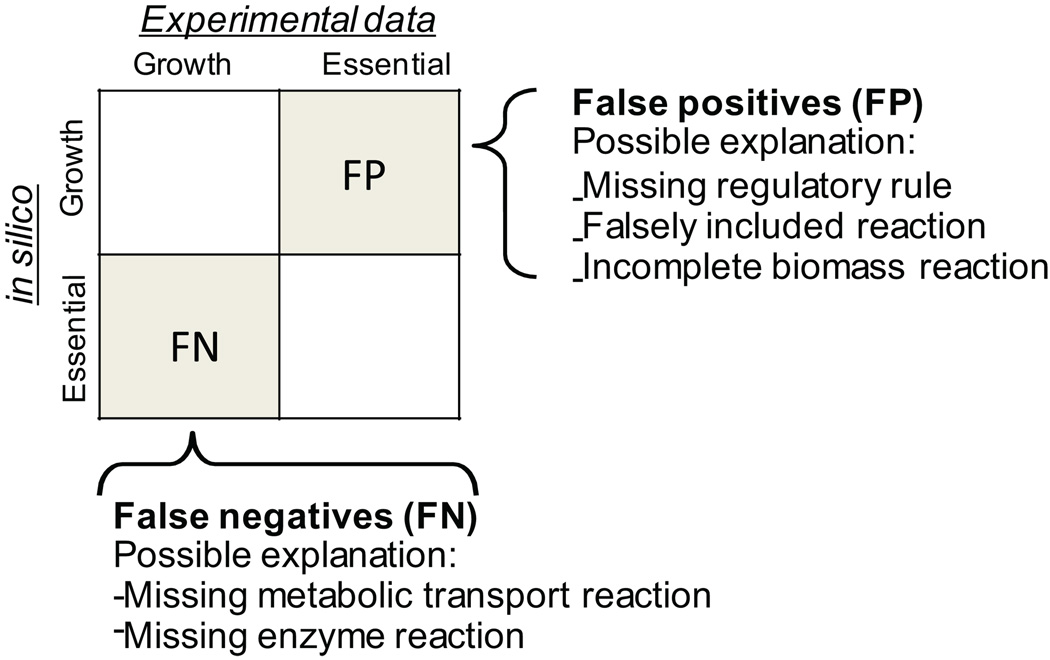
Single gene deletion phenotypes (Step 79 and 80)
Analysis of false positive and false negative predictions will help to further refine the network content if the information is available or provides a basis for experimental studies otherwise (Figure 9). Numerous reconstructions relied on phenotyping data (e.g., biolog data) or gene essentiality data to improve the network content and thus the predictive potential74, 75.
Figure 9. in silico gene essentiality study as network evaluation tool.
While agreement of gene essentiality between experimental and in silico data is very helpful to validate the reconstruction content and model setup, analysis of the inconsistencies will enable discovery of new biological knowledge
Known incapabilities (Step 81 and 82)
So far we compared whether the model could reproduce growth on certain substrate, secrete a particular by-product, etc. In this step, it should be tested if known incapabilities of the organism can also be reproduced by the model. For example, Helicobacter pylori is known to be auxotroph for certain amino acids, subsequently, their lack in the medium should abolish in silico growth76. It is important to use those “negative” data (incapabilities) as well as to correct for errors. Error cases can be removed by analyzing the confidence score associated with the reactions along the pathway. In the example of H. pylori, this would be the biosynthetic reactions leading to amino acid synthesis76. In a more algorithmic approach, a single reaction deletion study can be carried out and the results can be analyzed in terms of which deletions disable growth. This smaller subset of reactions needs to be manually evaluated. Note that the deletion of a single function may not be sufficient when alternate pathways exist in the network. Missing incapabilities may not only be caused by falsely added reactions in the metabolic network, but may be a consequence of missing regulatory information. Literature may provide the necessary data.
Comparison of predicted physiological properties with known properties (Step 83)
The model should be also tested for known capabilities, beside the aforementioned growth performance and secretion capability. For instance, this test can include known carbon splits in central metabolic pathways as observed with a recently published Pseudomonas putida network57. The P/O ratio was investigated for Methanosarcina barkeri77, Saccharomyces cerevisiae78 and compared to known growth data. Many more examples exist and the suite of necessary tests depends on the available data as well as the properties of the network.
Quantitative evaluation of growth rate (Step 84–94)
Too slow growth means that at least one precursor of the biomass function cannot be synthesized sufficiently. This implies that the model’s biomass production is either carbon-, nitrogen-, oxygen-, sulfur-, or phosphate-limited. Since there are generally less active uptake reactions for a particular element than biomass precursors, it is faster to test if any of the medium components are growth limiting. If the biomass reaction value increases when the uptake reaction flux is increased, it means that this compound is limiting. This gives you a hint as to, where in the network something must be missing or constraining. Furthermore analysis of shadow prices and reduced costs, which are associated with the LP solution, can be of great help to identify metabolites or reactions that limit the biomass rate. For example, the P. putida network57 that is not able to grow as fast as reported experimentally in silico when toluene is the carbon source. In silico analysis suggested that oxygen is rate limiting and that more oxygen-efficient reactions are missing in the network. Whether this discrepancy can be resolved by iterative network refinement depends on the specific case, and thus, no general solution can be proposed. As in the case of P. putida’s oxygen restriction, such error cases can lead to further experimental investigation that will ultimately increase our biological insight and the reconstruction’s quality.
When the predicted growth rate is higher than expected, many explanations are possible. 1. The optimization for growth assumes that microbial cells maximize their growth. However, as aforementioned, many other objective functions are possible and more appropriate depending on the experimental setup and growth conditions of the target organism6, 18–20, 79–82. 2. The GAM, which is part of the biomass reaction, may be estimated wrongly and needs adjustment. 3. It can indicate that constraints are missing or incorrect (e.g., NGAM, missing regulation). 4. Falsely included reactions increase growth rate. Knowledge about the model and the expected flux map is crucial for identifying the errors. Proton shuttling reactions may be present that circumvent the ATP synthetase (e.g., due to a futile cycle). Note that this is only the case in aerobic growth conditions. Such shuttling reactions may be enabled by many reversible transport reactions. Reactions associated with such loops can be readily identified (see Step 51–59). Also, looking at the flux through the reactions of oxidative phosphorylation may indicate if they are used under the aerobic condition or not. Alternatively, one can investigate if there is one reaction that enables the model to grow too fast. In this case, a single reaction deletion study will push you towards the right solution. Another approach could be to investigate the directionality of network reactions. As indicated earlier, reaction directionality may play a role in the fast growth. Therefore, improving reaction directionality assignments may be helpful. Make sure that only those reactions which are known to produce ATP are allowed for ATP synthesis, while all other reactions are set irreversible (ATP utilization). Similarly, reactions using quinones as electron acceptor should not run reversibly. This might cause problems and may allow circumventing the electron transport chain. These examples are very specific to a model and problem, and no general rule for corrections can be proposed.
Stage 5: Prospective use
Once the necessary content and desired in silico capability is reached, one can start to use the reconstruction in a prospective manner, which represents a fifth step in the reconstruction process that is not addressed here.
MATERIALS
EQUIPMENT
-
-
A standard personal computer that can run Matlab.
-
-
Matlab, version 6.0 or above (Mathwork Inc.), a numerical computation and visualization software (http://mathworks.com).
-
-
COBRA Toolbox (version 1.3.4 or above) is provided at http://systemsbiology.ucsd.edu/downloads/COBRAToolbox.
-
-
The SBML Toolbox for Matlab which allows reading models in SBML format (http://sbml.org/Software/SBMLToolbox).
-
-A linear programming (LP) solver. Multiple solvers are currently supported by the COBRA Toolbox:
- glpk (freeware): http://www.gnu.org/software/glpk/
- LINDO (LINDO Systems Inc.) Matlab API (commercial): http://www.lindo.com
- CPLEX (ILOG Inc.) through the Tomlab (Tomlab Optimization Inc.) optimization environment (commercial, but best LP solver available) http://tomopt.com/
- Mosek (MOSEK ApS) (commercial): http://www.mosek.com
-
-
Extreme pathway software package, X3, provided at http://systemsbiology.ucsd.edu/downloads/Extreme_Pathway_Analysis.
-
-
Excel (Microsoft Inc., http://office.microsoft.com/en-us/excel/default.aspx) or similar database programs can be used for collecting reconstruction information.
EQUIPMENT SETUP
COBRA Toolbox
The COBRA Toolbox16 should be downloaded and copied in a local folder on the user's computer. Extract the .zip file. After opening Matlab, a path should be set to the local folder, containing the COBRA Toolbox (Matlab → File → Set Path → Add with Subfolder, choose the corresponding folder and save). All working files (SBML and xls files) should also be stored in the local folder, in order to allow access to the reconstruction and models. A full documentation of the COBRA Toolbox can be found in the "doc" subfolder within the main Toolbox folder, which has all help files as html files. Furthermore, help for Matlab and COBRA Toolbox functions can be accessed via Matlab's "help" facility by typing "help function_name" on Matlab command line. See also Becker et al.16.
SBML Toolbox
Comprehensive documentation on SBML, the file format, and model setup, can be found at the official SBML website (http://sbml.org/documents/, level 2 version 1). The SBML file describing the model has to include at least the following information: stoichiometry of each reaction, upper/lower bounds of each reaction, and objective function coefficients for each reaction. Additionally, gene–reaction associations can be added to the "Notes" section.
Spreadsheet
The first two reconstruction steps are illustrated in this protocol using spreadsheets. It is important that the order of the columns in the spreadsheet match the example given in Supplemental methods 2.
Variables
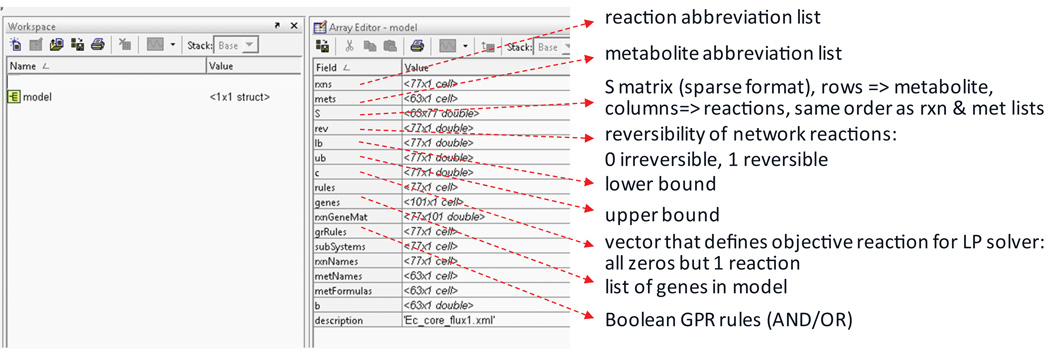
The imported model from the spreadsheets is contained in a model structure (see Figure 10 for details on this structure). All functions in the COBRA Toolbox access the information stored in the model structure. The values computed by the COBRA Toolbox are fluxes, which represent reaction rates for all model reactions. The units for fluxes used throughout this protocol are mmol/gDW/h, where gDW is the dry weight of the cell in grams.
Figure 10. Components of the model structure in Matlab.
The reconstruction is imported into Matlab (Step 39). The entire reconstruction content is stored in a structure array. The screen shot illustrates the main fields contained in the model structure. The information is stored in subarrays in these fields. Note that the order of the reactions and metabolites corresponds to the order of columns and rows in the S matrix, respectively.
Installation
The Matlab software, SBML Toolbox, and one or more of the suggested LP solvers should be installed following the instructions of the software providers. Note that the SBML Toolbox and the LP solver also need to be accessible in the Matlab path (see above). Sample installation instructions for the lp_solve LP solver on Windows can be found in Becker et al.16. The SBML Toolbox is downloaded and installed. Follow the installation instructions. Choose ‘libsbml’ in the dialog field. Once installed, open Matlab and type ‘install’. If you get an error with ‘libsbml’ (when opening Matlab again), go to set path and add the folder ‘libsbml’ with subfolders.
The COBRA Toolbox is initiated by typing in the Matlab command window:
changeCobraSolver(solverName); where ‘solverName’ is, e.g.,

initCobraToolbox;
 SBML Toolbox and the LP solver should be tested for functionality following the software provider's instructions before attempting to use the COBRA Toolbox.
SBML Toolbox and the LP solver should be tested for functionality following the software provider's instructions before attempting to use the COBRA Toolbox.
X3
X3 is the software package used to determine stoichiometrically unbalanced cycles, or Type III pathways. X3.exe needs to be placed and extracted in the local folder. The help can be accessed by opening the DOS command line, changing to the local folder, and typing X3 –h. The extreme pathway tool will be called from Matlab by the COBRA Toolbox.
KEGG
We will illustrate many steps of the protocol using KEGG41 because it is freely accessible and very helpful for the illustrated pathway-by-pathway reconstruction process. However, one has to keep in mind three properties of KEGG41: 1. It is NOT organism-specific data; hence, not all reactions associated with an enzyme may be catalyzed by the enzyme of the target organism, and 2. KEGG41 may not update the genome annotation of the target organism on a regular basis; hence, the information may be outdated and need a “second opinion” from another more recent resource. 3. Not all reactions in the KEGG41 database are mass- and charge-balanced as they omit protons and water molecules although the KEGG database is continuously updated and improved83, 84.
PROCEDURE
Stage 1: Creating a draft reconstruction
-
1|
Obtain genome annotation. The genome annotation can be obtained from various sources, including sequencing centers (e.g., TIGR) and the National Center for Biotechnology Information (NCBI) depository. The following information should be retrieved for each gene: genome position, coding region, strand, locus name, alias, gene function (i.e., current annotation), protein classification (e.g., Enzyme Commission (E.C.) number40).
 In eukaryotic organisms, information regarding alternate transcripts must also be collected, since different splice forms may have distinct functionality or cellular localization.
In eukaryotic organisms, information regarding alternate transcripts must also be collected, since different splice forms may have distinct functionality or cellular localization. -
2|
Identify candidate metabolic functions. This step is straight-forward once the genome annotation has been obtained. Apply different approaches to collect candidate metabolic functions including searching for E.C. numbers (complete and partial)40 and for metabolic terms (e.g., dehydrogenase, kinase, etc.) (Supplemental methods 1, Figure S1). If gene ontology (GO)39 or cluster of orthologous groups of proteins (COG)85 information is obtained with the genome annotation, they can be used as well to find metabolic enzymes.
-
3|
Obtain candidate metabolic reactions for these functions (e.g., from KEGG41). Use comprehensive reaction databases such as KEGG41, Brenda42, and publically available reconstructions as a resource to combine the gene functions with metabolic reactions.
-
4|
Assemble draft reconstruction. Collect all candidate metabolic genes and their potential reactions in a spreadsheet. This spreadsheet will serve as a starting point for the manual curation process (see Figure 2, and Supplemental data 1, for an example).
-
5|
Collect experimental data. The manual curation process relies heavily on experimental, organism-specific information. All possible information needs to be retrieved. The following steps will include reviewing scientific literature during which the information listed in Table 5 should be collected. Alternatively, additional experimental data can be generated by growing and measuring various metabolic capabilities and properties of the target organism.
Table 5.
List of experimental data commonly used for reconstruction, modeling and network evaluation.
| Data type | purpose | Literature | Databases | Genome | Growth experiments | Phenotyping | Protein structures | Comparative genomics* | Microarray data | Proteomic data | Exometabolmic data | Metabolomic data | Flucomic data | Single gene deletion | Biochemical essays |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene function | Reconstruction refinement | X | X | X | X | X | |||||||||
| Protein function | Reconstruction refinement | X | X | X | X | X | X | ||||||||
| Reaction mechanism | Reconstruction refinement | X | X | X | X | ||||||||||
| Growth media | Transport, simulations | X | X | X | X | X | |||||||||
| Carbon sources | Transport reactions, simulations | X | X | X | X | X | |||||||||
| Gene/protein presence/absence | Condition-specific models, cell-type models | X | X | X | X | ||||||||||
| Reaction constraints | Simulations | X | X | X | X | X | X | ||||||||
| Network evaluation | Debugging | X | X | X | X | X | X | X | |||||||
| Gap filling | Debugging | X | X | X | X | X | X | X | |||||||
| Cofactor/substrate specificity | Reconstruction refinement | X | (X) | X | X | ||||||||||
| Reaction directionality | Reconstruction refinement | X | (X) | X |
Comparative genomics can be done using, e.g., SEED32.
Stage 2: Manual reconstruction refinement
-
6|
Determine and verify substrate and cofactor usage. Use primary literature, and to a lesser extend KEGG41 and Brenda42, to determine and verify substrate and cofactor specificity of the enzyme in the target organism. As a rule of thumb, one can assume that enzymes, which have only one reaction associated in KEGG41, for example, do not require organism refinement.
 Often only biochemical data can reveal the correct cofactor and substrate as binding sites may not be distinguishable in gene sequence for related metabolites.
Often only biochemical data can reveal the correct cofactor and substrate as binding sites may not be distinguishable in gene sequence for related metabolites. -
7|
Obtain a neutral formula for each metabolite in the reaction. The neutral formula can be readily obtained from various resources, including KEGG41, Brenda42, and PubChem86. While PubChem86 is more comprehensive, KEGG41 is certainly the most accessible resource, especially, when KEGG41 is used for obtaining the reactions.
 Check that the formula is correct (i.e., verify with other databases and textbooks).
Check that the formula is correct (i.e., verify with other databases and textbooks). -
8|
Determine the charged formula for each metabolite in the reaction. Retrieve the molecular structure for each metabolite, if you have not already done so in Step 7. Determine the charged formulae (e.g., for pH 7.2) based on the pKa value of the functional groups (Figure 3). This can also be done using software packages such as Pipeline Pilot and pKa DB can predict pKa values for a given compound (Table 1).
-
9|
Calculate reaction stoichiometry. Count every element and the charge on each side of the equation. On each side, the same number of elements and charge must be present. Protons and water may need to be added to the reaction. This step is easy for many central metabolic reactions but may become challenging for more complex reactions.
-
10|
Determine reaction directionality. Use biochemical data and literature if available. Alternatively, the standard Gibbs free energy of formation (ΔfGo) and of reaction (ΔrGo) can be calculated based on group contribution theory for most KEGG41 reactions from Web GCM44, 45. If data reaction of interest is not available, the following rule of thumb may be applied: 1) reactions involving transfer of phosphate from ATP to an accepter molecule should be irreversible (with the exception of the ATP synthetase, which is known to occur in reverse); 2) reactions involving quinones are generally irreversible.
-
11|
Add information for gene and reaction localization. This information may be difficult to obtain from primary literature. Consider to use algorithms such as PSORT47 and PASUB48 if no experimental data is available.
 In the absence of appropriate data, proteins should be assumed to reside in the cytosol.
In the absence of appropriate data, proteins should be assumed to reside in the cytosol. -
12|
Add subsystem information to reaction. This will be of great help for the debugging and network evaluation work. The subsystem assignment can be done either based on biochemical textbooks or KEGG41 maps. Note that a reaction or an enzyme can appear in multiple KEGG41 maps; therefore, the subsystem should reflect its primary function.
-
13|
Verify gene-protein-reaction (GPR) association. Determine if the functional protein is a heteromeric enzyme complex; if the enzyme (complex) can carry out more than one reaction; and if more than one protein can carry out the same functions (i.e., isozymes exist). Use KEGG41, organism-specific databases and primary literature.
 Mistakes or mis-assignments in the GPR associations will change results of in silico gene deletion studies.
Mistakes or mis-assignments in the GPR associations will change results of in silico gene deletion studies. -
14|
Add metabolite identifier. Associate each metabolite with at least one of the following identifiers: ChEBI52, Kegg41, and PubChem53. In addition, associate database-independent representations of metabolites such as SMILES54 and InCHI strings55, 56 with each metabolite.
-
15|
Determine and add confidence score. Use the proposed confidence score system listed in Table 2.
-
16|
Flag reactions for which information from other organisms was used.
-
17|
Add references and notes based on experimental information. In Steps 6 to 13 many organism-specific, experimental data is collected that needs to be associated with the reconstruction in the form of references and notes. This allows other users of the reconstruction to easily retrace the evidence and supporting material for reaction and gene inclusion.
-
18|
Repeat Steps 6 to 17 for all genes identified in the draft reconstruction. Also repeat these steps for metabolic functions that were identified from bibliomic sources during the reconstruction process and whose genes could not determined.
-
19|
Add spontaneous reactions to the reconstruction. Use biochemical literature and databases (KEGG41 and Brenda42) to identify candidate spontaneous reactions to include. Only include those reactions which have at least one metabolite present in the reconstruction to minimize the number of dead-end. Associate the spontaneous reactions with an artificial gene (s0001) and protein (S0001).
-
20|
Add extracellular and periplasmic transport reactions to the reconstruction. This addition is done based on experimental data. The rule here is that for every metabolite that is known to be taken up from the medium or that is known to be secreted into the medium, a transport reaction should exist (from extracellular space to periplasm and from periplasm to cytoplasm). Include transport reactions for metabolites that can diffuse through the membranes. Small, hydrophilic compounds can diffuse through the outer membrane87.
-
21|
Add exchange reactions to the reconstruction. Exchange reactions need to be added for all extracellular metabolites. The exchange reactions represent the systems boundaries (Figure 7).
-
22|
Add intracellular transport reactions to the reconstruction. (For multi-compartment reconstructions only). Use biochemical and physiological information, however, finding experimental data is often not easy. Only include intracellular transport reactions that really need to be there to avoid futile cycles, or Type III pathways.
-
23|
Draw metabolic map (optional). If appropriate drawing software is available, the creation of organism-specific maps is very useful for gap analysis, network evaluation, and data mapping.
Determine biomass composition
-
24|
Determine the chemical composition of the cell, i.e., protein, RNA, DNA, lipids, Cofactor content (see also supplemental methods 1, Figure S3A). This information can be retrieved from experimental data or primary literature.
-
25|Determine the amino acid content either experimentally (option A) or by estimation (option B).
- Determination of amino acid content experimentally.
- Obtain data for each amino acid.
- Estimation of amino acid composition from genome information. Use, for example, CMR database (Figure 11).
- The amino acid content can be determined by selecting the Genome Tools tab, followed by Analysis Tools, and finally Codon Usage.
-
26|
Use the molar percentage and molecular weight of each amino acid to calculate the weight per mol protein. Sum the individual amino acid values to give a total molecular weight of the protein content. Subsequently, calculate the weight percent per amino acid. Then multiply the calculated weight percent by the cellular content percentage of the macromolecule and divide by the molecular weight of the individual monomer (Figure 11 and Supplemental methods 1, Figure S3B).
-
27|Determine the nucleotide content either experimentally (option A) or by estimation (option B).
- Determination of nucleotide content experimentally.
- Obtain data for each deoxynucleotide triphosphate (dATP, dCTP, dGTP, dTTP) and each nuvleotide triphosphate (ATP, CTP, GTP, UTP).
- Estimation of nucleotide composition from genome information. Use, for example, CMR database (Figure 11).
- From the Genome Tools tab (see Step 25), select Summary Information, followed by DNA Molecule Info. The number of each dNTP (i.e., dATP, dCTP, dGTP, and dTTP) present in the genome is listed on the summary page.
- In order to determine the RNA composition of the cell, use the codon usage that was accessed for the amino acid content (Step 25). Remember that RNA incorporates uracil instead of thymine; therefore, the codon usage needs to be read with every T replaced by a U.
- Tabulate the frequency of each RNA monomer.
-
28|
Calculate the fractional distribution of each nucleotide to the biomass composition by repeating Step 26.
-
29|
Determine the lipid content. Determine the contributions from fatty acids and phospholipids. Therefore, determine the average molecular weight of a fatty acid in the cell by incorporating the average fatty acid composition of the cell (requires experimental data, e.g., from literature). Use the average molecular weight of each fatty acid and sum the weight contributions of each to determine the average molecular weight for a fatty acid chain. Use this weight to calculate the average molecular weight of various lipids within the cell. Perform such computation by summing the molecular weight of the core structure of the molecule and the molecular weight of the fatty acids attached to the core structure based on the average molecular weight of one fatty acid that was determined above. The molar percentages of the three major phospholipids, phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL), may be found in the literature. Thus, determine the phospholipid contributions to the biomass function (Supplemental methods 1, Figure S3C).
-
30|
Determine the content of the soluble pool (polyamines and vitamins and cofactors). The soluble pool contains, for example, spermidine, coenzyme A, and folic acid (see supplemental methods 1, Table S4, for a more comprehensive list). Use Figure 12 as a template to determine the composition of the soluble pool for your target organism and to calculate their fractional distributions to the biomass reaction.
-
31|Determine the ion content. The calculation of the molar fraction of the ions is illustrated in Supplemental methods 1, Table S5. It assumes that concentration data are available or can be estimated for each ion. Information about the ion content can be obtained from different resources, including primary literature and databases (e.g., CyberCell Database88). Convert the reported concentration (ci) for each ion species i, into mM. Add all ion species (total ion concentration, ctotal). Calculate the molar fraction (fi) of each ion species i by dividing ci with ctotal:
-
32|
Determine growth associated maintenance (GAM). Use experimental data to determine the GAM. Alternatively, part of GAM can be estimated by the energy required for macromolecular synthesis, e.g., proteins. Figure 13 illustrates how to calculate the GAM using the total amount (mmol) of macromolecule (Protein, DNA, and RNA) and known amount of phosphate bonds necessary to synthesize a macromolecule. Note that this estimate will be too low as other growth-associated cellular processes also require ATP.
-
33|
Compile and add biomass reaction to the reconstruction. In this step, all precursors are assembled in one single reaction - the biomass reaction - which is then added to the reaction list of the reconstruction. Add GAM to biomass reaction as follows: x ATP + x H2O → x ADP + x Pi + x H+, where x is the number of required phosphate bond.
 Note that some metabolites might be produced. For instance, in the E. coli biomass reaction, proton (H+), orthophosphate (Pi) and some other metabolites are produced65. These metabolites originate mainly from the growth associated ATP hydrolysis (Step 32).
Note that some metabolites might be produced. For instance, in the E. coli biomass reaction, proton (H+), orthophosphate (Pi) and some other metabolites are produced65. These metabolites originate mainly from the growth associated ATP hydrolysis (Step 32). -
34|
Add non-growth associated ATP maintenance reaction (NGAM). Add the following reaction to the reconstruction reaction list: 1 ATP + 1 H2O → 1 ADP + 1 Pi + 1 H+.
-
35|
Add demand reactions to the reconstruction. Add demand functions for compounds that are known to be produced by the organism, e.g., certain cofactors, lipopolysaccharide, and antigens, but i) for which information is available about their fractional distribution to the biomass or ii) which may be only produced in some environmental conditions.
-
36|
Add sink reactions to the reconstruction. Sink reactions are of great use for compounds that are produced by non-metabolic cellular processes but needed to be metabolized.
 Adding too many sink reactions may enable the model to grow without any resources in the medium. Therefore, sink reactions have to be added with care.
Adding too many sink reactions may enable the model to grow without any resources in the medium. Therefore, sink reactions have to be added with care. -
37|
Determine growth medium requirements. Use experimental data and primary literature to retrieve essential nutrients and defined medium composition. Compile a list of growth requirements.
Figure 11. Flow chart to calculate the fractional contribution of a precursor to the biomass reaction.
This approach can be used for amino acids, nucleotide triphosphates (ATP, GTP, CTP, UTP), and deoxy-nucleotide triphosphates (dATP, dGTP, dCTP, dTTP). The steps are illustrated for L-alanine (Ala). (A) The fractional contribution of alanine to the proteome is obtained from experimental data or estimated from genome sequence. (B) To convert the molar percentage into weight of alanine per mole protein, the molar percentage is multiplied by the molecular weight of alanine. Note that the polymerization of amino acid leads to the loss of a water molecule, which needs to be considered when calculating the molecular weight. Once the weight of amino acid per mole protein is obtained for all amino acids, they are summed to obtain the weight of protein per mole protein. (C) The weight of alanine per mole protein is converted into weight alanine per weight protein by multiplying with the sum of all amino acids’ weight. (D) Finally, the weight of alanine is multiplied by the cellular content of protein (see Figure 13A) and divided by its molecular weight to obtain the mole alanine per cell dry weight. Multiplying this molar contribution by a factor of 1000 will result in a final unit of mmol alanine per gram dry weight.
Figure 12. Determination of the content of soluble pool.
Depending on the available information from literature, measurements or database entries the conversion into mmol/gDW and g/gDW is shown. The value in the purple box corresponds to the stoichiometric coefficient in the biomass reactions for the precursor. a Information was obtained from Cybercell Database (CCDB, see Table 1 for the link).75
Figure 13. Determination of growth associated maintenance (GAM) cost.
A. Calculation of growth-associated maintenance cost. B. Sample calculation for E. coli65. The energy necessary for the synthesis of the macromolecules from the building blocks were obtained from Table 5 – 6 of Chapter 3 in Neidhardt et al.64. The coefficient cP, cD, cR were calculating the total energy necessary for the macromolecules divided by the total number of building blocks (See Neidhardt et al.64).
Stage 3: Conversion from reconstruction to mathematical model
-
38|
Initialize the COBRA Toolbox. Install Matlab, the required Toolboxes (SBML Toolbox and COBRA Toolbox), and a LP solver16. Start Matlab as described in the installation instruction. Within Matlab, change to the directory where the COBRA Toolbox was installed. Initiate the COBRA Toolbox by entering the command
 in the Matlab command line. Note that the default LP solver can be changed by editing the initCobraToolbox script or at any time during a Matlab session by using the
in the Matlab command line. Note that the default LP solver can be changed by editing the initCobraToolbox script or at any time during a Matlab session by using the  function included in the Toolbox.
function included in the Toolbox.A list of frequently used COBRA Toolbox functions is given in Supplemental methods 1, Table S6. See also the Nature protocol on the COBRA Toolbox for details on initializing and using the Toolbox16.

-
39|Load reconstruction into Matlab. Save the reaction list in a spreadsheet with the same order of columns as shown in supplemental methods 2 (‘RxnFileName’). A second file containing metabolite information needs to be saved as well (‘MetFileName’). The following COBRA Toolbox function should be used to read the reconstruction into Matlab:
The loaded metabolic model is stored in a structure named ‘model’ in Matlab. This structure contains all the information about the reconstruction in the different fields of the structure. Figure 10 provides a description of the individual fields and their content.

-
40|Verify S matrix. Use
-
41|Set objective function. Use the following COBRA Toolbox function to set the objective function of the model:
The reaction(s) that should be set as the objective function is given by ‘rxnNameList’. It will receive a corresponding coefficient ‘objectiveCoeff’. This means that a single reaction or a linear combination of multiple reactions can be chosen as objective function.
 The COBRA Toolbox is set up in a way that the coefficient(s) for the objective function has to be a positive number. When minimizing, the input option to the COBRA Toolbox function optimizeCBmodel.m can be set to ‘min’. The default option of the ‘optimizeCBmodel’ function is maximizing (‘max’) (see Supplemental methods 1, Table S6).
The COBRA Toolbox is set up in a way that the coefficient(s) for the objective function has to be a positive number. When minimizing, the input option to the COBRA Toolbox function optimizeCBmodel.m can be set to ‘min’. The default option of the ‘optimizeCBmodel’ function is maximizing (‘max’) (see Supplemental methods 1, Table S6). -
42|Set simulation constraints. Use the following function to set the constraints of the model:The list of reactions for which the bounds should be changed is given by ‘rxnNameList’, while an array contains the new boundary reaction rates (‘value’). The type of bound can be set to lower bound (‘l’), upper bound (‘u’). Alternatively, both bounds can be changed (‘b’). Use the following command to lists all constrained reactions that are greater than a minimal value (‘MinInf’) and smaller than a maximal value (‘MaxInf’):Additionally, there is a function available that lists all reactions and their flux values in a solution:
Stage 4: Network evaluation = ‘Debugging mode’
Test if network is mass- and charge balanced
-
43|Check for stoichiometrically unbalanced reactions. All, or a subset, of the network reactions can be given as input (‘RxnList’) along with the model structure (‘model’):
In case of unbalanced reactions, the function returns a structure containing the name of the unbalanced reaction and which elements are unbalanced (‘UnbalancedRxns’).
-
44|
Evaluate stoichiometrically unbalanced reactions. Looking at the reaction equations and the charged formula for each metabolite will help to balance the reactions. Normally, there are two common errors causing unbalanced reactions: Missing proton and/or water or the stoichiometric coefficient of at least one metabolite is wrong. If it is the latter error, repeat Step 9. If a proton as substrate is missing, a proton donor may be necessary (e.g., NADH, NADPH). This will require a literature search to identify a candidate proton donor. If a water molecule is missing, keep in mind that after adding water to the equation the proton and oxygen will need to be balanced again.
 A few network reactions are always unbalanced. These reactions include the biomass reaction, demand, sink, and exchange reactions.
A few network reactions are always unbalanced. These reactions include the biomass reaction, demand, sink, and exchange reactions. -
45|Identify metabolic dead-ends. Use
-
46|
Identify candidate reactions to fill gaps. Use primary literature and genome annotation tools to find candidate genes and reactions to fill the gap (see Table 1 and 8 for some example tools). Also, use KEGG41 maps, biochemical textbooks, or other available biochemical maps to identify the metabolic ‘environment’ of the dead-end metabolite. If the genome annotation of the target organism is present in KEGG41, one can highlight the dead-end metabolite on the map. This may give an indication of which enzyme(s) may be able to produce or synthesize the dead-end metabolite and thus provide a good starting point for literature and/or genome search.
-
47|
Add gap reactions to the reconstruction. If experimental and/or annotation data support gap reactions or they are needed for modeling purposes, the reaction(s) should be added to the reconstruction by repeating Steps 6 to 17.
 Keep in mind that adding new reactions to the network may cause new gaps. Therefore, when adding reactions you should make sure that all metabolites are connected to the network. Repeat Step 45, if necessary.
Keep in mind that adding new reactions to the network may cause new gaps. Therefore, when adding reactions you should make sure that all metabolites are connected to the network. Repeat Step 45, if necessary. -
48|
Add notes and references to dead-end metabolites. Each dead-end metabolite should be documented. The note should distinguish between knowledge and scope gap for future reference (Figure 8A).
 The more detailed and carefully the gap filling steps are done (Steps 46 to 48) the easier and faster the debugging process will be.
The more detailed and carefully the gap filling steps are done (Steps 46 to 48) the easier and faster the debugging process will be. -
49|
Add missing exchange reactions to model. The gap filling process may have resulted in the inclusion of further transport reactions. Exchange reactions thus need to be added to the reconstruction. Repeat Step 21.
-
50|Set exchange constraints for a simulation condition. Determine an environmental condition in which most network evaluation tests should be carried out initially (‘standard condition’). Use
Test for stoichiometrically balanced cycles, or Type III pathways (optional)
-
51|Test for Type III pathways. Therefore, use the following function:
A list of indices of the exchange reactions in the S matrix (‘ListExch’) has to be provided to the function. These exchange reactions will be set to zero and then the flux variability of the closed model is calculated. This function requires that X3.exe is in the working directory. The function will return if there are Type III pathways in the model.

-
52|
Analyze output if Type III pathways found. If Type III pathways have been identified, there are two output files: one file (‘ModelTestTypeIII_myT3.txt’) has all Type III pathways as a matrix, where the rows are the different pathways and the columns correspond to the network reaction (in the same order as given in ‘ModelTestTypeIII_myRxnMet.txt’). Note that the extreme pathway package converts network reactions into elementary reactions (i.e., irreversible reactions). A second file (‘ModelTestTypeIII_myT3_Sprs.txt’) contains the Type III pathways in a sparse format, which is easier to analyze by hand.
-
53|
Identify Type III pathways. Note that reversible reactions form Type III pathways as well. In general, you are looking for Type III pathways that contain three or more reactions. It is possible that multiple, complicated Type III pathways exist in the model. Listing the corresponding reaction formulas or even drawing a map might be helpful to understand how the reactions form the loop(s).
-
54|
Analyze directionality of each reaction participating in a Type III pathway. Re-investigate the thermodynamic information if available (Step 10).
-
55|
Analyze if any reaction participating in a Type III pathway may be falsely included in the reconstruction by reviewing the supporting evidence.
-
56|
If none of the reactions or reaction directions can be corrected based on experimental or thermodynamic information, you can try to iteratively limit the directionality of the loop reactions. A more elaborate procedure has been described elsewhere67.
-
57|
Adjust directionality for all reactions identified in Step 54 to 55, note the change and reasons.
-
58|
After eliminating a reaction direction or a deletion of a reaction, repeat the Type III pathway analysis. Also, make sure that the removal of directionality or reaction does not affect growth.
 Keep in mind that such a change to the network is a hypothesis and may cause problems under different simulation conditions (e.g., environmental conditions).
Keep in mind that such a change to the network is a hypothesis and may cause problems under different simulation conditions (e.g., environmental conditions). -
59|
Re-compute gap list.
 . Again, the list ‘Gaps’ contains remaining gaps in the network. It will be helpful to have an overview of the remaining dead-end metabolites.
. Again, the list ‘Gaps’ contains remaining gaps in the network. It will be helpful to have an overview of the remaining dead-end metabolites.
Test if biomass precursors can be produced in standard medium (set in Step 42)
-
60|Obtain the list of biomass components:
-
61|Add demand function for each biomass precursor (‘metaboliteNameList’):
Note that ‘metaboliteNameList’ should be identical to ‘BiomassComponent’, obtained in Step 60. The new model is returned (‘modelNew’), which has additional demand reactions for every precursor whose abbreviations are listed in ‘rxnNames’.
For each biomass component i, perform the following test:
-
62|Change objective function to the demand function (‘rxnName’):
-
63|Maximize (‘max’) for new objective function (Demand function)
The structure ‘FBAsolution’ contains the optimal solution vector (‘FBAsolution.x’) and also the value for the objective reaction (‘FBAsolution.obj’). If it is Case 1, the model can produce biomass component (FBAsolution.obj > 0), proceed with the next biomass component. If it is Case 2, the model cannot produce biomass component (FBAsolution.obj = 0). Follow Steps 64 and 65.
-
64|
Identify reactions that are mainly responsible for synthesizing the biomass component.
-
65|
For each of these reactions, follow the wire diagram given in Figure 14.
-
66|
Test if biomass precursors can be produced in other growth media. Repeat Step 60 to 65.
Figure 14. Flow chart on debugging network reactions that cannot carry flux.
‘rxn ‘ stands for reaction. ‘conf’ stands for confidence score. ‘met’ stands for metabolite.
Test if model can produce known secretion products
-
67|
Collect list of known secretion products and medium conditions.
-
68|Set the constraints to the desired medium condition (e.g., minimal medium + carbon source). For changing the constraints use the following function:Reactions whose bounds should be changed are listed in ‘rxnNameList’. The new value for each reaction is contained in the array ‘value’. Finally, define the type of constraint in the list ‘boundType’. The possible types are: ‘l’ for lower bound, ‘u’ for upper bound, and ‘b’ if both reaction bounds should be set to the specified value. If the model shall be required to grow in addition to producing the by-product, set the lower bound (boundType = ‘l’) of the biomass reaction (‘rxnNameList ‘) to the corresponding value (‘value’).
-
69|Change the objective function to the exchange reaction of your secretion product:
The reaction(s) that should be set as the objective function is given by ‘rxnNameList’. They will receive a corresponding coefficient ‘objectiveCoeff’.
-
70|Maximize (‘max’) for the new objective function (as a secretion is expected to have a positive flux value, see Figure 7):
If the product can be produced (FBAsolution.obj > 0), proceed with the next by-product. If the product cannot be produced (FBAsolution.obj = 0), the corresponding pathway is missing or incomplete and thus gap analysis must be performed (Steps 45 to 49).
Test if model can produce a certain ratio of two secretion products
-
71|Set the constraints to the desired medium condition (e.g., minimal medium + carbon source). For changing the constraints use the following function:
-
72|
Verify that both by-products can be produced independently. Repeat Steps 67–70.
-
73|Add a row to the S matrix (see Figure 8B for an example of a S matrix) to couple the by-product secretion reactions:The two reactions that should be set to a certain ratio are listed in ‘ListOfRxns’. Their ratio is given in ‘RatioCoeff’ by listing the corresponding coefficients in this array. For example, 1:2 is given as [12]. If the model is required to growth while producing the by-product, set the lower bound of the biomass reaction to the corresponding value.
-
74|Change the objective function to the exchange reaction of one of your secretion products:
-
75|Maximize for the new objective function (as a secretion is expected to have a positive flux value, see Figure 7):
If the product can be produced (FBAsolution.obj > 0), the second by-product can be produced in the defined ratio. If the product cannot be produced (FBAsolution.obj = 0, or problem is infeasible), i.e., the ratio cannot be matched. The debugging is less straight-forward in this case as multiple reasons may apply. One very likely reason is that the organism (or cell) in the experimental condition under which the ratio was determined did not grow optimally. However, if you set in Step 71 a lower bound on the growth rate that may cause the discrepancy (due to competition for, e.g., carbons in by-products and biomass reaction). You could try to set this bound lower. Alternatively, some more elaborate tools that are currently not in the COBRA Toolbox can be used to identify missing genes/reactions (Supplemental methods 1, Table S3).
Check for blocked reactions
-
76|Change simulation conditions to rich medium or open all exchange reactions:
Note that the value of the exchange reactions (‘rxnNameList’) does not matter as this step is testing a qualitative not quantitative property. Therefore, one can set the value to – infinity (e.g., −1000) and + infinity (e.g., +1000). Since we are changing upper and lower bound the boundType is ‘b’.
-
77|Run analysis for blocked reactions. The function returns a list of blocked reactions (‘BlockedReactions’).
-
78|
Connect reaction to remaining network (optional). Depends on the function of the blocked reaction. Follow the diagram in Figure 14.
Compute single gene deletion phenotypes
-
79|Compute single gene deletion phenotypes. Use the following function in the COBRA Toolbox:
This function allows the use of different methods (‘method’) for optimization, e.g., FBA, minimization of metabolic adjustment (MOMA)6, or linear MOMA16. The list of genes that shall be deleted is given by ‘geneList’. If no gene list is given or the string is empty, all genes in the reconstruction will be deleted and tested for growth capabilities of the knock-out mutant. The function calculates the growth rate of the wild-type strain (‘grRateWT’) of each deletion strain (‘grRateKO’) as well as the relative growth rate ratios (‘grRatio’).
-
80|
Compare with experimental data. The evaluation of inconsistencies will lead to further reconstruction refinement (Figure 9). Repeat the gap analysis as necessary (Steps 45 to 49).
Test for known incapabilities of the organism
-
81|
Set simulation condition. Change objective function. Test for incapability by maximizing for objective function. If incapable, no solution or zero flux should be returned.
-
82|Use single reaction deletion to identify candidate reactions that enable the model’s capability despite known incapability:
This smaller subset of reactions needs to be manually evaluated. Note that the deletion of a single function may not be sufficient when alternate pathways exist in the network.
 Missing incapabilities may not only be caused by falsely added reactions in the metabolic network, but may be a consequence of missing regulatory information. Literature may provide the necessary data.
Missing incapabilities may not only be caused by falsely added reactions in the metabolic network, but may be a consequence of missing regulatory information. Literature may provide the necessary data.
Test if the model can predict the correct growth rate or other quantitative properties
-
83|
Compare predicted physiological properties with known properties. Use the suite of functions in the COBRA Toolbox along with experimental data (e.g., phenotypic, physiological, genetic data).
Test if the model can grow fast enough
-
84|Optimize for biomass reaction in different medium conditions and compare with experimental data. If the model does not grow at all, follow option A. If the model does not grow fast enough, follow option B.
- If the model does not grow at all.
- Check your boundary constraints. If these are correct, it is possible that the simulated condition does not support growth (compare with experimental data) or your network is incomplete. In the latter case, return to Steps 45 to 48 to identify missing links in the network.
- If the model does not grow fast enough.
- Check your boundary constraints. If these are correct, the possibilities of error modes are quite numerous. It is advised to verify the constraints applied to the model. Use the function which lists all constrained reactions that are greater than a minimal value (‘MinInf’) and smaller than a maximal value (‘MaxInf’):
-
85|Test if any of the medium components are growth limiting. Therefore, increase the uptake rate (‘value’) of one substrate (‘rxnNameList ‘) at a time by using:
 and setting the bound type to lower bound ‘l’ (‘boundType’).
and setting the bound type to lower bound ‘l’ (‘boundType’).
-
86|
Maximize for biomass. If the biomass reaction value increases, it means that this compound is limiting. This gives you a hint as to, where in the network something must be missing.
-
87|Determine reduced cost associated with network reactions when optimizing for objective function. Use
Set primalOnlyFlag to ‘false’ to get the reduced cost returned with the optimal solution. When maximizing the objective function ‘osenseStr’ will be ‘max’ while minimization is defined by ‘min’.
Find reactions with lowest reduced cost values. Increase flux through those reactions, if possible, by removing upper bounds. This will lead to increase flux through the objective reaction.
Test if the model grows too fast
-
88|
Optimize for biomass reaction in different medium conditions and compare with experimental data.
-
89|Verify that the model constraints are set as intended. Use the function which lists all constrained reactions that are greater than a minimal value (‘MinInf’) and smaller than a maximal value (‘MaxInf’):
Perform one or more of the following test, to identify possible errors in the network:
-
90|
Verify that all fractions and precursors in the biomass reaction are consistent with current knowledge. This may include that the GAM in the biomass reaction is not correct.
-
91|
Identify shuttling reactions, e.g., proton shuttling, by repeating Step 51–58. You are looking for reactions associated with loops.
-
92|
Re-investigate the thermodynamic information associated with the network reaction, i.e., reaction directionality, supporting evidence, and uncertainty associated with thermodynamic data.
-
93|Use single reaction deletion to identify single reactions that enables the model to grow too fast. Use the following function by setting the ‘method’ to ‘FBA’ and the ‘rxnList’ should contain one or more reactions to be deleted. If all network reactions shall be tested ‘rxnList’ does not need to be defined:
The function will return the wild-type growth rate (‘grRateW’), the growth rate of the reaction deleted network (‘grRateKO’), and the relative growth rate ratio (‘grRatio’). However, it is most likely that multiple reactions contribute to this observation and thus they are not identified by this method.
-
94|Reduced cost. The reduced cost analysis can be used to identify those reactions that can reduce the growth rate (positive cost value). Use:
Set primalOnlyFlag to ‘false’ to get the reduced cost returned with the optimal solution. When maximizing the objective function ‘osenseStr’ will be ‘max’ while minimization is defined by ‘min’.
 Changes to the model may be condition-specific and should be well documented.
Changes to the model may be condition-specific and should be well documented. An unconstrained ATPM reaction can change the model prediction in some cases. For example, if the computed growth rate of the model is too high, check the flux value through the ATPM in the optimal solution.
An unconstrained ATPM reaction can change the model prediction in some cases. For example, if the computed growth rate of the model is too high, check the flux value through the ATPM in the optimal solution.
Data assembly and Dissemination
-
95|Print Matlab model content. Make the final reconstruction available to the research community in at least 2 formats: 1. as a spreadsheet containing all information collected during the reconstruction process (as shown in supplemental methods 2); and 2. in SBML format which is a transportable format of the models and can be used with other modeling tools. To export the reconstruction from Matlab into Excel format, use:
To export a model in SBML format, use the same function but change the format to ‘sbml’. The output file name is defined by ‘FileName’.
 Note that the SBML format will not contain all identifiers, references and notes. It is therefore crucial to distribute the reconstruction in a different format. Ideally, the reconstruction content is made available through a web page, such as BiGG (See Table 1), which facilitates queries.
Note that the SBML format will not contain all identifiers, references and notes. It is therefore crucial to distribute the reconstruction in a different format. Ideally, the reconstruction content is made available through a web page, such as BiGG (See Table 1), which facilitates queries. -
96|
Add gap information to the reconstruction output. In Steps 45 to 48 information regarding the remaining and resolved network gaps were collected. These should be associated with the output of the final reconstruction (e.g., in Excel format).
TIMING
The timing of the entire reconstruction process depends on the properties of the target organism (prokaryote vs. eukaryote, genome size), the quality of the genome annotation, and the availability of experimental data. The timing listed below represents an average and can be used to plan the different stages. All COBRA Toolbox functions described in this protocol finish with a couple of seconds to some few hours on a newer personal computer (Intel Core 2 Duo 6600 2.4 GHz with 4Gb of memory running Windows Vista).
Step 1| through 4| (Stage 1, draft reconstruction): days to a week.
Step 5| (Stage 1, collection of experimental data): ongoing throughout the reconstruction process
Step 6| through 23| (Stage 2, reconstruction refinement): months to a year (if debugging and gap filling is done along the way)
Step 24| through 32| (Stage 2, biomass determination): days to weeks, depending on data availability
Step 34| through 36| (Stage 2, biomass determination): days to a week.
Step 37| (Stage 2, growth requirements): days to weeks, depending on data availability
Step 38| through 42| (Stage 3, conversion): days to a week.
Step 43| through 94| (Stage 4, network evaluation/debugging): week to months.
Step 95| and 96| (Data assembly): days to weeks, depending how much and in which format data was collected.
TROUBLESHOOTING
Step 38| See installation instructions of the COBRA Toolbox16 for details on how to install and setup Matlab, SBML and COBRA Toolbox.
- Step 39| The script may fail during the loading of the model from the xls files. Check:
-
-if headers are correct (supplemental methods 2)
-
-if all necessary information is available
-
-if metabolic reaction is written correctly → example; if multiple spaces in the reaction, the script does not work. Separator for left hand side and right hand side can be -->, ->, <==>, <=>
-
-Mixing number and string can cause problems as well. See Ecoli_core.xls as example on how the input file should look like.
-
-
Step 51| Make sure that you are working in the directory were the X3.exe script was copied to. The .expa file produced by the function must be in the same directory as X3.exe.
ANTICIPATED RESULTS
This protocol will result in a reconstruction that covers most of the known metabolic information of the target organism and represents a knowledge database. This reconstruction can be used as a resource for information (query tool), high-throughput data mapping (context for content), and a starting point for mathematical models. Table 6 lists a subset of published reconstructions which were constructed based on the presented protocol.
Table 6. Extract of reconstructions and their key properties that were constructed in accordance with this protocol.
For a complete list of reconstructions, constructed in part or in full in accordance with this protocol can be found at http://gcrg.ucsd.edu/In_Silico_Organisms/Other_Organisms. This website is continuously updated. GR – genes in reconstruction. Mets – metabolites, Rnxs – reactions, Comp – compartments, Ref – reference. Please refer to Table S1 for compartment abbreviations.
| Organism | Strain | Genes | Version | GR | Mets | Rxns | Comp | Ref |
|---|---|---|---|---|---|---|---|---|
| Bacillus subtilis | 4,225 | model_v3 | 844 | 988 | 1,020 | 2 (c,e) | 75 | |
| Escherichia coli | K12 MG1655 | 4,405 | iAF1260 | 1,260 | 1,039 | 2,077 | 3 (c,e,p) | 65 |
| Helicobacter pylori | 26695 | 1,632 | iIT341 | 341 | 485 | 476 | 2 (c,e) | 76 |
| Pseudomonas putida | KT2440 | 5,350 | iNJ746 | 746 | 911 | 950 | 3 (c,p,e) | 57 |
| Pseudomonas putida | KT2440 | 5,350 | iJP815 | 815 | 886 | 877 | 2 (c,e) | 96 |
| Pseudomonas aeruginosa | PA01 | 5,640 | iMO1056 | 1,056 | 760 | 883 | 2 (c,e) | 97 |
| Mycoplasma genitalium | G-37 | 521 | iPS189 | 189 | 274 | 262 | 2 (c,e) | 98 |
| Lactobacillus plantarum | WCFS1 | 3,009 | 721 | 531 | 643 | 2 (c,e) | 73 | |
| Streptomyces coelicolor | A3(2) | 8,042 | 700 | 500 | 700 | 2 (c,e) | 99 | |
| Leishmania major | Friedlin | 8,370 | iAC560 | 560 | 1,101 | 1,112 | 8 (a,f,y,c,e,m,r,n) | 100 |
| Saccharomyces cerevisiae | Sc288 | 6,183 | iMM904 | 904 | 713 | 1,412 | 8(c,e,m,x,n,r,v,g) | 101 |
| Homo sapiens | 28,783 | Recon 1 | 1,496 | 2,766 | 3,311 | 8 (c,e,m,x,n,r,v,g) | 15 |
To facilitate the use of the presented COBRA Toolbox commands (Steps 43 to 94), we listed examples of their use in the Supplementary Method 1.
Box 1: Glossary.
Bibliome – A bibliome is a collection of primary and review literature as well as textbooks.
Biochemical, Genetic and Genomic (BiGG) knowledge base – A BiGG knowledge base is a genome-scale reconstruction, which incorporates in a structured manner genomic, proteomic, biochemical and physiological information of a particular organism or cell.
Biomass reaction – The biomass reaction lumps all known biomass precursors and their fractional distribution to a cell into one network reaction.
Blocked reactions – Network reactions that cannot carry any flux in any simulation condition are called blocked reactions. Generally, these blocked reactions are caused by missing links in the network.
Constraint-based reconstruction and analysis (COBRA) – COBRA is a modeling approach in which manually curated, stoichiometric network reconstructions are constructed. Subsequently, models can be obtained and analyzed by applying equality and inequality constraints and by computing functional states. Constraints include mass conservation and thermodynamics (for directionality) as well as constraints reflecting experimental conditions and regulatory constraints
Dead-end metabolite A dead-end metabolite that is only produced or consumed in the network.
Demand reaction – When the consumption reaction(s) of a metabolite is not known or outside the scope of the reconstruction it can be represented by this unbalanced, intracellular reaction (e.g., 1 A -->).
Exchange reactions These reactions are unbalanced, extra-organism reactions that represent the supply to or removal of metabolites from the extra-organism “space”. (See Box 3).
Extreme pathways (ExPa’s) – ExPa’s are a unique and minimal set of flux vectors which lie at the edges of the bounded null space. Biochemically meaningful steady-state solutions can be obtained by nonnegative linear combination of ExPa’s.
Flux-balance analysis (FBA) – FBA is a formalism that defined the metabolic network as a linear programming optimization problem. The main constraints in FBA are imposed by the steady state mass conservation of metabolites.
Futile cycles – Stoichiometrically unbalanced cycles, which are associated with energy consumption.
Gene-protein-reaction (GPR) association – GPR association connect genes, proteins and reactions in a logical relationship (AND, OR).
Genome-scale model (GEM) – A GEM is derived from a GENRE, by converting it into a mathematical form (i.e., an in silico model) and by assessing computationally its phenotypic properties.
Genome-scale network reconstruction (GENRE) – A GENRE formed based on an organism-specific BiGG knowledge base. A GENRE is a collection biochemical transformation derived from the genome annotation and the bibliome of the target organism. A network GENRE is unique to an organism, as its genome is.
Flux variability analysis (FVA) – FVA is a frequently used computational tool for investigating more global capabilities under a given simulation condition (e.g., network redundancy). Therefore, every network reaction will be chosen as an objective function and the minimal and maximal possible flux value through the reaction is determined by minimizing and maximizing the objective function.
Linear programming (LP) – LP is an optimization technique, in which a linear objective function is optimized (i.e., minimized or maximized) subject to linear equality and inequality constraints.
Network gap – A network gap is a missing reaction or function in the network, which can connect one or more dead-end metabolites with the remainder of the network.
Objective function – An objective function is a network reaction, or a linear combination of network reactions, for which is optimized in the linear programming problem.
Sink reaction – When the synthesis reaction(s) of a metabolite is not known or outside the scope of the reconstruction its discharge can be represented by this unbalanced, intracellular reaction (e.g., 1 A <-->)
P/O ratio – This ratio represents the number of ATP molecules (P) which are formed per oxygen atom (O) consumed during respiration.
Reduced cost A parameter associated with linear programming. It can be used to investigate properties associated with the calculated optimal solution. Each network reaction has a reduced cost values associated, which represents the amount the objective value would increase if the flux through the reaction would be increased by one unit. Note that by definition reduced costs values that can increase the objective value are negative numbers.
Type III extreme pathway – These stoichiometric balanced cycles (SBC) are a subset of ExPa’s that are only composed of intracellular reactions, i.e., that all exchange reactions (i.e., systems boundaries) have zero flux.
Supplementary Material
ACKNOWLEDGEMENT
We would like to acknowledge R.M.T. Fleming, A. Feist, and N. Jamshidi, for valuable discussions. We are thankful to M. Abrahams, S.A. Becker, and F.-C. Cheng for reading the manuscript. We would like to thank S. Burning for preparing the biomass reaction manual as well as A. Bordbar and R.M.T.Fleming for providing Matlab code. I.T. was supported by National Institutes of Health (NIH) grant R01 GM057089.
Footnotes
Competing interest statement: The authors declare that they have no competing financial interests.
Author contribution: IT and BOP designed concept and wrote manuscript. IT developed protocol.
REFERENCES
- 1.Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabasi AL. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- 2.Thiele I, Price ND, Vo TD, Palsson BO. Candidate metabolic network states in human mitochondria: Impact of diabetes, ischemia, and diet. J Biol Chem. 2005;280:11683–11695. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
- 3.Pal C, et al. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 4.Barrett CL, Herring CD, Reed JL, Palsson BO. The global transcriptional regulatory network for metabolism in Escherichia coli attains few dominant functional states. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:19103–19108. doi: 10.1073/pnas.0505231102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 6.Segre D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotech. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nature reviews. 2009;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nature reviews. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 10.Notebaart RA, van Enckevort FH, Francke C, Siezen RJ, Teusink B. Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics. 2006;7:296. doi: 10.1186/1471-2105-7-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durot M, Bourguignon PY, Schachter V. Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS microbiology reviews. 2009;33:164–190. doi: 10.1111/j.1574-6976.2008.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price ND, Papin JA, Schilling CH, Palsson B. Genome-scale microbial in silico models: the constraints-based approach. Trends in biotechnology. 2003;21:162–169. doi: 10.1016/S0167-7799(03)00030-1. [DOI] [PubMed] [Google Scholar]
- 13.Schilling CH, Edwards JS, Letscher D, Palsson BO. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnology and Bioengineering. 2000;71:286–306. [PubMed] [Google Scholar]
- 14.Varma A, Palsson BO. Metabolic Flux Balancing: Basic concepts, Scientific and Practical Use. Nat Biotechnol. 1994;12:994–998. [Google Scholar]
- 15.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker SA, et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox. Nat. Protocols. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 17.Savinell JM, Palsson BO. Network analysis of intermediary metabolism using linear optimization. I. Development of mathematical formalism. Journal of theoretical biology. 1992;154:421–454. doi: 10.1016/s0022-5193(05)80161-4. [DOI] [PubMed] [Google Scholar]
- 18.Burgard AP, Maranas CD. Optimization-based framework for inferring and testing hypothesized metabolic objective functions. Biotechnology and bioengineering. 2003;82:670–677. doi: 10.1002/bit.10617. [DOI] [PubMed] [Google Scholar]
- 19.Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Molecular systems biology. 2007;3:1–15. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gianchandani EP, Oberhardt MA, Burgard AP, Maranas CD, Papin JA. Predicting biological system objectives de novo from internal state measurements. BMC Bioinformatics. 2008;9:43. doi: 10.1186/1471-2105-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papin JA, Palsson BO. The JAK-STAT Signaling Network in the Human B-Cell: An Extreme Signaling Pathway Analysis. Biophysical journal. 2004;87:37–46. doi: 10.1529/biophysj.103.029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Thiele I, Jamshidi N, Palsson BO. Identification of potential pathway mediation targets in Toll-like receptor signaling. PLoS Comput Biol. 2009;5:e1000292. doi: 10.1371/journal.pcbi.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiele I, Jamshidi N, Fleming RM, Palsson BO. Genome-scale reconstruction of Escherichia coli's transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput Biol. 2009;5:e1000312. doi: 10.1371/journal.pcbi.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gianchandani EP, Papin JA, Price ND, Joyce AR, Palsson BO. Matrix Formalism to Describe Functional States of Transcriptional Regulatory Systems. PLoS Comput Biol. 2006;2:e101. doi: 10.1371/journal.pcbi.0020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gianchandani EP, Joyce AR, Palsson BO, Papin JA. Functional States of the genome-scale Escherichia coli transcriptional regulatory system. PLoS Comput Biol. 2009;5:e1000403. doi: 10.1371/journal.pcbi.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mobley HLT, Mendz GL, Hazell SL. Helicobacter Pylori. Washington, D.C.: ASM Press; 2001. [PubMed] [Google Scholar]
- 27.Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. Edn. 2nd. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 28.Dickinson JR, Schweizer M. The metabolism and molecular physiology of Saccharomyces cerevisiae. Edn. 2nd. London ; Philadelphia: Taylor & Francis Ltd; 2004. [Google Scholar]
- 29.Ramos JL. Pseudomonas. New York Kluwer: Academic/Plenum Publishers; 2004. [Google Scholar]
- 30.Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics (Oxford, England) 2002;18 Suppl 1:S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 31.Pinney JW, Shirley MW, McConkey GA, Westhead DR. metaSHARK: software for automated metabolic network prediction from DNA sequence and its application to the genomes of Plasmodium falciparum and Eimeria tenella. Nucleic Acids Res. 2005;33:1399–1409. doi: 10.1093/nar/gki285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein L. Genome annotation: from sequence to biology. Nature reviews. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- 34.Aziz RK, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overbeek R, Bartels D, Vonstein V, Meyer F. Annotation of bacterial and archaeal genomes: improving accuracy and consistency. Chemical reviews. 2007;107:3431–3447. doi: 10.1021/cr068308h. [DOI] [PubMed] [Google Scholar]
- 36.Manichaikul A, et al. Metabolic network analysis integrated with transcript verification for sequenced genomes. Nature methods. 2009;6:589–592. doi: 10.1038/nmeth.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boneca IG, et al. A revised annotation and comparative analysis of Helicobacter pylori genomes. Nucleic Acids Res. 2003;31:1704–1714. doi: 10.1093/nar/gkg250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karp PD, et al. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 2007;35:7577–7590. doi: 10.1093/nar/gkm740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.(NC-IUBMB), N.C.o.t.I.U.o.B.a.M.B. Enzyme Nomenclature. Edn. 6th. San Diego, California: Academic Press; 1992. [Google Scholar]
- 41.Kanehisa M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barthelmes J, Ebeling C, Chang A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA: the enzyme information system in 2007. Nucleic Acids Res. 2007;35:D511–D514. doi: 10.1093/nar/gkl972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karp PD, et al. The EcoCyc Database. Nucleic Acids Res. 2002;30:56–58. doi: 10.1093/nar/30.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophysical journal. 2008;95:1487–1499. doi: 10.1529/biophysj.107.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleming RMT, Thiele I, Nasheuer HP. Quantitative assignment of reaction directionality in constraint-based models of metabolism: Application to Escherichia coli. Biophys Chem. 2009 doi: 10.1016/j.bpc.2009.08.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kümmel A, Panke S, Heinemann M. Systematic assignment of thermodynamic constraints in metabolic network models. BMC Bioinformatics. 2006;7:1–12. doi: 10.1186/1471-2105-7-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardy JL, et al. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics (Oxford, England) 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 48.Lu Z, et al. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics (Oxford, England) 2004;20:547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- 49.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 50.Ross-Macdonald P, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 51.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 52.Brooksbank C, Cameron G, Thornton J. The European Bioinformatics Institute's data resources: towards systems biology. Nucleic Acids Res. 2005;33:D46–D53. doi: 10.1093/nar/gki026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. Journal of Chemical Information and Computer Sciences. 1988;28:31–36. [Google Scholar]
- 55.Coles SJ, Day NE, Murray-Rust P, Rzepa HS, Zhang Y. Enhancement of the chemical semantic web through the use of InChI identifiers. Organic & biomolecular chemistry. 2005;3:1832–1834. doi: 10.1039/b502828k. [DOI] [PubMed] [Google Scholar]
- 56.Williams AJ. Internet-based tools for communication and collaboration in chemistry. Drug discovery today. 2008;13:502–506. doi: 10.1016/j.drudis.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 57.Nogales J, Palsson BO, Thiele I. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC systems biology. 2008;2:79. doi: 10.1186/1752-0509-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Izard J, Limberger RJ. Rapid screening method for quantitation of bacterial cell lipids from whole cells. Journal of Microbiological Methods. 2003;55:411–418. doi: 10.1016/s0167-7012(03)00193-3. [DOI] [PubMed] [Google Scholar]
- 59.Benthin S, Nielsen J, Villadsen J. A simple and reliable method for the determination of cellular RNA content. Biotechnology Techniques. 1991;5:39–42. [Google Scholar]
- 60.Herbert D, Phipps PJ, Strange RE. Chemical analysis of microbial cells. Methods in Microbiology. 1971;5:209–344. [Google Scholar]
- 61.Lindahl L, Zengel JM. Ribosomal genes in Escherichia coli. Annu Rev Genet. 1986;20:297–326. doi: 10.1146/annurev.ge.20.120186.001501. [DOI] [PubMed] [Google Scholar]
- 62.Sawada M, Osawa S, Kobayashi H, Hori H, Muto A. The number of ribosomal RNA genes in Mycoplasma capricolum. Mol Gen Genet. 1981;182:502–504. doi: 10.1007/BF00293942. [DOI] [PubMed] [Google Scholar]
- 63.Hui I, Dennis PP. Characterization of the ribosomal RNA gene clusters in Halobacterium cutirubrum. J Biol Chem. 1985;260:899–906. [PubMed] [Google Scholar]
- 64.Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the bacterial cell: a molecular approach. Sunderland, Mass: Sinauer Associates; 1990. [Google Scholar]
- 65.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Molecular systems biology. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schilling CH, Letscher D, Palsson BO. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. Journal of theoretical biology. 2000;203:229–248. doi: 10.1006/jtbi.2000.1073. [DOI] [PubMed] [Google Scholar]
- 67.Price ND, Thiele I, Palsson BO. Candidate states of Helicobacter pylori's genome-scale metabolic network upon application of loop law thermodynamic constraints. Biophysical journal. 2006;90:3919–3928. doi: 10.1529/biophysj.105.072645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palsson BO. Systems biology: properties of reconstructed networks. New York: Cambridge University Press; 2006. [Google Scholar]
- 69.Gutnick D, Calvo JM, Klopotowski T, Ames BN. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. Journal of bacteriology. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schroeder C, Selig M, Schoenheit P. Glucose fermentation to acetate, CO 2 and H 2 in the anaerobic hyperthermophilic eubacterium Thermotoga maritima: involvement of the Embden-Meyerhof pathway. Archives of Microbiology. 1994;161:460–470. [Google Scholar]
- 71.Satish Kumar V, Dasika MS, Maranas CD. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reed JL, Palsson BO. Genome-Scale In Silico Models of E. coli Have Multiple Equivalent Phenotypic States: Assessment of Correlated Reaction Subsets That Comprise Network States. Genome Res. 2004;14:1797–1805. doi: 10.1101/gr.2546004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teusink B, et al. Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J Biol Chem. 2006;281:40041–40048. doi: 10.1074/jbc.M606263200. [DOI] [PubMed] [Google Scholar]
- 74.Reed JL, et al. Systems approach to refining genome annotation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem. 2007;282:28791–28799. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- 76.Thiele I, Vo TD, Price ND, Palsson B. An Expanded Metabolic Reconstruction of Helicobacter pylori (iIT341 GSM/GPR): An in silico genome-scale characterization of single and double deletion mutants. J Bacteriol. 2005;187:5818–5830. doi: 10.1128/JB.187.16.5818-5830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feist AM, Scholten JCM, Palsson BO, Brockman FJ, Ideker T. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Molecular systems biology. 2006;2:1–14. doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Famili I, Forster J, Nielsen J, Palsson BO. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13134–13139. doi: 10.1073/pnas.2235812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knorr AL, Jain R, Srivastava R. Bayesian-based selection of metabolic objective functions. Bioinformatics (Oxford, England) 2007;23:351–357. doi: 10.1093/bioinformatics/btl619. [DOI] [PubMed] [Google Scholar]
- 80.Holzhutter HG. The principle of flux minimization and its application to estimate stationary fluxes in metabolic networks. Eur J Biochem. 2004;271:2905–2922. doi: 10.1111/j.1432-1033.2004.04213.x. [DOI] [PubMed] [Google Scholar]
- 81.Shlomi T, Berkman O, Ruppin E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7695–7700. doi: 10.1073/pnas.0406346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schuster S, Pfeiffer T, Fell DA. Is maximization of molar yield in metabolic networks favoured by evolution? Journal of theoretical biology. 2008;252:497–504. doi: 10.1016/j.jtbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 83.Ott MA, Vriend G. Correcting ligands, metabolites, and pathways. BMC Bioinformatics. 2006;7:517. doi: 10.1186/1471-2105-7-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jarlier V, Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS microbiology letters. 1994;123:11–18. doi: 10.1111/j.1574-6968.1994.tb07194.x. [DOI] [PubMed] [Google Scholar]
- 88.Sundararaj S, et al. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res. 2004;32:D293–D295. doi: 10.1093/nar/gkh108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klamt S, Saez-Rodriguez J, Gilles ED. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC systems biology. 2007;1:2. doi: 10.1186/1752-0509-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klamt S, Stelling J, Ginkel M, Gilles ED. FluxAnalyzer: exploring structure, pathways, and flux distributions in metabolic networks on interactive flux maps. Bioinformatics (Oxford, England) 2003;19:261–269. doi: 10.1093/bioinformatics/19.2.261. [DOI] [PubMed] [Google Scholar]
- 92.Luo RY, Liao S, Zeng SQ, Li YX, Luo QM. FluxExplorer: A general platform for modeling and analyses of metabolic networks based on stoichiometry. Chinese Science Bulletin. 2006;51:689–696. [Google Scholar]
- 93.Lee DY, Yun H, Park S, Lee SY. MetaFluxNet: the management of metabolic reaction information and quantitative metabolic flux analysis. Bioinformatics (Oxford, England) 2003;19:2144–2146. doi: 10.1093/bioinformatics/btg271. [DOI] [PubMed] [Google Scholar]
- 94.Lee SY, et al. Systems-level analysis of genome-scale in silico metabolic models using MetaFluxNet. Biotechnol. Bioproc. Eng. 2005;10:425–431. [Google Scholar]
- 95.Chhabra SR, et al. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J Biol Chem. 2003;278:7540–7552. doi: 10.1074/jbc.M211748200. [DOI] [PubMed] [Google Scholar]
- 96.Puchalka J, et al. Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput Biol. 2008;4:e1000210. doi: 10.1371/journal.pcbi.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oberhardt MA, Puchalka J, Fryer KE, Martins dos Santos VA, Papin JA. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. Journal of bacteriology. 2008;190:2790–2803. doi: 10.1128/JB.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suthers PF, et al. A genome-scale metabolic reconstruction of Mycoplasma genitalium, iPS189. PLoS Comput Biol. 2009;5:e1000285. doi: 10.1371/journal.pcbi.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Borodina I, Krabben P, Nielsen J. Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res. 2005;15:820–829. doi: 10.1101/gr.3364705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chavali AK, Whittemore JD, Eddy JA, Williams KT, Papin JA. Systems analysis of metabolism in the pathogenic trypanosomatid Leishmania major. Molecular systems biology. 2008;4:177. doi: 10.1038/msb.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mo ML, Palsson BO, Herrgard MJ. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC systems biology. 2009;3:37. doi: 10.1186/1752-0509-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.