Abstract
Most species within the genus Conus are considered to be specialists in their consumption of prey, typically feeding on molluscs, vermiform invertebrates or fish, and employ peptide toxins to immobilize prey. Conus californicus Hinds 1844 atypically utilizes a wide range of food sources from all three groups. Using DNA- and protein-based methods, we analyzed the molecular diversity of C. californicus toxins and detected a correspondingly large number of conotoxin types. We identified cDNAs corresponding to seven known cysteine-frameworks containing over 40 individual inferred peptides. Additionally, we found a new framework (22) with six predicted peptide examples, along with two forms of a new peptide type of unusual length. Analysis of leader sequences allowed assignment to known superfamilies in only half of the cases, and several of these showed a framework that was not in congruence with the identified superfamily. Mass spectrometric examination of chromatographic fractions from whole venom served to identify peptides corresponding to a number of cDNAs, in several cases differing in their degree of posttranslational modification. This suggests differential or incomplete biochemical processing of these peptides. In general, it is difficult to fit conotoxins from C. californicus into established toxin classification schemes. We hypothesize that the novel structural modifications of individual peptides and their encoding genes reflect evolutionary adaptation to prey species of an unusually wide range as well as the large phylogenetic distance between C. californicus and Indo-Pacific species.
Keywords: Conotoxins, Conus californicus, feeding diversity, cDNA library, RT-PCR, signal peptides
1. Introduction
Cone snails are carnivorous molluscs that are justly acclaimed for their potent venoms used in prey capture. The genus Conus contains more than 700 species (Röckel et al., 1995; Daly and Craik, 2009), each of which may elaborate over 100 peptides within their venom. Over 3,000 entries for these substances have been listed in the useful web-based database, ConoServer (http://research1t.imb.uq.edu.au/conoserver/), with additional accessions added on a virtually daily basis (Kaas et al., 2008). The term conopeptide refers to venom peptides and includes cysteine (Cys)-rich peptides as well as those that possess two Cys residues or none (Halai and Craik, 2009). The Cys-rich peptides, known as conotoxins, are typically 10 to 40 amino acids in length and contain up to 10 cysteines (Terlau and Olivera, 2004; Daly and Craik, 2009).
Conotoxins are categorized into frameworks according to the number and respective positions of their Cys residues, and they are further classified by the signal sequence of the prepro-peptide (leader). Because this leader is cleaved during biosynthesis, its amino acid sequence must be determined from the corresponding cDNA. The signal sequences comprise the initial 18–22 amino acids of the leader and tend to be highly conserved among toxins having identical Cys-frameworks. These features has been used to define at least 15 superfamilies of conotoxins (Terlau and Olivera, 2004; Loughnan et al., 2009). Conopeptides also typically contain multiple posttranslational modifications (PTMs) that may be modulated by sequences within the propeptide region (Buczek et al., 2005).
At present, 22 Cys-frameworks have been identified, with most conotoxins falling into eight categories (Norton and Olivera, 2006). Within a given framework, the specificity of action may vary, depending on amino acid composition and intercysteine spacing (Halai and Craik, 2009). For example, certain μ-type toxins (M superfamily, Cys-framework 3) block different isoforms of voltage-gated Na channels (McIntosh and Jones, 2001; Norton and Olivera, 2006).
Most Conus species are restricted in their hunting to one prey type: worm-like invertebrates (annelids and enteropneusts), molluscs or fish (Röckel et al., 1995), and interspecific competition within this genus is thought to have led to feeding-niche specialization and toxin diversification (Duda et al., 2001; Duda and Palumbi, 2004; Remigio and Duda, 2008). Most studies supporting this conclusion have focused on Conus species from the tropical Indo-Pacific region where species diversity is by far the greatest. Relatively few conotoxins have been identified from Eastern Pacific (Hopkins et al., 1995) or Atlantic (Aguilar et al., 2009; Zamora-Bustillos et al., 2009) species, but these generally fit within the conceptual framework described above.
Conus californicus Hinds 1844 is an atypical member of the genus. Although probably descended from vermivorous ancestors (Duda et al., 2001), it has evolved into a generalist feeder that consumes at least 56 different species of all three prey types, in addition to scavenging (Kohn, 1966; Saunders and Wolfson, 1961; Stewart and Gilly, 2005). C. californicus is endemic to the temperate northeast Pacific Ocean, and over most of its range from the Farallon Islands of central California to Bahia Magdalena in Baja California Sur, Mexico, it is the only representative of the genus (Morris et al., 1980), a situation that may have existed since the late Miocene (Stanton, 1966). Sympatric congeneric species occur only in the southernmost portion of its range (Magdalena Bay to Cabo San Lucas).
Phylogenetic studies indicate a distant relationship between C. californicus and the rest of the genus, including other eastern Pacific members (Espiritu et al., 2001; Duda and Kohn, 2005). This outlying position and the potentially relaxed interspecies competition in C. californicus presumably underlie the extreme dietary diversity of this species. These features suggest that C. californicus may express novel peptide toxins, and yet this species has been largely overlooked, with initial studies involving semipurified venom components (Cottrell and Twarog, 1972; Elliott and Raftery, 1979). The present study describes the identification of a large assortment of conotoxin types expressed by C. californicus, many of which are difficult to accommodate within the conventional conopeptide classification scheme.
2. Materials and methods
2.1. General
Specimens of Conus californicus (shell length 1.5–3.5 cm) were collected from shallow subtidal areas in southern Monterey Bay, CA and La Jolla, CA. Intertidal specimens were collected in Bahia Asuncion, BCS, Mexico. Snails were dissected, and venom ducts weighing 2.5–5 mg were separated from the attached muscular bulb and radular sac. In some cases venom was manually extruded before proceeding with RNA isolation.
Total RNA was isolated from venom ducts using either RNaqueous (Applied Biosystems/Ambion, Austin, TX, USA) or Perfect RNA, Eukaryotic (Eppendorf, Westbury, NY, USA) kits and quantified by UV absorbance at 260 nm. For construction of a cDNA library, mRNA was prepared and fractionated using the SuperScript Plasmid System with Gateway Technology for cDNA Synthesis and Cloning (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. PCR was carried out using Pfu Turbo or Pfu Ultra DNA polymerases (Stratagene, La Jolla, CA, USA) on reverse-transcribed templates or with Taq DNA polymerase on bacterial clones according to the manufacturers’ instructions. Amplifications were performed using a PTC Thermal Cycler (MJ Research, Waltham, MA, USA) with 1.5–2 mM MgCl2, 200 μM dNTP and 250 nM primers. After an initial denaturation of 7 min at 94°C, reaction mixtures were subjected to 40 cycles of 94°C (10 s) followed by 46°C (30 s), to 72°C at a rate of 0.6°C/min and 1 min at 72°C. After cycling was complete, an additional extension time of 7 min at 72°C was allowed. Purification of PCR products utilized Wizard Prep (Promega, Madison, WI, USA) or Qiaquick (Qiagen, Valencia, CA, USA) kits. Sequencing was accomplished with Big Dye terminator chemistry (Applied Biosystems, Foster City, CA, USA) utilizing either ABI Model 377 or 3100 automatic sequencers to give electropherograms that were examined visually and edited as required. Sequence data were imported into either Gene Works 2.4 (Intelligenetics, Inc., Mountain View, CA, USA) or Sequencher 4.1 (Gene Codes Corporation, Ann Arbor, MI, USA) and examined manually for peptide sequences typical of conopeptides.
2.2. Identification of conotoxin sequences using a cDNA library
RNA (12 μg) was extracted from pooled venom duct tissue (less venom) from 12 Monterey snails, size fractionated, and applied to an oligo-dT column to give 0.12 μg of mRNA. This was then used for cDNA synthesis with the SuperScript II kit and primers containing Sal I and Not I restriction sites. Products were ligated into the polylinker of the pSport1 vector between Sal I and Not I sites and introduced into Top10 Electrocompetent cells (Invitrogen) by electroporation. After growth on ampicillin medium (100 μg/ml), random colonies were inoculated into ampicillin L-Broth for overnight growth. Plasmids were purified with a conventional mini-prep procedure and assayed for an insert length of ca. 300–600 bp by digestion with Mlu I (one site of which is contained within the Sal I adapter of the cDNA insert) followed by gel electrophoresis. Alternatively, direct PCR on unpurified plasmids using the flanking M-13 recognition sites was used to estimate insert size.
Sequencing was carried out on purified plasmids or PCR products using the T7 priming region of the vector that is located upstream of the 5-prime end of the cDNA insert. This approach yielded the full-length sequence (signal, propeptide and toxin-coding region) encoding a number of putative peptide toxins.
2.3. Identification of additional peptide toxins by RT-PCR
Pooled total RNA from 12 snails (from ducts after removal of venom) was used in first-strand reverse transcription with either SuperScript II or AccuScript High Fidelity (Stratagene) enzyme employing an oligo (dT)12–18 primer. Alternatively, venom ducts from individual snails were processed for total RNA without removal of venom because this substantially increased RNA yield. PCR with either Pfu or Pfu Ultra DNA polymerase was then carried out on this first strand material using primers designed from full-length sequences of the cDNA library (see Supplementary Material, Table S1). Forward primers were located at the putative start site of the signal sequence (or slightly downstream) for the four-Cys conotoxins and near the end of the propeptide region for the remainder. Reverse primers were within the 3′-UTR of all sequences.
PCR products were cloned into the plasmid pCR4 Blunt-TOPO (Invitrogen) and transformed into competent DH5αT1 cells for subsequent plating on ampicillin medium. Individual colonies were picked, suspended in 15μl dH2O and directly subjected to PCR using the M13F/R primer pair flanking the insert region of the vector. PCR products were purified and sequenced using nested T3/T7 primers. For clones showing single nucleotide polymorphic substitutions, repeated PCR analyses or PCR on different isolates were carried out to confirm the sequences.
2.4. Sequence data analysis
The cleavage points of the hydrophobic signal sequences were determined with the SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/) (Bendtsen et al., 2004). The sequence analysis program of the ConoServer database (Kaas et al., 2008) provided detailed information on proteolysis and other PTMs of predicted peptide sequences. Comparisons with the deposited sequences in GenBank for homology were carried out using BLASTX and BLASTN queries. Details of conotoxins in the O1-superfamily with a six-cysteine framework were determined from the Knottin Database (http://knottin.cbs.cnrs.fr/) (Gracy et al., 2008).
2.5. Venom components and mass spectrometric analysis
Total venom was expelled from single venom ducts (~5 mg per duct, with 10 individual ducts examined via MS) and suspended in 150 μL H2O by sonication and vortexing followed by centrifugation at 10K rcf for 5 min. Capillary high performance liquid chromatography (HPLC) (0.3mm ×15 cm) was carried out on 2.5 μL volumes of these venom extracts with analysis by matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS). Disulfide linkages were reduced by treating part of the venom sample with dithiothreitol, and the resulting free sulfhydryl groups were alkylated with iodoacetamide. After purification, this material was subjected to capillary HPLC with analysis in parallel by electrospray ionization and MALDI MS, in general as described previously (Jakubowski et al., 2006; Kelley et al., 2006). For full details see Mass Spectral Analysis, Supplementary Material.
3. Results and discussion
Analysis of the conotoxins found in C. californicus will be presented in a way that primarily considers the number and disposition of Cys residues in the mature peptide (Cys-framework). In some cases affinity with an established superfamily through signal sequence homology is evident, but in many cases the assignment of a superfamily is ambiguous at best. We follow the nomenclature (Walker et al., 1999) in which cDNAs have the initial species letter capitalized, whereas isolated peptides are designated by lower case letters. Arabic numerals correspond to the disulfide framework. While much of the information on sequence and Cys-framework is based on sequence data, PTMs for many peptides have been confirmed via MS (Jakubowski et al., 2006).
3.1. Four-Cys peptide types
3.1.1. Framework 1
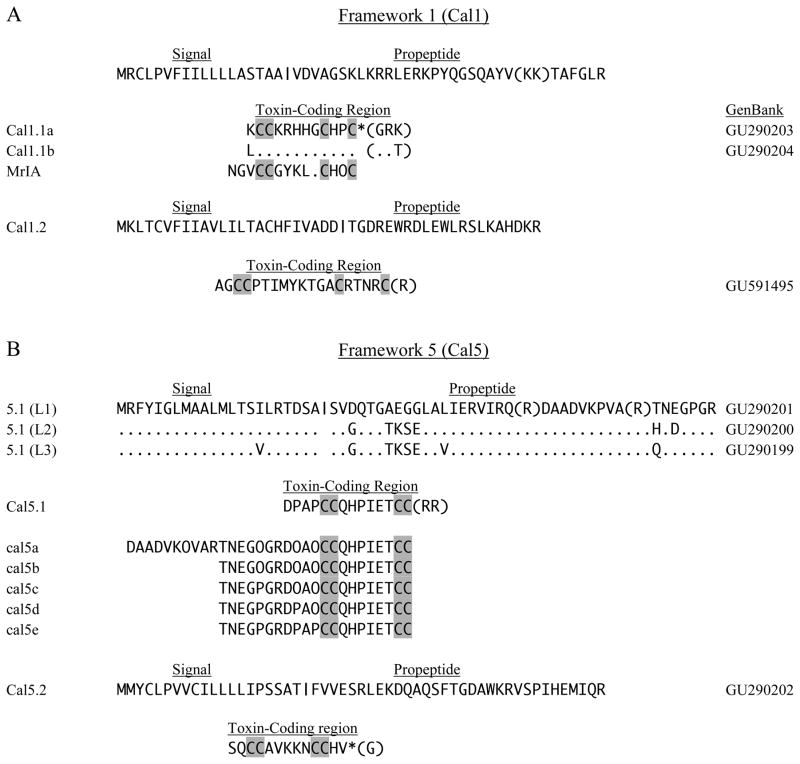
Mature peptide-toxins corresponding to Cal1.1a and Cal1.1b (Fig. 1A) have a Cys-framework (CC-CHPC) that is characteristic of the type 1 framework that resembles that of χ-conotoxins (T-superfamily). Their common signal sequence has homology with those of T-superfamily members (McIntosh et al., 2000; Nilsson et al., 2005; Peng et al., 2007). Two predicted sites of cleavage (KK, as shown in parentheses, or R) are present within the propeptide region of the Cal1.1 cDNAs (Fig. 1A), and the mature toxins may thus have an N-terminal extension of six amino acids. C-terminal amidation is predicted for the mature peptides corresponding to Cal1.1 (Buczek et al 2005; Fricker 2005).
Fig. 1.
Four-Cys sequences of frameworks 1 and 5. In this (and subsequent figures), cDNA clones have the initial species letter capitalized and isolated peptides are designated by all lower case letters (Walker et al., 1999). Vertical bars indicate potential signal peptide cleavage regions, and parentheses show predicted sites of proteolysis in propeptide and mature toxin regions. Predicted C-terminal amidation is indicated by an asterisk (*). A. The framework 1 toxins, Cal1.1a and 1.1b, share a common precursor region (signal and propeptide). Conotoxin Mr1A is shown for comparison. B. For the framework 5 toxin Cal5.1, three leader sequences (L1–L3) were observed. Peptides cal5a–cal5e were identified in venom and correspond to leader sequence L1. O designates hydroxyproline.
These two Cal1.1 toxins share the C-terminal -CHOC motif (O indicates hydroxyproline) with the χ-conotoxins MrIA and MrIB from a snail-eating species, C. marmoreus (Sharpe et al., 2001), and we assign framework 1 by analogy. The intercysteine connectivity in this case has been shown to be (I–IV, II–III). This arrangement has also been termed framework 10, but this assignment has been withdrawn, and conopeptides having the pattern –CC-C-C- are now considered to be members of framework 1 (Jiminez et al., 2007).
Alpha conotoxins (A-superfamily) also possess the same CC-C-C disposition of cysteines, but in this case, the Cys-Cys connectivity is different (I–II, III–IV) compared to (I–IV, II–III) for the χ-conotoxins, and the signal sequences are also distinct from those of the T-superfamily. Alpha conotoxins are classified into subtypes based on inter-cysteine spacing, CC-m-C-n-C, where m and n are the number of amino acids. Although numerous αm/n subtypes with different affinities for nicotinic acetylcholine receptors in a variety of taxa have been described (α3/5-fish; α4/3-polychaete worms; α4/7-relatively nonselective) (Terlau and Olivera, 2004), there have been no reports of a 〈5/2 subtype that would fit the primary structures of Cal1.1a and Cal1.1b.
Lack of similarity to A-superfamily members, and the excellent homology of the signal sequence with that of the T-superfamily, leads us to conclude that Cal1.1 toxins are members of the T-superfamily. As such, they may be χ-conotoxins, which inhibit norepinephrine transporters (Sharpe et al., 2001; Han et al, 2008), but we have no data on the function or PTMs of the Cal1.1 peptides as these were not isolated in our study. An additional conotoxin cDNA, Cal1.2 (Fig. 1A) may also be formally included within framework 1 on the basis of Cys spacing. There is also no correlation in Cys (II–III, III–IV) spacing (9/4) with known α- or χ-conotoxins. This seemingly incongruous combination of framework and leader sequence not previously associated with defined conotoxin superfamilies is common in C. californicus and makes classification of these toxins into the existing nomenclature schema difficult. We have tried to be conservative with our nomenclature and recognize that future work may require a major evaluation of C. californicus toxin nomenclature.
3.1.2. Framework 5
Three distinct leader sequences (L1–L3 in Fig. 1B) were identified as precursors of a single predicted Cal5.1 toxin. Framework 5 peptides have previously been found exclusively within the T1-superfamily (Peng et al., 2007; Zamora-Bustillos et al., 2009), but the signal sequences of Cal5.1 show less than 25% identity with this superfamily. Assignment to another superfamily based on leader sequence identity was not possible. Although a new superfamily might be warranted for these toxins, the singular position of C. californicus might make such a superfamily relevant only within this species. We therefore chose to designate only the framework.
Examination of total venom by HPLC-MS revealed five peptides, cal5a–5e, that exhibited various PTMs. These peptides were longer than the sequence predicted by the Cal5.1 cDNA, presumably due to proteolytic cleavage at alternative sites. The peptide cal5a appears to be cleaved at residue R43 of the Cal5.1 (L1) sequence, and the remaining four cal5 peptides are cleaved at R53. Peptides derived from the L2 and L3 leader sequences were not observed. These peptides show the predicted C-terminal truncation predicted by the final arginine residues in the cDNA sequence.
Interestingly, the degree of conversion of proline into hydroxyproline differs among these peptides, with the most complete hydroxylation only observed for cal5a and cal5b. In no case was the proline residue between the Cys pairs hydroxylated. It is possible that PTM (including proteolysis at position 53) is incomplete in some of these peptides. We previously noted that the HPLC profile of ‘milked venom’, obtained by inducing snails to inject venom into artificial substrates, is less complex than that of total duct venom (Marshall et al., 2002), consistent with the idea that differences in the cal5 peptides (and potentially many others) reflect different degrees of PTM.
A second predicted peptide with an apparent type 5 framework (Cal5.2 in Fig. 1B) has a signal sequence that is consistent with the T1-superfamily, although the MYCL motif of the leader has been found in only one other conotoxin, Im5.1 (Walker et al., 1999). The mature peptide is predicted to have a C-terminal amide. Limited studies on the mode of action of the T-1 toxins suggest interaction with calcium channels or with a G-protein-coupled receptor affecting these channels (Rigby et al., 1999; Walker et al., 1999). T-1 toxins are found in all Conus feeding types.
3.1.3. Frameworks 14 and 16
The conotoxins of framework 14 have four cysteines separated from each other and have been found in vermivorous species from both the Indo-Pacific region, the Western Pacific and Western Atlantic Oceans (Möller et al., 2005) (Fig. 2B). Based on the cysteine pattern in their primary structures (Loughnan et al., 2009; Kaas, et al., 2010), framework 14 conotoxins have been placed in the same category; however, they exhibit diverse intercysteine spacings and connectivities, as well as peptide sequences and modes of action. The peptide lt14a (L-superfamily) from C. litteratus inhibited neuronal-type nicotinic acetylcholine receptors in frog muscle and was a highly effective analgesic in mice (Peng et al., 2006). Peptide pl14a (J-superfamily) from C. planorbis was unusual in inhibiting Kv1.6 channels as well as neuronal and muscle subtypes of nAChRs (Imperial et al., 2006). Peptide vil14a from C. villepinii was shown in preliminary studies to block potassium channels (type unspecified) in PC12 cells (Möller et al., 2005). Interestingly, the Cys-Cys connectivity of vil14a is the (I–IV, II–III), whereas pl14a has the (I–III, II–IV) configuration.
Fig. 2.
Four-cysteine sequences of Frameworks 14 and 16. A. The framework 14 toxins, 14.1a–c, were observed as cDNAs only. Toxin Cal14.2 was also observed as peptides cal14a and cal16a in venom. B. Framework 14 toxins from other Conus species. C. Framework 16 toxin Cal16.1 showed two leader sequences (L1and L2) and was also identified as peptide cal16a. Conotoxin Lt16.1 is shown for comparison. O designates hydroxyproline, and pE indicates N-terminal pyroglutamate.
We identified Cal14.1 cDNAs that encode three framework 14 toxins of 27 amino acids that share a common signal and propeptide sequence (Fig. 2A). C-terminal amidation of the mature toxins is predicted. The signal sequence only vaguely resembles those of the J- or L-superfamilies, and there is little similarity between the mature regions of Cal14.1 toxins and the framework 14 conotoxins discussed above, other than the framework itself.
A second toxin of framework 14, Cal14.2, does not resemble either Cal14.1a–c or other framework 14 sequences, nor shows significant similarity in its signal sequence to other reported superfamilies. The mature toxin of Cal14.2 is predicted to be C-terminal amidated, but it does not resemble Cal14.1a–c or other framework 14 sequences. The corresponding peptide, cal14a, observed in venom, has a C-terminal amide plus a hydroxyproline residue at position-18. Again, our remarks concerning the undesirability of establishing new superfamilies within C. californicus apply here.
A single framework 16 toxin, Cal16.1 (Fig. 2B, C), was observed to have two nearly identical precursor variants (L1 and L2) that can be identified with the T-superfamily. The sequence for Cal16.1 predicts C-terminal amidation with formation of an 18-amino acid toxin that has little resemblance to the only other reported framework 16 toxin, lt16.1 of the M-superfamily, from C. litteratus (Pi et al., 2006), with a considerably different framework spacing. The corresponding peptide identified in venom, cal16a, has a C-terminal amide and also is modified at the N-terminal position with formation of a pyroglutamate residue and apparent loss of the initial three amino acids. No mode of action has been reported for lt16.1.
3.2. Six-Cys peptide types
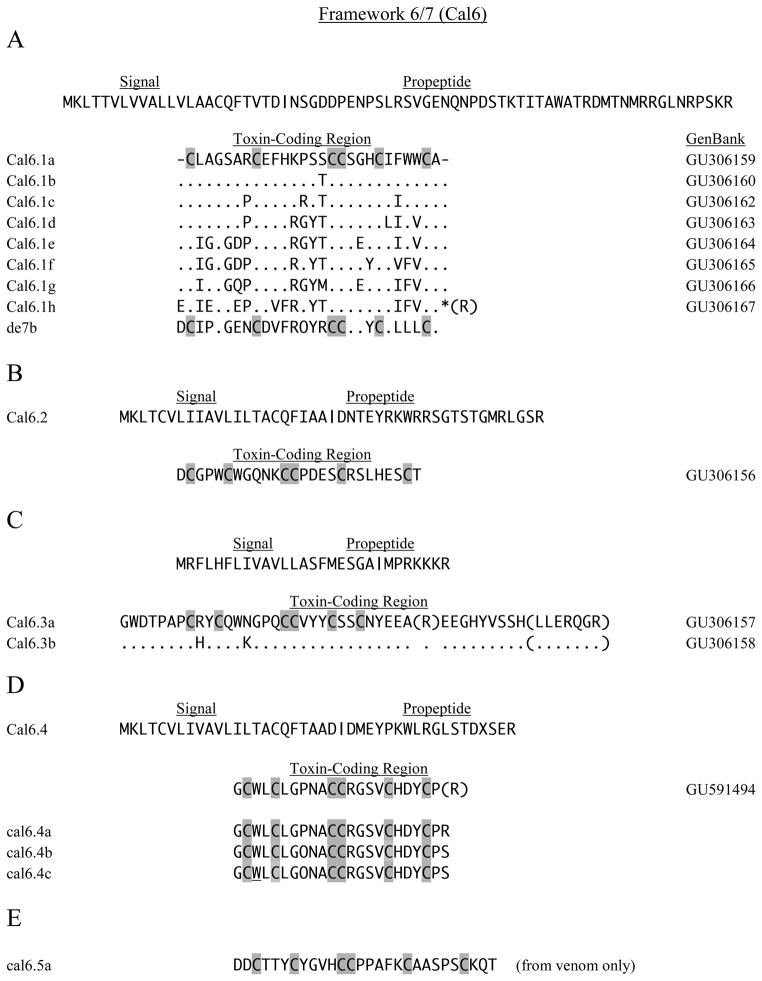
3.2.1. Framework 6/7
The cDNA encoding Cal6.1a, and its precursor peptide, can be assigned to the O1-superfamily on the basis of its signal sequence. The other members of this series (Fig. 3A) are assumed to be the same by analogy, but the location of the forward RT-PCR primer (between amino acid residues 55–63) did not include the signal portion of these sequences. The predicted amino acid composition of the Cal6.1 series varies somewhat between the invariant cysteines, but certain groupings can be distinguished (Fig. 3A). The mature region of Cal6.1h is probably amidated after C-terminal processing, whereas the other members are predicted to have free carboxy termini.
Fig. 3.
Six-cysteine sequences of Framework 6/7. In all panels O indicates hydroxyproline, and W indicates 6-bromotryptophane. A. – C. Sequence data for Cal6.1, Cal6.2 and Cal6.3 cDNAs are shown in the three respective panels. The sequence for de7b (Aguilar, et al., 2009) is shown for comparison to Cal6.1h. D. Peptides cal6.4a–c were identified in venom. E. The peptide cal6.5a did not correspond to any sequence of the cDNA library.
O-superfamily conotoxins generally are 25–35 amino acids in overall length and have 3–6 residues (mainly six) between Cys(I–II), 5–10 residues between Cys(II–III), 2–8 between Cys(IV–V), and 3–10 between Cys(V–VI) (Knottin Database, Jan., 2010). An important aspect of O-conotoxins is the possibility for formation of the inhibitory cystine-knot (ICK) motif in which two sets of disulfide-bonded segments form a ring through which a third disulfide-linked strand is threaded (Craik et al., 2001). This cystine-knot structure has been found in a large number of proteins and peptides from plants, animals and fungi that, in many cases, act as toxins and feeding deterrents. The knot structure is generally essential for activity, and the lengths and composition of the intercysteine residues serve to determine and modulate activity. A consensus (Craik et al., 2001) showed the following intercysteine spacings for ICK formation: CX3–7CX3–6CX0–5CX1–4 CX4–13C, but a recent report indicated that additional amino acid residues can occur between various cysteines (Gracy et al., 2008). On this basis, it is probable that the Cal6.1 conotoxins can form ICK structures.
O-conotoxins act on voltage-gated ion channels according to the following general patterns: μO-toxins block Nav channels; δ-toxins modify inactivation of certain Nav channels; ω-toxins block Ca+2 channels; κ-toxins block potassium channels; and γ-toxins antagonize pacemaker channels (McIntosh and Jones, 2001; Heinemann and Leipold, 2007).
A comparison of Cal6.1h with de7b, a putative delta conotoxin from C. delessertii, a vermivorous Atlantic species (Aguilar et al., 2009), reveals considerable amino acid homology, with 18 identities (including O/P and the six cysteines) out of a total 28 (Fig. 3A). Charge and hydrophobicity of residues also tends to be conserved in other positions of the two sequences. Furthermore, the unique seven-amino-acid Cys(II–III) spacing of the Cal6.1 series is identical to that of de7b, and a high degree of sequence homology exists in this region. Although electrophysiological studies were not carried out on de7b, a detailed analysis of its structure (Aguilar et al., 2009) suggests that it is a δ-conotoxin. Because de7b was isolated from a vermivorous species, presumably this peptide is active against worms, and we therefore suggest that Cal6.1h might also encode a δ-conotoxin active against worms. Differences between the other seven members of the Cal6.1 series may reflect varying activity toward Nav channels of the different taxa on which C. californicus preys, but this must be regarded as speculative at present.
Another member of the O1-superfamily identified by signal homology is Cal6.2 (Fig. 3B), but it bears little resemblance to the Cal6.1 set. Again, ICK formation would appear to be possible based on inter-Cys spacings, although the Cys(I–II) spacing does not correspond to the canonical six of the O1-conotoxins. This three-residue spacing is typical of the O3-conotoxins that are otherwise quite different from Cal6.2 (ConoServer). Differences between Cal6.1a–h and Cal6.2 certainly imply a difference in activity, but the specific mode of action cannot be predicted without additional data.
Conotoxins Cal6.3a and 6.3b (Fig. 3C) show the Cys framework of the O-superfamily, but the common signal sequence shows greater similarity to those of the T-superfamily. The overall length of the predicted mature peptides is atypical for O-conotoxins, and although alternative cleavage (in parentheses) could yield a processed size more usual for O-type toxins (Heinemann and Leipold, 2007), the short Cys(I–II) and Cys(V–VI) spacings are clearly different from other Cal6 toxins and from conventional O-conotoxins. Moreover, an ICK structure for the Cal6.3 peptide may not be possible with the unusually short Cys(I–II) and Cys(V–VI) spacings. We feel that Cal6.3a, b are not actually O-conotoxins and should not be assigned to that superfamily.
The predicted sequence of Cal6.4 (Fig. 3D) has a signal sequence clearly placing it within the O1-superfamily, but the Cys-Cys spacing, particularly the short Cys(I–II) segment, is different from other O-conotoxins, as well as Cal6.1 and Cal6.2 members. Intercysteine spacings are consistent with ICK formation. Three peptides were identified that are related to the predicted mature toxin. In cal6.4a, the final arginine is preserved, although truncation before this residue was predicted. The peptides cal6.4b and cal6.4c have C-terminal serine residues that clearly differ from the cDNA sequence of Cal6.4, and they also differ from each other in bromination of Trp at position-3 of the mature toxin (further details available in the Supplementary Material, Table S2 and Fig. S1). The proline of cal6.4a at position-8 is not hydroxylated, whereas it is in the other sequences; this feature, along with variation in Trp bromination, may again reflect incomplete or differential processing, as in the cal5 examples.
The peptide cal6.5a (Fig. 3E) provides another example of the 6/7 framework, but it was obtained by de novo sequencing of an HPLC fraction; therefore, an assignment to a superfamily cannot be made as there is no cDNA information (see Supplementary Material, Fig. S2, showing the de novo sequence alignment). The intercysteine spacing of Cys(II–III) does not correspond to any members of the O-superfamily (it is too short), and it is also different from other cal6 peptides.
3.2.2. Framework 9
A different Cys arrangement characterizes the Cal9.1 conotoxins (Fig. 4A), a closely related group that shows little affinity to other reported superfamilies. The small number of framework 9 conotoxins so far reported lie within the P-superfamily (Miles, 2002; Pi, 2006). It is of considerable interest that a Kunitz domain is present in the Cal9.1 series, suggesting a possible relationship with the conkunitzins, Conk-S1 and -S2, that are reported to be potassium channel blockers from the piscivorous C. striatus (Bayrhuber et al., 2005). The conkunitzins have about the same length as the Cal9.1 toxins, but they have a framework with four-Cys residues rather than six.
Fig. 4.
Six-cysteine sequences of framework 9. A. Sequences of the Cal9.1 family show a similarity to a variety of toxins and protease inhibitors (see text for details). B. Alignment of Kunitz-type toxins and protease inhibitors from diverse species compared to Cal9.1a. Abbreviations are: DTxE, dendrotoxin E (Dendroaspis polylepis, Black Mamba); As-fr-19, wasp venom (Anoplius samariensis); AsKC3, anemone toxin (Anemonia sulcata); AnnTx, anntoxin (Hyla annectens); Conk-S-1, conkunitzin S1 (Conus striatus). BPTI, bovine pancreatic trypsin inhibitor is shown at bottom for comparison. Black boxes in DTx-1 indicate the residues critical to block of K channels. N-terminal pyroglutamate in DTx-1 is indicated by pE. C. Another family of toxins, Cal9.2, has little similarity with the Cal9.1 family.
A BLAST search in GenBank showed that Cal9.1a had 49% identity to dendrotoxin-E (DtxE), a six-Cys peptide of 59 amino acids containing a Kunitz domain, found in the venom of Dendroaspis polylepis, the black mamba (Fig. 4B). DtxE is strongly inhibitory toward both trypsin and chymotrypsin (Schweitz and Moinier, 1999). Many other toxins that are homologs of the Kunitz serine protease-inhibitors have been described (Yuan et al., 2008). Some examples, shown in Fig. 4B, include the venom component, As-fr-19, from the solitary wasp, Anoplius samariensis (Hisada et al., 2005), the anemone toxins isolated from Anemonia sulcata (kalicludines) AsKC1-2 and -3 (Schweitz et al., 1995, Honma and Shiomi, 2005) and an amphibian toxin, anntoxin, isolated from skin secretions from the tree frog Hyla annectens (You et al., 2009). Anntoxin is similar to the conkunitzins, having the same four-Cys framework and spacing, but these features clearly differentiate these sequences from the Cal9.1 family.
Certain dendrotoxins such as DTx-1, as well as the anemone kalicludines, block potassium channels (Katoh et al., 2000; Harvey, 2001); however, kalicludines also inhibit trypsin with high affinity, whereas DTx-1 does not inhibit trypsin (Schweitz et al., 1995). The six-Cys toxins, Cal9.1, dendrotoxins, kalicludines and wasp venom, can form the standard three disulfide-bridged system of the Kunitz domain, but the conkunitzins and anntoxin can only form two bridges.
Structure-activity relationships for DTx-1 have indicated that a cationic domain consisting of lysine residues at positions 5, 28 and 29 (spatially close in the folded structure of the peptide), and a triad composed of Lys 19, Tyr 17 and Trp 37, are critical to block of Kv1 channels (Katoh et al., 2000). Charge reversal or neutralization of all of these lysine residues in Cal9.1 members suggests that these peptides are not likely to be potent KKv1blockers. Whether they are protease inhibitors is also uncertain.
Cal9.2 toxins (Fig. 4C) are presented here as mature peptides of 45 or 46 amino acids that are predicted to have N-terminal pyroglutamic acid residues. Five additional amino acids may exist at the N-terminal end, because an additional putative cleavage site (KK) occurs earlier in the propeptide sequence than the canonical arginine residue. The signal sequence is not similar to that of the P-superfamily, nor does it resemble that of the Cal9.1 toxins. No homology was observed with any other conotoxin, and a BLAST search in GenBank did not reveal any significant similarity to reported proteins.
3.3. Eight-Cys peptide types
3.3.1. Framework 12
We have previously identified a large and highly diverse family of framework 12 toxins (not shown here, but see Table 1), Cal12.1.1–12.1.4, and a related toxin with the same framework but less variability, Cal12.2a–d (Gilly et al., 2011). Two of the Cal12.1.1 series (12.1.1a, b) block voltage-gated Na channels in cephalopod molluscs, but the actions of other Cal12.1 and Cal12.2 variants are not known. As discussed elsewhere, these unusual toxins are allied with the O1-superfamily through signal sequence homology, and by the fact that the type 6 framework is ‘embedded’ within type 12 (there are two extra cysteines appended to the N-terminal region). No other commonality between these Cal12 toxins and any other conotoxins appears to exist, and we suggested the use of the new term O (+2) for the Cal12 toxins. GenBank accession numbers for the framework 12 sequences are provided in Supplementary Material, Table S3.
Table 1.
Summary of superfamily assignments for cDNAs encoding conopeptides in Conus californicus. The abbreviated signal sequence is the first five amino acids of the hydrophobic leader. Suggested superfamilies are based on framework type (indicated by #) and signal sequences (indicated by &), respectively (ConoServer). Gray boxes indicate assignments that we consider reasonably well established by the congruence of superfamilies deduced from framework and signal sequence. An inability to make any identification with a superfamily is indicated by a lone question mark (?). An asterisk (*) indicates a framework not previously reported for other Conus species. O(+2) designates a putative superfamily described in detail elsewhere (Gilly et al., 2011).
| Toxin | Cys-Cys arrangement | Framework type | Super- family# | Signal Sequence | Super- family& |
|---|---|---|---|---|---|
| Cal1.1a-b | CC-CXP-C | 1 | T | MRCLP | T |
| Cal1.2 | CC-C-C | 1 | T or A | MKLTC | O |
| Cal5.1 | CC-CC | 5 | T | MRFYI | ? |
| Cal5.2 | CC-CC | 5 | T | MMYCL | T |
| Cal6.1a-h | C-C-CC-C-C | 6/7 | O | MKLTT | O |
| Cal6.2 | C-C-CC-C-C | 6/7 | O | MKLTC | O |
| Cal6.3a-b | C-C-CC-C-C | 6/7 | O? | MRFLH | ? |
| Cal6.4 | C-C-CC-C-C | 6/7 | O? | MKLTC | O |
| Cal9.1a-d | C-C-C-C-C-C | 9 | P? | MTFLL | ? |
| Cal9.2a-f | C-C-C-C-C-C | 9 | P? | MNCYL | ? |
| Cal12.1-4 | C-C-C-C-CC-C-C | 12 | O(+2) | MKLTC | O |
| Cal12.2 | C-C-C-C-CC-C-C | 12 | O(+2) | MKLTC | O |
| Cal14.1 | C-C-C-C | 14 | L or J ? | KLCMV | L? |
| Cal14.2 | C-C-C-C | 14 | L or J ? | MKFLL | ? |
| Cal16.1 | C-C-CC | 16 | M? | MRCLS | T |
| Cal22a-f | C-C-C-C-C-C-C-C | 22* | ? | MMSTK | ? |
| CalMKLL-1,2 | (C-) -C-C-C-C-C-C-C-C-C | ?* | ? | MKLLL | ? |
3.3.2. Framework 22
An apparently new eight-Cys framework is evident in the Cal22 series, and we have designated this as framework 22 (Fig. 5A). The signal sequence for cal22 does not correlate well with that of any of the reported superfamilies, although it does show weak similarity with the M-superfamily that mainly includes the six-Cys framework 3. Six variants with predicted lengths of 47 or 45 amino acids were found for the mature toxin regions, which divide into two subgroups. Deletions of two amino acids in Cal22e and Cal 22f, each between the second and third cysteines, set these sequences apart. As shown in Fig. 5A, C-terminal proteolysis is indicated by (RR) to yield free carboxyl termini.
Fig. 5.
Unusual conopeptide sequences. A. Eight-cysteine sequences of the proposed new framework 22 comprise the Cal22 family. B. Predicted precursor sequences for CalMKLL types showing (in parentheses) possible alternative proteolytic cleavage positions within the putative propeptide region that could generate a conotoxin with either 9 or 10 cysteines.
3.4. Nine (ten)-Cys peptide types
Two peptide precursors with a leader sequence beginning with MKLL do not conform to any existing framework (Fig. 5B). If proteolytic cleavage of the propeptide occurs at either R-28 or R-37, then the resulting mature peptide would have nine cysteines. Alternatively, cleavage immediately after the signal sequence would give rise to a predicted ten-Cys peptide having 112 amino acids. Conopeptides with such long sequences include conophysin-R from C. radiatus, an 84 amino acid peptide with 14 cysteines (Lirazan et al., 2002), as well as conodipine-M from C. magus that occurs as two linked peptide-chains (McIntosh et al., 1995). Neither of these peptides bears similarity to the MKLL peptides from C. californicus.
3.5. Conclusions
In this paper we describe cDNAs encoding 17 basic varieties of conotoxins in Conus californicus, several of which have multiple members (see Table 1). Although these conotoxins occur mainly as types that can be accommodated within nine of the standard Cys-frameworks, two frameworks were found that have not been reported from other Conus species. Assignment of framework based only on amino acid sequence is somewhat arbitrary, and in some cases conotoxins within a given framework show considerable variability in intercysteine spacings. This sort of mismatch is pronounced for some of C. californicus conotoxins, which makes definitive framework assignment difficult. Therefore, we have endeavored to place these sequences into established frameworks in a conservative manner.
Assignment of C. californicus toxins to superfamilies is also difficult. As indicated in Table 1, relatively solid assignments of conotoxins to known superfamilies can be made in only four cases (darker gray boxes), with two more being somewhat questionable (Cal14.1 and Cal6.4; lighter gray boxes). In nine other cases, the signal sequence either did not match the superfamily normally associated with the Cys-framework (Cal5.1 and Cal16.1) or did not match known superfamilies. As previously discussed, we have chosen not to assign new superfamily designations to these anomalous conotoxins from C. californicus.
Both the apparent mismatch between the signal sequence and Cys-framework and the generally unusual signal sequences in C. californicus may involve the presence of an intron in the pro-region of conotoxin genes that separates exon I (encoding the signal sequence) from the remaining portion of the gene (Yuan et al., 2007). In general, this has allowed the mutation rate for exon I to be considerably lower than that for the toxin-coding region, which in turn has resulted in the observed conservation of the signal sequences and divergence of the mature peptide region. In C. californicus, a species showing a distant phylogenetic relationship to the rest of the genus, evolution of exon I appears not to have been constrained in the same manner as for other Conus species. This may have led to an unusually high level of diversity in toxin precursors as well as mature toxins.
Our results suggest that the evolution of the extremely diverse nature of these conotoxins in C. californicus has proceeded in a fundamentally different way than in the Indo-Pacific complex of species, namely in the virtual absence of significant selection-pressure linked to competition with congeneric species for specific prey types. Rather, lack of competition appears to have led to a different evolutionary strategy for toxin diversification that may serve to maximize the variety of prey types. In addition, the concept of conotoxin superfamilies may prove to be problematic when applied to species that are only distantly related to the well-studied Indo-Pacific clade.
Supplementary Material
Acknowledgments
We thank Mike Morris, Eddie Kisfaludy, Charles Hanifin and Clayton Gilly for assistance with snail collection and Alex Norton for animal husbandry. Sequence data for Cal12.2 and Cal6.4 were obtained by Joseph Schulz. The project described was supported by the National Science Foundation (NSF) by Grant No. IBN-0131788-002 to W.F.G. and by Award No. P30DA018310 from the National Institute On Drug Abuse (NIDA) and Award No. 5RO1NS031609 from the National Institute of Neurological Disorders and Stroke (NINDS) to J.V.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of NSF, NIDA, NINDS or the National Institutes of Health.
Abbreviations
- Cys
cysteine
- HPLC
high performance liquid chromatography
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- PTM
posttranslational modification
Appendix: Supplementary Materials
Note: Supplementary material associated with this paper can be found in the online version.
References
- Aguilar MB, Flores-Torres A, Batista CVF, Falcon A, Lopez-Vera E, Heimer de la Cotera EP. Structural characterization of five post-translationally modified isomorphs of a novel putative δ-conotoxin from the vermivorous snail Conus delessertii from the Mexican Caribbean Sea. Peptides. 2009;30:458–466. doi: 10.1016/j.peptides.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Bayrhuber M, Vijayan V, Ferber M, Graf R, Korokottu JI, Garrett JE, Olivera BM, Terlau H, Zweckstetter M, Becker S. ConkunitzinS-1 is the first member of a new Kunitz-type neurotoxin family. J Biol Chem. 2005;280:23766–23770. doi: 10.1074/jbc.C500064200. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.3. J Mol Biol. 2004;340:793–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–3069. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell GA, Twarog BM. Active factors in the venom duct of Conus californicus. Br J Pharmacol. 1972;44:365P–366P. [PMC free article] [PubMed] [Google Scholar]
- Craik DJ, Daly NL, Waine C. The cystine knot motif in toxins and implications for drug design. Toxicon. 2001;39:43–60. doi: 10.1016/s0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- Daly NL, Craik NL. Structural studies of conotoxins. Life. 2009;61:144–150. doi: 10.1002/iub.158. [DOI] [PubMed] [Google Scholar]
- Duda TF, Jr, Kohn AJ, Palumbi SR. Origins of diverse feeding ecologies within Conus, a genus of venomous marine gastropods. Biol J Linn Soc Lond. 2001;73:391–409. [Google Scholar]
- Duda TF, Jr, Palumbi SR. Gene expression and feeding ecology: evolution of piscovery in the venomous gastropod genus Conus. Proc Roy Soc Lond B. 2004;271:1165–1174. doi: 10.1098/rspb.2004.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Jr, Kohn AJ. Species-level phylogeography and evolutionary history of the hyperdiverse marine gastropod genus Conus. Mol Phylogenet Evol. 2005;34:257–272. doi: 10.1016/j.ympev.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Elliott EJ, Raftery MA. Venom of marine snail Conus californicus: Biochemical studies of a cholinomimetic component. Toxicon. 1979;17:259–268. doi: 10.1016/0041-0101(79)90216-2. [DOI] [PubMed] [Google Scholar]
- Espiritu DJ, Watkins M, Dia-Monje V, Cartier GE, Cruz LJ, Olivera BM. Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon. 2005;39:1899–1916. doi: 10.1016/s0041-0101(01)00175-1. [DOI] [PubMed] [Google Scholar]
- Fricker LD. Neuropeptide-processing enzymes: applications for drug discovery. AAPS J. 2005;7:E449–E455. doi: 10.1208/aapsj070244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilly WF, Richmond TA, Duda T, Elliger C, Lebaric Z, Schulz J, Bingham JP, Sweedler JV. A diverse family of novel peptide toxins from an unusual cone snailConus californicus. J Exp Biol. 2011 doi: 10.1242/jeb.046086. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracy J, Le-Nguyen D, Gelly J, Kaas Q, Heitz A, Chiche L. KNOTTIN: the knottin or inhibitor cystine knot scaffold in 2007. Nucleic Acids Res. 2008;36:D314–319. doi: 10.1093/nar/gkm939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halai R, Craik DJ. Conotoxins: natural product drug leads. Nat Prod Rep. 2009;26:526–536. doi: 10.1039/b819311h. [DOI] [PubMed] [Google Scholar]
- Han TS, Teichert RW, Olivera BM, Bulaj G. Conus venoms - a rich source of peptide-based therapeutics. Curr Pharm Des. 2008;14:2462–2479. doi: 10.2174/138161208785777469. [DOI] [PubMed] [Google Scholar]
- Harvey AL. Twenty years of dendrotoxins. Toxicon. 2001;39:15–26. doi: 10.1016/s0041-0101(00)00162-8. [DOI] [PubMed] [Google Scholar]
- Heinemann SH, Leipold E. Conotoxins of the O-superfamily affecting voltage-gated sodium sodium channels. Cell Mol Life Sci. 2007;64:1329–1340. doi: 10.1007/s00018-007-6565-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisada M, Satake H, Masuda K, Aoyama M, Murata K, Shinada T, Iwashita T, Ohfune Y, Nakajima T. Molecular components and toxicity of the venom of the solitary wasp, Anoplius samariensis. Biochem Biophys Res Comm. 2005;330:1048–1054. doi: 10.1016/j.bbrc.2005.03.087. [DOI] [PubMed] [Google Scholar]
- Honma T, Shiomi K. Peptide toxins in sea anemones: structural and functional aspects. Mar Biotechnol. 2005;8:1–10. doi: 10.1007/s10126-005-5093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C, Grilley M, Miller C, Shon K, Cruz LJ, Gray WR, Dykert J, Rivier J, Yoshikami D, Olivera BM. A new family of Conus peptides targeted to the nicotinic acetylcholine receptor. J Biol Chem. 1995;270:22361–22367. doi: 10.1074/jbc.270.38.22361. [DOI] [PubMed] [Google Scholar]
- Imperial JS, Bansal PS, Alewood PF, Daly NL, Craik DJ, Sporning A, Terlau H, Lopex-Vera E, Bandyopadhyay PK, Olivera BM. A novel conotoxin inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochem. 2006;45:8331–8340. doi: 10.1021/bi060263r. [DOI] [PubMed] [Google Scholar]
- Jakubowski JA, Kelley WP, Sweedler JV. Screening for post-translational modifications in conotoxins using liquid chromatography/mass spectrometry: an important component of conotoxin discovery. Toxicon. 2006;47:688–99. doi: 10.1016/j.toxicon.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Jiminez EC, Olivera BM, Teichert RW. αC-conotoxin PrXA: A new family of nicotinic acetylcholine receptor antagonists. Biochemistry. 2007;46:8717–8724. doi: 10.1021/bi700582m. [DOI] [PubMed] [Google Scholar]
- Kaas Q, Westermann JC, Halai R, Wang CK, Craik DJ. ConoServer, a database for conopeptide sequences and structures. Bioinformatics. 2008;24:445–446. doi: 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]
- Kaas Q, Westermann JC, Craik DJ. Conopeptide characterization and classification: An analysis using ConoServer. Toxicon. 2010;55:1491–1509. doi: 10.1016/j.toxicon.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Katoh E, Nishio H, Inui T, Nishiuchi Y, Kimura T, Sakakibara S, Yamazaki T. Structural basis for the biological activity of dendrotoxin-1, a potent potassium channel blocker. Biopolymers. 2000;54:44–57. doi: 10.1002/(SICI)1097-0282(200007)54:1<44::AID-BIP50>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Kelley WP, Schulz JR, Jakubowski JA, Gilly WF, Sweedler JV. Two toxins from Conus striatus that individually induce tetanic paralysis. Biochemistry. 2006;45:14212–22. doi: 10.1021/bi061485s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn AJ. Food specialization in Conus in Hawaii and California. Ecology. 1966;B47:1041–1043. [Google Scholar]
- Lirazan M, Jimenez EC, Craig AG, Olivera BM, Cruz LJ. Conophysin-R, a Conus radiatus venom peptide belonging to the neurophysin family. Toxicon. 2002;40:901–908. doi: 10.1016/s0041-0101(02)00079-x. [DOI] [PubMed] [Google Scholar]
- Loughnan ML, Nicke A, Lawrence A, Lewis RJ. Novel aD-conopeptides and their precursors identified by cDNA cloning define the D-conotoxin superfamily. Biochemistry. 2009;48:3717–3729. doi: 10.1021/bi9000326. [DOI] [PubMed] [Google Scholar]
- Marshall J, Kelley WP, Rubakhin SS, Bingham J, Sweedler JV, Gilly WF. Anatomical correlates of venom production in Conus californicus. Biol Bull. 2002;203:27–41. doi: 10.2307/1543455. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Ghomashchi F, Gelb MH, Dooley DJ, Stoehr SJ, Giordani AB, Naisbitt SR, Olivera BM. Conodipine-M, a novel phospholipase A2 isolated from the venom of the marine snail Conus magus. J Biol Chem. 1995;270:3518–3526. doi: 10.1074/jbc.270.8.3518. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Corpuz GO, Layer RT, Garrett JE, Wagstaff JD, Bulaj G, Vyazovkina A, Yoshikami D, Cruz LJ, Olivera BM. Isolation and characterization of a novel Conus peptide with apparent antinociceptive activity. J Biol Chem. 2000;275:32391–32397. doi: 10.1074/jbc.M003619200. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Jones RM. Cone venom - from accidental stings to deliberate injection. Toxicon. 2001;39:1447–1451. doi: 10.1016/s0041-0101(01)00145-3. [DOI] [PubMed] [Google Scholar]
- Miles LA, Dy CY, Nielsen J, Barnham KJ, Hinds MG, Olivera BM, Bulaj G, Norton RS. Structure of a novel P-superfamily spasmodic conotoxin reveals an inhibitory cystine knot motif. J Biol Chem. 2002;277:43033–43040. doi: 10.1074/jbc.M206690200. [DOI] [PubMed] [Google Scholar]
- Möller C, Rahman S, Lauer-Fields J, Bubis J, Fields GB, Mari F. A novel conotoxin framework with a Helix-loop-helix (Cs α/α) fold. Biochem. 2005;44:15986–15996. doi: 10.1021/bi0511181. [DOI] [PubMed] [Google Scholar]
- Morris RH, Abbot DP, Haderlie EC. Intertidal invertebrates of California. Stanford University Press; Palo Alto, CA: 1980. [Google Scholar]
- Nilsson KPR, Lovelace ES, Caesar CE, Tynngard N, Alewood PF, Johansson HM, Sharpe IA, Lewis RJ, Daly NL, Craik DJ. Solution structure of χ-conopeptide MrIA, a modulator of the human norepinephrine transporter. Pep Sci. 2005;80:815–823. doi: 10.1002/bip.20302. [DOI] [PubMed] [Google Scholar]
- Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–798. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Peng C, Tang S, Pi C, Liu J, Wang F, Wang L, Zhou W, Xu A. Discovery of a novel class of conotoxin from Conus litteratus, lt14a with a unique cysteine pattern. Peptides. 2006;27:2174–2181. doi: 10.1016/j.peptides.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Peng C, Wu X, Han Y, Yuan D, Chi C, Wang C. Identification of six novel T-1 conotoxins from Conus pulicarius by molecular cloning. Peptides. 2007;28:2116–2124. doi: 10.1016/j.peptides.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Pi C, Liu J, Peng C, Liu Y, Jiang X, Zhao Y, Tang S, Wang L, Dong M, Chen S, Xu A. Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics. 2006;88:809–819. doi: 10.1016/j.ygeno.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Remigio EA, Duda TF., Jr Evolution of ecological specialization and venom of a predatory marine gastropod. Mol Ecol. 2008;17:1156–1162. doi: 10.1111/j.1365-294X.2007.03627.x. [DOI] [PubMed] [Google Scholar]
- Rigby AC, Lucas-Meunier E, Kalume DE, Czerwiec E, Hambe B, Dahlqvist I, Fossier P, Baux G, Roepstorff P, Baleja JD, Furie BC, Furie B, Stenflo J. A conotoxin from Conus textile with unusual posttranslational modifications reduces presynaptic Ca2+ influx. Proc Natl Acad Sci. 1999;96:5758–5763. doi: 10.1073/pnas.96.10.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röckel D, Korn W, Kohn AJ. Manual of the Living Conidae. Verlag Christa Hemmen; Wiesbaden, Germany: 1995. [Google Scholar]
- Saunders PR, Wolfson F. Food and feeding behavior in Conus californicus Hinds 1844. Veliger. 1961;3:73–76. [Google Scholar]
- Schweitz H, Bruhn T, Guillemare E, Moinier D, Lancelin JM, Beress L, Lazdunski E. Kalicludines and Kaliseptine. Two different classes of sea anemone toxins for voltage-sensitive K+ channels. J Biol Chem. 1995;270:25121–25126. doi: 10.1074/jbc.270.42.25121. [DOI] [PubMed] [Google Scholar]
- Schweitz H, Moinier D. Mamba toxins. Perspectives in Drug Discovery and Design. 1999;15/16:83–110. [Google Scholar]
- Sharpe IA, Gehrmann J, Loughnan ML, Thomas L, Adams DA, Atkins A, Palant E, Craik DJ, Alewood PF, Lewis RJ. Two new classes of conopeptides inhibit the α1-adrenoceptor and noradrenaline transporter. Nat Neurosci. 2001;4:902–907. doi: 10.1038/nn0901-902. [DOI] [PubMed] [Google Scholar]
- Stanton RJ. Megafauna of the upper Miocene Castaic Formation, LosAngeles County, California. J Paleontol. 1966;40:21–40. [Google Scholar]
- Stewart J, Gilly WF. Piscivorous behavior of a temperate cone snail, Conus californicus. Biol Bull. 2005;209:146–153. doi: 10.2307/3593132. [DOI] [PubMed] [Google Scholar]
- Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Walker CS, Steel D, Jacobsen RB, Lirazan MB, Cruz LJ, Hooper D, Shetty R, DelaCruz RC, Nielsen JS, Zhou LM, Bandyopadhyay P, Craig AG, Olivera BM. The T-superfamily of conotoxins. J Biol Chem. 1999;274:30664–30671. doi: 10.1074/jbc.274.43.30664. [DOI] [PubMed] [Google Scholar]
- You D, Hong J, Rong M, Yu H, Liang S, Ma Y, Yang H, Wu J, Lin D, Lai R. The first gene-encoded amphibian neurotoxin. J Biol Chem. 2009;284:22079–22086. doi: 10.1074/jbc.M109.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D, Han Y, Wang C, Che C. From the identification of gene organization of α conotoxins to the cloning of novel toxins. Toxicon. 2007;49:1135–1149. doi: 10.1016/j.toxicon.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Yuan C, He Q, Peng K, Diao J, Jiang L, Tang X, Liang S. Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS ONE. 2008;3:e3414. doi: 10.1371/journal.pone.0003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Bustillos R, Aguilar MB, Falcon A, Heimer de la Cotera EP. Identification, by RT-PCR, of four novel T-1-superfamily conotoxins from the vermivorous snail Conus spurius from the Gulf of Mexico. Peptides. 2009;30:1396–1404. doi: 10.1016/j.peptides.2009.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.