Abstract
A near-infrared light-responsive drug delivery platform based on Au-Ag-nanorods (Au-Ag NRs) coated with DNA-crosslinked polymeric shells was constructed. DNA complementarity has been applied to develop a polyacrylamide-based sol-gel transition system to encapsulate anticancer drugs into the gel scaffold. The Au-Ag NR based nanogels can also be readily functionalized with targeting moieties, such as aptamers, for specific recognition of tumor cells. When exposed to NIR irradiation, the photothermal effect of the Au-Ag NRs leads to a rapid rise in the temperature of the surrounding gel, resulting in the fast release of the encapsulated payload with high controllability. In vitro study confirmed that aptamer functionalized nanogels can be used as drug carriers feasible for targeted drug delivery with remote control capability by NIR light with high spatial/temporal resolution.
Keywords: near-infrared, light responsive, nanorods, DNA-crosslinked, nanogels, targeted drug delivery
Strategies for controlled pharmaceutical release with polymeric materials have been intensively investigated in recent years for applications in such areas as biomedicine and tissue engineering.1,2 Smart hydrogels, which are susceptible to physicochemical changes in response to stimuli such as pH,3,4 molecular recognition,5–7 and temperature,8–10 have attracted particular attention. Hydrogels are cross-linked hydrophilic polymers able to imbibe large amounts of water or biological fluids, and, as a result, swelling occurs. Stimuli trigger de-swelling or degrading of the elastic network, thereby making hydrogels responsive to environmental changes and, in turn, enabling the release of pre-loaded effectors. However, although smart hydrogels can undergo conformational or chemical changes based on changes in the surrounding medium, there are still major challenges in functionality and efficiency when stimuli are subtle, especially for biochemical signals or biomarkers present at subnanomolar concentrations. Therefore, triggers which can be controlled exogenously by irradiation with light or exposure to electrical and magnetic fields are increasingly studied for applications in sensors11,12 and drug delivery systems.13,14 In addition to an externally actuatable property, the capability of remote control also enables the release of pre-loaded effectors from polymeric materials with remarkable spatial/temporal resolution.
Photo-sensitive nanoparticles (NPs), such as gold nanoshells,15 gold nanorods (NR),16–18 and gold nanocages,14 have been widely developed and recently applied in photo-thermal therapy. These nanostructures possess intense absorption bands in the near-infrared (NIR) range. Upon exposure to a NIR laser beam, the absorbed photon energy is converted to heat with high efficiency. Once the heat dissipates into the surroundings (i.e., the target tissue), the rise in temperature increases membrane permeability and/or promotes the destruction of biological samples. Photo-induced hyperthermia is considered a relatively noninvasive and benign adjuvant/alternative for cancer treatment. Gold NPs are highly efficient photo-thermal convectors, but their nonporous nanostructures exhibit low loading capacities and limited elasticity, restricting their use in effective drug/gene delivery. Gold nanocages possess hollow interiors and porous walls. Although they have been applied to controllable payload release using NIR lasers, the nonspecific leakage of pre-loaded effectors may deteriorate the targeting efficacy.14 Other photothermal strategies using photons to produce heat in a similar manner display comparable low loading and releasing efficiency, and the loads have to be specialized.19 Therefore, hydrogel-coated nanoparticles, or NP-based nanogels, which can be easily functionalized by manipulating both the properties of hydrogel and the composite nanomaterials, have been demonstrated as promising emerging candidates and are having a major impact on controlled drug delivery and soft actuator application.13
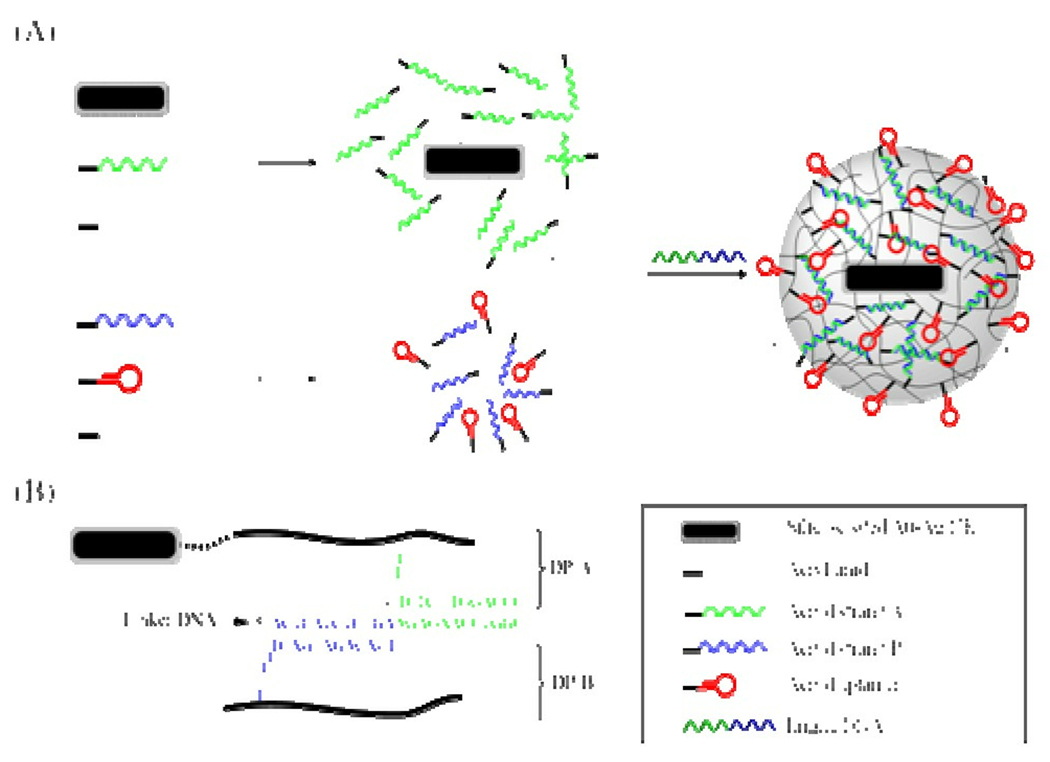
Here we report an interesting approach for engineering a novel light-responsive drug delivery system (Figure 1A). This as-prepared nanostructure is constructed of a photothermal unit, oligonucleotides and polymers. The double-stranded oligonucleotide crosslinked hydrogel layer can be dissolved at elevated temperature, ensuring a rapid melting process and an effective pay-load release. Specifically, gold-silver-based nanorods (Au-Ag NRs),20,21 which can absorb NIR photon energy more efficiently than spherical gold and silver NPs, were used as templates for colloid-based polymerization. The methacryl groups on the particle surface could initiate the growth of linear polymeric chains through copolymerization with the acrylamide monomer, as well as acrydite-modified oligonucleotides (acryd strand A). Therefore, multiple linear polyacrylamide polymers (DP-A) grafted with DNA strand A were formed on the surface of the NRs. Similarly, a second piece of acrydite-modified oligonucleotide (acryd strand B) has also been incorporated into another DNA polyacrylamide chain (DP-B). The sequences of DNA strand A and B are each complementary to an adjacent area of a cross-linking oligonucleotide (linker DNA). By mixing the DP-A-modified NRs, the DP-B solutions and the linker DNA in stoichiometric ratios, linker DNA will hybridize to both DP-A and DP-B, resulting in the formation of core-shell nanogels. The detailed DNA sequences and linkages are shown in Figure 1B. Note that other acrydite-modified oligonucleotides, such as aptamers, can also be copolymerized within either DP-A or DP-B for additional functionalities.
Figure 1.
(A) A schematic diagram illustrating the formation of an aptamer-functionalized core-shell nanogel. B) The DNA sequences and linkages in the nanogel (not to scale).
To prevent NRs from serious aggregation during the polymerization process, poly(ethylene glycol)-modified NRs were first coated with a silica shell by hydrolysis of tetraethyl orthosilicate (TEOS) in an ethanol/water mixture. The thickness of the silica shell was 23±4 nm, as observed by transmission electron microscopy (TEM) (Figure 2A). Because of an increase in local refractive index around the nanorods produced by the silica shells, spectra of the silica-coated NRs also demonstrated a red shift of the surface plasmon band (corresponding to the longitudinal oscillation mode) (Figure 2B).
Figure 2.
(A) Transmission electron microscopy (TEM) images and (B) UV-visible spectra of (a) Au-Ag NRs, (b) silica-coated Au-Ag NRs, and (c) NR-based nanogels (the inserted image is a single nanogel). Scale bars: (a) 100 nm, (b) 500 nm, (c) 1 µm.
The silane-coupling agent 3-(methacryloxy)propyl triethoxysilane (MPS) was used to modify the silica-coated NRs, and the methacryl groups initiated the growth of multiple DP-A chains on the surface. After crosslinking, the successful NR-based nanogel formation was demonstrated by the blue shift of the longitudinal surface plasmon resonance (SPR) band caused by the change in the local refractive index produced by the DNA-crosslinked polymeric shell. TEM imaging shows that the core-shell nanogels had a relatively uniform size of 284±18 nm in the dry state (Figure 2A). This result is in reasonable agreement with the mean hydrodynamic diameter (315±25nm) estimated from the dynamic light scattering analysis shown in Supplementary Figure S1(c).
To demonstrate the controlled release capability of our nanogel system, fluorescein (FAM) molecules were added to the solution mixture and encapsulated within the DNA-crosslinked polymeric layer during gel formation. After a series of centrifugation/washing steps to remove uncaptured dye molecules, the release profile of the captured FAM was examined by monitoring the increasing fluorescence signals at 520 nm from the supernatants at known time intervals after the nanogels were exposed to light irradiation and then spun down (Figure 3). It should be noted that the concentration of FAM molecules encapsulated inside the nanogels (2 nM) was about 3.8 µM.
Figure 3.
Controlled release of the entrapped fluorescein from the core-shell nanogels (10 nM) by visible and NIR light irradiations.
Only slight leaking of FAM molecules (less than 5 % of the entire payload) from the nanogels occurred during the first 30 min under visible light (450 nm) exposure, demonstrating the stable encapsulating capability of the core-shell nanogels. When the suspension of dye-loaded nanogels was exposed to a NIR laser (808 nm, 600 mW), a burst release of FAM molecules (65 ± 7%, with 100% corresponding to the concentration when the coated hydrogels were totally dissolved by heating the solution to 80 °C) occurred within 10 min. The payload kept releasing and leveled off at 74 ± 4% with prolonged exposure (50 min). The burst release of FAM from the nanogels was ascribed to the rapid dissolution of the DNA-crosslinked gel layer on the NR surface. Dynamic light scattering measurement (Figure S1(d)) also demonstrated a significant decrease (~44%) in the hydrodynamic diameter of the nanogel in response to NIR exposure. Compared to the frequently used thermo-responsive hydrogel nanocomposites, e.g., poly(N-isopropylacrylamide),14,22,23 this is the first demonstration utilizing NIR light to trigger the payload release from a NR-based DNA-crosslinked nanogel. Furthermore, the remote heating generated by Au-Ag NRs through photo-thermal conversion drives an efficient thermal dehybrization of the linker oligonucleotides from their complementary sequences grafted onto the polyacrylamide chains (DP-A and DP-B; refer to Figure 1). This leads to a rapid gel-to-sol transition of the gel layer. Although these core-shell nanogels exhibit controlled release over a short period, the release kinetics, which is correlated with the temperature, mechanical properties, and pore size of the gel, can be easily modulated by altering the length, nucleotide composition, and base stacking parameters of the DNA sequence. In this case, the melting temperatures between the linker DNA and the two complementary DNA segments on DP-A and DP-B were in the range of 35–41 °C. The nature of the same polymeric backbone has been characterized previously in the format of bulk hydrogel.24 The chemical and physical properties were maintained while increasing the temperature to 100°C, and there was no obvious evidence that the photothermal effect of NR would induce any chemical reactions on the polymers.
In order to achieve active tumor targeting, site-specific ligands, such as small peptides and antibodies that bind to specific markers on the surfaces of tumor cells, have been covalently linked to drug carriers for targeted cancer therapy. Recently, a cell-based SELEX (systematic evolution of ligands by exponential enrichment) selection method has been developed to generate a variety of single-stranded oligonucleotides called aptamers.25–27 Aptamers are highly specific for different types of tumor cells and have excellent binding affinity. They also possess advantageous characteristics, including small size, ease of synthesis and modification, low toxicity or immunogenicity, as well as high stability. As a consequence, they have emerged as a novel class of molecular probes in diagnostic and therapeutic applications.28–31
To prove that our DNA-crosslinked nanogels are functional for drug delivery when tethered to aptamers, we selected acrydite-modified sgc8c, an aptamer specific to CCRF-CEM cells (a T-cell acute lymphoblastic leukemia cell line), and polymerized it into the linear DP-B chain. After crosslinking, it was first necessary to test if the aptamers on the nanogel surface maintained their specific binding and high affinity to target cancer cells. To investigate the binding characteristics of these aptamer-tethered nanogels toward different types of cells, the 3’- end of aptamer sgc8c was labeled with a fluorophore, FAM. After incubation with target cells, the sample was analyzed by flow cytometry. A separate batch of core-shell nanogels was prepared with a FAM-labeled random DNA library (lib-NGs) to serve as a negative control to show the specificity of the FAM-labeled sgc8c-conjugated core-shell nanogels (sgc8c-NGs). Figure 4 displays the flow cytometric comparison of target CEM cells and control Ramos cells after 1 h of incubation (4 °C) with sgc8c-NGs and lib-NGs. A noticeable change in the fluorescence signal is observed between lib-NG and (sgc8c-NG)-labeled CEM cells, indicating that the binding capability of the aptamer probes is maintained well after the conjugation with nanogels. No significant change in fluorescence intensity occurred on Ramos cells, which lack the target proteins of sgc8c, further confirming the specific recognition of the sgc8c-NGs for target cells.
Figure 4.
Flow cytometric assay to monitor the binding of fluorescein (FAM)-labeled sgc8c–nanogel (sgc8c-NG) conjugates (2 nM) to (A) CCRF-CEM (target) cells and (B) Ramos (control) cells at 4 °C for 1 h. The red curve represents the cells only; the green curves represent the background binding of nanogel tethered with FAM-labeled random DNA library (lib-NG); and the light blue and dark blue curves represent the FAM-labeled sgc8c-NG conjugates incubated with CEM and Ramos cells, respectively.
Similar results were obtained by confocal microscopy for different types of cells incubated with sgc8c-NGs (Figure S2). After multiple centrifugation/washing cycles to remove unbound conjugates, CEM cells presented very bright fluorescence, whereas no fluorescence was displayed for the control Ramos cells. Furthermore, compared with the cellular intensity after an incubation of 2h was carried out at 4 °C (Figure S2A), a stronger fluorescence signal from target CEM cells was observed after 2 h at 37° (Figure S2B). This result is quite consistent with our previous internalization study, showing that endocytosis was inhibited and cellular uptake was completely eliminated at 4 °C.32 No significant fluorescence signal change was observed from nontarget Ramos cells, indicating that sgc8c-NGs can be specifically taken up by the CEM cells after receptor binding.
Next, investigations of the controllable release system were extended to an in vitro study for targeted drug delivery by introducing a chemotherapeutic agent, doxorubicin (Dox). Dox is the most utilized anticancer drug against a range of neoplasms, including acute lymphoblastic and myeloblastic leukemias, as well as malignant lymphomas.33 To confirm the toxicity of Dox to leukemia cells used in this study, two cell lines were incubated with Dox in culture medium without FBS at 37°C, 5% CO2. After 2h, the unbound Dox molecules were removed by centrifugation, and fresh medium (10% FBS) was added for further cell growth (48 h). The relative viability of cells with different treatment was determined by MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy-phenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay. The results shown in Figure 5 (b) demonstrated that Dox at micromolar levels possessed high toxicity toward both CEM and Ramos cells and could greatly inhibit cell proliferation.33 In addition, the viability of cells incubated with sgc8c-NGs was also compared to that of untreated cells. Only 3 (±2)% of CEM cells and 8 (±3)% of Ramos cells were killed, respectively (Figure 5(c)). This result supported the idea that the sgc8c-NG conjugate itself shows little or no toxicity to the cells.
Figure 5.
Cytotoxicity assays of (A) CCRF-CEM (target) cells and (B) Ramos (control) cells in the absence of nanogels (a), and those incubated with (b) Dox (0.75 µM), (c) sgc8c–NGs (2 nM), (d) Dox-loaded lib-NGs (2 nM), and (e) Dox-loaded sgc8c–NGs (2 nM) in culture medium without FBS at 37 °C, 5% CO2 for 2h. Cells were exposed to a near infrared laser at 808 nm (600 mW) for 0, 5, and 10 min, respectively (blue, red and green). After treatment, cells were subsequently grown in fresh medium (10% FBS) for 48 h. The cytotoxicity was then measured by an MTS assay.
Having established that our drug release system is NIR-controllable, we next tested the cytotoxic effect induced by laser irradiation after treatment. To accomplish this, cells were incubated with different conjugates and then exposed to a laser light of 808 nm at 600 mW for 5 and 10 min, respectively. Figure 5(a) demonstrates that direct irradiation of the cells alone (10 min) had little effect on cell viability (less than 10% loss). The low light absorption by natural endogenous cytochromes of these cells accounts for their high survival.21 In contrast, cells which had been incubated with Dox-loaded sgc8c-NGs, and then irradiated, were killed (Figure 5(e)). Moreover, as irradiation time increased, a significant dead cell percentage (67±5%) was observed for CEM cells, while little effect (less than 8%) was observed from the experiments carried out with sgc8c-NG conjugates (Figure 5(c). These results correlate with the release profile plotted in Figure 3, demonstrating that a burst release of the payload from nanogels can be observed when the sample is subjected to NIR irradiation for 10 min.
The specific killing efficiency was also investigated by incubating target cells with Dox-loaded nanogel conjugated with random DNA (lib-NG)) under experimental conditions identical to those for sgc8c-NGs. The percentage of dead cells before and after illumination was 2 (±2) % and 10 (±3) %, respectively (Figure 5(d)). Furthermore, Ramos (control) cells were incubated with Dox-loaded sgc8c-NGs at 37°C for 2 h, followed by exposure to the laser at 600 mW (Figure 5(e)). The percentage of dead cells remained the same (less than 10%) after a 10-min irradiation. These results indicate that our aptamer-conjugated nanogel system shows both minimal nonspecific binding and remote controllable release capability, making this system a good candidate for selective cell recognition and targeted drug delivery.
In summary, we have demonstrated a light-responsive drug delivery platform based on Au-Ag-NRs coated with DNA-crosslinked polymeric shells. DNA complementarity has been applied to develop a polyacrylamide-based sol-gel transition system to encapsulate anticancer drugs into the gel scaffold. The nanogels can also be readily functionalized with targeting moieties, such as aptamers, for specific recognition of tumor cells. When exposed to NIR irradiation, the photothermal effect of Au-Ag NRs leads to a rapid rise in the temperature of the surrounding gel. The heat dissolves the coated gel shells and releases the encapsulated payload with high controllability. The in vitro study confirmed that linking the cell-SELEX-selected aptamers with our novel drug nanocarriers is feasible for targeted drug delivery and that the remote control capability with NIR light also shows their potential effectiveness and flexibility in drug release with high spatial/temporal resolution.
MATERIALS AND METHODS
Reagents and Cells
All the chemicals used to synthesize core-shell nanogels were purchased from Sigma-Aldrich Chemical, Inc. Doxorubicin hydrochloride (Dox) was purchased from Fisher Scientific (Houston, TX, USA). The materials for DNA synthesis using an ABI 3400 DNA/RNA synthesizer (Applied Biosystems, Foster City, CA), including CPG columns and reagents for DNA modification and coupling, were purchased from Glen Research Co. CEM-CCRF cells (CCL-119 T-cell, human acute lymphoblastic leukemia) and Ramos cells (CRL-1596, B-cell line, human Burkitt`s Lymphoma) were obtained from ATCC. All cell lines were cultured in RPMI 1640 medium purchased from ATCC and supplemented with 10% fetal bovine serum (heat-inactivated, GIBCO) and 100 IU/mL penicillin-streptomycin (Cellgro). The flow cytometry results were obtained from a FACScan cytometer (Becton Dickinson Immunocytometry Systems). The TEM data were obtained using transmission electron microscopy (TEM) analysis (Hitachi H7100, Tokyo, Japan).
DNA Preparation
DNA products were synthesized by solid-state phosphoramidite chemistry at a 1 µmol scale, deprotected in AMA (ammonium hydroxide/40% aqueous methylamine 1:1) at 65 °C for 30 min, and further purified by reversed-phase HPLC on a C-18 column with acetonitrile/aqueous triethylammonium acetate as the mobile phase. For Acryd DNAs (strand A, strand B and sgc8c), acrydite phosphoramidite was dissolved in acetonitrile and loaded into the DNA synthesizer. The synthesized acrydite-modified oligonucleotide monomers were further purified by reversed-phase HPLC and quantitatively characterized by absorption at 260 nm.
Sequences of the Synthesized DNA
| Name | Sequence |
|---|---|
| Acryd strand A | 5’-Acrydite-TTT TTC ACA GAT GAG T-3’ |
| Acryd strand B | 5’-Acrydite-TTT TCC CAG GTT CTC T-3’ |
| Linker DNA | 5’-ACT CAT CTG TGA AGA GAA CCT GGG-3’ |
| Acryd sgc8c | 5′- Acrydite-ATC TAA CTG CTG CGC CGC CGG GAA AAT ACT GTA CGG TTA GA-3′ |
Synthesis of the Au-Ag NR
Cetyl trimethylammonium bromide (CTAB) aqueous solution (0.2 mM, 5.0 mL) was mixed with 0.5 mM NaAuCl4 (5.0 mL). Fresh 0.01 mM NaBH4 (0.6 mL) was added to this solution, which was sonicated for 3 minutes, resulting in a brownish-yellow seed solution. Another aliquot of CTAB (0.2 mM, 50.0 mL) was mixed with 1.0 mM NaAuCl4 (50.0 mL) and AgNO3 (4.0 mM, 2.5 mL). After gentle mixing of the solution (growth solution), 78.8 mM ascorbic acid (0.7 mL) was added. The color of the growth solution changed rapidly from dark-yellow to colorless. Finally, a portion of the seed solution (0.12 mL) was added to the growth solution. The solution gradually changed color to dark pink over a period of 30 minutes. The sizes and the absorptions of the as-prepared Au-Ag NRs were verified through transmission electron microscopy (TEM) and UV-Vis absorption analysis, respectively. These nanocomposites appeared to be monodisperse, with average length of 56 ± 7 nm and width of 12 ± 3 nm. The transverse and longitudinal absorption bands of the Au-Ag NRs were centered at wavelengths of 509 and 857 nm, respectively. A solution of Au-Ag nanorods containing CTAB was centrifuged at 14,000 g for 10 min and resuspended in water to remove excess CTAB. One milliliter of the gold nanorod solution (1 nM) was added to 200 µL of 1 mM PEG5000-SH. The mixture was stirred overnight and dialyzed for 3 days.
Silica Coating of the Au-Ag NRs
The PEG-modified Au-Ag NR solution was concentrated by centrifugation. One hundred micro-liters of the PEG-modified gold nanorod solution (10 nM) was diluted with 780 µL of ethanol. The mixture was added to 20 µL of 5% ammonia solution and 100 µL of 50 mM tetraethyl orthosilicate (TEOS). The reaction was allowed to proceed for 20 h at room temperature. The reaction mixture was added to 100 µL of 100 mM 3-(methacryloxy)propyl triethoxysilane (MPS) to graft methacryl groups on the NR surface. The mixture was stirred for 24 h and dialyzed against an ethanol/water solution (1/1) to remove excess MPS.
Synthesis of the NR-based Core-shell Nanogels
Solution A: The 1 nM silica-coated Au-Ag NRs solution was centrifuged and resuspended in 50 µL of water. At room temperature, 2.35 µL of 853 µM strand A, 2 µL of 10 mM acrylamide, 1 µL of 1% solution of the initiator I2959 (Sigma-Aldrich Chemical), 10 µL of buffer (100 mM Tris-HCl, 500 mM NaCl, 100 mM MgCl2) and 34.65 µL of water were mixed with the Au-Ag NRs suspension. The solution was kept under UV irradiation (350 nm, 6W portable UV lamp) for 19 minutes. Solution B: At room temperature, 52.15 µL of 767 µM strand B, 4 µL of 2M acrylamide, 4 µL of 1% solution of the initiator I2959, 20 µL of buffer (100 mM Tris-HCl, 500 mM NaCl, 100 mM MgCl2) and 119.85 µL of water were mixed together. Solution B was irradiated with UV light for 19 minutes. At room temperature, 100 µL of solution A, 10 µL of solution B and 10µL of 200 µM linker DNA were mixed together and kept in a water bath at 65 °C for 5 minutes, followed by slow cooling to room temperature.
Fluorescein and Dox Loading
During the gel-shell formation, 100 µM fluorescein or doxorubicin was added to the mixed solution to encapsulate fluorescent dye or drug molecules inside the nanogels. The uncaptured molecules were removed by several centrifugation/washing steps.
NIR Controlled Release
A CWdiode laser (Thorlabs, Newton, NJ) with wavelength of 808 nm (600 mW) was used for the laser irradiation experiment. The fluorescein-loaded nanogel solution (10 nM) was put into a quartz well and illuminated by NIR light. The quartz well for all the tests was a standard micro-cuvette (1cm × 1cm in diameter, 4cm in length). The laser source was positioned 1cm from one transparent side of the cuvette. After the nanogels had been spun down, the fluorescence signal (520 nm) in the supernatant was determined in order to monitor the release kinetics of the dye molecules.
Cytotoxicity Assay
The cell viability of different cell lines to nanogels was determined using the CellTiter 96 cell proliferation assay (Promega, Madison, WI, USA). The cells (5×104 cells/well) were incubated with nanogels in culture medium without FBS at 37°C, 5 % CO2. After 2 h, the media were removed and fresh media (10 % FBS) were added for further cell growth (48 h). To measure cytotoxicity, CellTiter reagent (20 µL) was added to each well and incubated for 2 h. The absorption was recorded at 490 nm using a plate reader (Tecan Safire microplate reader, AG, Switzerland).
Supplementary Material
Acknowledgment
This work was supported by NIH GM 066137 and GM079359, the State of Florida Center for Nano-Biosensors and China Grand Grant 2009ZX10004-312. Y.F. Huang acknowledges support from National Tsing Hua University (99N2947E1) and the National Science Council (98-2113-M-007-003, 99-2113-M-007-006-MY2) of Taiwan, ROC. XZ thanks the National Nature Science Foundation of China (No. 20975012) and the 111 Project in China (B07012) grant for supporting this research.
Footnotes
Supporting Information Available. Dynamic light scattering measurements of NRs and nanogels and confocal microscopic images of the binding of FAM-labeled sgc8c-NG conjugates with target and non-target cells. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Hoffman AS. Hydrogels for Biomedical Applications. Adv.Drug Deliv. Rev. 2002;54:3–12. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- 2.Stuart MAC, Huck WTS, Genzer J, Muller M, Ober C, Stamm M, Sukhorukov GB, Szleifer I, Tsukruk VV, Urban M, et al. Emerging Applications of Stimuli Responsive Polymer Materials. Nat. Mater. 2010;9:101–113. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- 3.Kopeček J, Vacík J, Lím D. Friedel-crafts Copolymerization of Thiophene and P-di(chloromethyl)benzene. I. Products, Kinetics, and Mechanism of the Early Stages of the Reaction. J. Polym. Sci. A1. 1971;9:2801–2815. [Google Scholar]
- 4.Siegel RA, Firestone BA. pH-dependent Equilibrium Swelling Properties of Hydrophobic Polyelectrolyte Copolymer Gels. Macromolecules. 1988;21:3254–3259. [Google Scholar]
- 5.Miyata T, Jige M, Nakaminami T, Uragami T. Tumor Marker-Responsive Behavior of Gels Prepared by Biomolecular Imprinting. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1190–1193. doi: 10.1073/pnas.0506786103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyata T, Asami N, Uragami T. A Reversibly Antigen-responsive Hydrogel. Nature. 1999;399:766–769. doi: 10.1038/21619. [DOI] [PubMed] [Google Scholar]
- 7.Lu ZR, Kopečkova P, Kopeček J. Antigen Responsive Hydrogels Based on Polymerizable Antibody Fab Fragment. Macromol. Biosci. 2003;3:296–300. [Google Scholar]
- 8.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Reversible Hydrogels from Self-Assembling Artificial Proteins. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Stewart RJ, Kopeček J. Hybrid Hydrogels Assembled from Synthetic Polymers and Coiled-coil Protein Domains. Nature. 1999;397:417–420. doi: 10.1038/17092. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida R, Uchida K, Kaneko Y, Sakai K, Kikuchi A, Sakurai Y, Okano TA, Sakurai Y, Okano T. Comb-type Grafted Hydrogels with Rapid Deswelling Response to Temperature Changes. Nature. 1995;374:240–242. [Google Scholar]
- 11.Bashir R, Hilt JZ, Elibol O, Gupta A, Peppas NA. Micromechanical Cantilever as an Ultrasensitive pH Microsensor. Appl. Phys. Lett. 2002;81:3091–3093. [Google Scholar]
- 12.Hilt JZ, Gupta AK, Bashir R, Peppas NA. Ultrasensitive Biomems Sensors Based on Microcantilevers Patterned with Environmentally Responsive Hydrogels. Biomed. Microdevices. 2003;5:177–184. [Google Scholar]
- 13.Satarkar NS, Biswal D, Hilt JZ. Hydrogel Nanocomposites: a Review of Applications as Remote Controlled Biomaterials. Soft Matter. 2010;6:2364–2371. [Google Scholar]
- 14.Yavuz MS, Cheng YY, Chen JY, Cobley CM, Zhang Q, Rycenga M, Xie JW, Kim C, Song KH, Schwartz AG, et al. Gold Nanocages Covered by Smart Polymers for Controlled Release with Near-Infrared Light. Nat. Mater. 2009;8:935–939. doi: 10.1038/nmat2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lal S, Clare SE, Halas NJ. Nanoshell-Enabled Photothermal Cancer Therapy: Impending Clinical Impact. Accounts Chem.Res. 2008;41:842–1851. doi: 10.1021/ar800150g. [DOI] [PubMed] [Google Scholar]
- 16.Murphy CJ, Gole AM, Stone JW, Sisco PN, Alkilany AM, Goldsmith EC, Baxter SC. Gold Nanoparticles in Biology: Beyond Toxicity to Cellular Imaging. Accounts Chem. Res. 2008;41:1721–1730. doi: 10.1021/ar800035u. [DOI] [PubMed] [Google Scholar]
- 17.Huang XH, Neretina S, El-Sayed MA. Gold Nanorods: From Synthesis and Properties to Biological and Biomedical Applications. Adv. Mater. 2009;21:4880–4910. doi: 10.1002/adma.200802789. [DOI] [PubMed] [Google Scholar]
- 18.Zhao WA, Karp JM. Tumour Targeting: Nanoantennas Heat up. Nat. Mater. 2009;8:453–454. doi: 10.1038/nmat2463. [DOI] [PubMed] [Google Scholar]
- 19.Chen JY, McLellan JM, Siekkinen A, Xiong YJ, Li ZY, Xia YN. Facile Synthesis of Gold-Silver Nanocages with Controllable Pores on the Surface. J. Am. Chem. Soc. 2006;128:14776–14777. doi: 10.1021/ja066023g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang YF, Chang HT, Tan W. Cancer Cell Targeting Using Multiple Aptamers Conjugated on Nanorods. Anal. Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 21.Huang YF, Sefah K, Bamrungsap S, Chang HT, Tan W. Selective Photothermal Therapy for Mixed Cancer Cells Using Aptamer-Conjugated Nanorods. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- 22.Satarkar NS, Hilt JZ. Hydrogel Nanocomposites as Remote-Controlled Biomaterials. Acta .Biomaterialia. 2008;4:11–16. doi: 10.1016/j.actbio.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Kawano T, Niidome Y, Mori T, Katayama Y, Niidome T. PNIPAM Gel-Coated Gold Nanorods for Targeted Delivery Responding to a Near-Infrared Laser. Bioconjugate Chem. 2009;20:209–212. doi: 10.1021/bc800480k. [DOI] [PubMed] [Google Scholar]
- 24.Akdemir ZS, Kayaman-Apohan N. Investigation of Swelling, Drug Release and Diffusion Behaviors of Poly(N-isopropylacrylamide)/ Poly (N-vinylpyrrolidone) Full-IPN Hydrogels. Polym. Adv. Technol. 2007;18:932–939. [Google Scholar]
- 25.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic Evolution of a DNA Aptamer Binding to Rat Brain Tumor Microvessels: Selective Targeting Endothelial Regulatory Protein Pigpen. J. Biol. Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 26.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A Tenascin-C Aptamer Identified by Tumor Cell SELEX: Systematic Evolution of Ligands by Exponential Enrichment. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shangguan D, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, Sefah K, Yang CYJ, Tan WH. Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acd. Sci. U. S. A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiel KW, Giangrande PH. Therapeutic Applications of DNA and RNA Aptamers. Oligonucleotides. 2009;19:209–222. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang XH, Tan WH. Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach. Acc. Chem. Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khati M. The Future of Aptamers in Medicine. J. Clin. Pathol. 2010;63:480–487. doi: 10.1136/jcp.2008.062786. [DOI] [PubMed] [Google Scholar]
- 31.Kang HZ, O'Donoghue MB, Liu HP, Tan WH. A Liposome-Based Nanostructure for Aptamer Directed Delivery. Chem. Commun. 2010;46:249–251. doi: 10.1039/b916911c. [DOI] [PubMed] [Google Scholar]
- 32.Xiao ZY, Shangguan DH, Cao ZH, Fang XH, Tan WH. Cell-Specific Internalization Study of an Aptamer from Whole Cell Selection. Chem. Eur. J. 2008;14:1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 33.Huang YF, Shangguan DH, Liu HP, Phillips JA, Zhang XL, Chen Y, Tan WH. Molecular Assembly of an Aptamer-drug Conjugate for Targeted Drug Delivery to Tumor Cells. Chembiochem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.