Abstract
TNF has been implicated in the progression of many chronic liver diseases leading to fibrosis; however, the role of TNF in fibrogenesis is controversial and the specific contribution of TNF receptors to HSC activation remains to be established. Using hepatic stellate cells (HSC) from wild-type, TNF-receptor-1 (TNFR1) knockout, TNF-receptor-2 (TNFR2) knockout, or TNFR1/R2 double knockout (TNFR-DKO) mice we show that loss of both TNF receptors reduced pro-Collagen-α1(I) expression, slowed down HSC proliferation, and impaired PDGF-induced pro-mitogenic signaling in HSC. TNFR-DKO HSC exhibited decreased AKT phosphorylation and in vitro proliferation in response to PDGF. These effects were reproduced in TNFR1 knockout but not TNFR2 knockout HSC. In addition, MMP-9 expression was dependent on TNF binding to TNFR1 in primary mouse HSC. These results were validated in the human HSC cell line LX2 using neutralizing antibodies against TNFR1 and TNFR2. Moreover, in vivo liver damage and fibrogenesis following bile duct ligation were reduced in TNFR-DKO and TNFR1 knockout mice compared to wild-type or TNFR2 knockout mice.
Conclusions
TNF regulates HSC biology through its binding to TNFR1, which is required for HSC proliferation and MMP-9 expression. These data indicate a regulatory role for TNF in extracellular matrix remodeling and liver fibrosis, suggesting that targeting TNFR1 may be of benefit to attenuate liver fibrogenesis.
Keywords: Fibrosis, MMP-9, PDGF, TIMP-1, bile duct ligation
INTRODUCTION
Tumor necrosis factor (TNF) is an inflammatory cytokine produced by macrophages/monocytes during acute inflammation and is responsible for a diverse range of signaling events within cells. TNF exerts its biological functions by interactions with two members of the TNF receptor superfamily, namely TNFR1 and TNFR2. The cytoplasmic tail of TNFR1 contains a death domain, which is essential for induction of apoptosis. However, this motif is missing in TNFR2 and the function of this latter receptor is poorly understood (1, 2). In the liver, TNF functions as double-edged sword through TNFR1, being required for normal hepatocyte proliferation during liver regeneration (3, 4) and induction of NF-kappaB, which is essential to elicit antiapoptotic defense and in the control of the immune response. Yet, on the other hand, TNF is the mediator of hepatotoxicity and inflammation in many animal models and has also been implicated as an important pathogenic player in patients with alcoholic liver disease, nonalcoholic steatohepatitis or viral hepatitis (5, 6). Human and animal studies suggest that hepatocellular injury followed by inflammation and activation of the innate immune system leads to early-stage liver fibrosis, ultimately resulting in hepatic stellate cell (HSC) activation and extracellular matrix (ECM) deposition (7, 8). While the contribution of TNF to hepatocellular injury and inflammation has been widely studied (5, 6, 9, 10) its specific contribution to HSC activation and liver fibrogenesis remains controversial. In this sense, experimental studies performed with knockout mice after CCl4 administration have shown that the absence of either TNFR1 (11) or TNFR1/R2 (TNFR-DKO) (12) inhibit liver fibrosis accompanied by reduced expression of pro-Collagen-α1(I) mRNA, without effect on hepatic injury, suggesting a profibrogenic role for TNF. In contrast, a recent study showed that the inhibition of TNF processing via TNF-alpha converting enzyme attenuated liver injury and inflammation following CCl4 administration, but increased collagen deposition, effects reproduced in the TNFR-DKO mice (13). Moreover, several reports using cultured HSC point to an anti-fibrogenic role of TNF via inhibition of the pro-Collagen-α1(I) gene expression (14–17) due, in part, via glutathione depletion (18).
Hence, while TNF has been implicated in the progression of many chronic liver diseases leading to fibrosis the specific involvement of TNF or its receptors, TNFR1 and TNFR2, in HSC activation remains to be established. The morphological and metabolic changes associated with HSC activation, reproduced by culturing isolated HSC on plastic (19, 20), were studied in HSC from wild-type, TNFR-DKO, TNFR1 and TNFR2 knockout mice to evaluate the impact of TNF signaling, and thus, its potential direct contribution to liver fibrosis. The results, validated in vitro in human activated LX2 cells, and in vivo using a bile duct ligation mice model, lead us to underscore the contribution of TNFR1 in liver fibrosis, and suggest that blockage of specific TNF receptors may be effective to reduce hepatic deterioration during fibrogenesis.
EXPERIMENTAL PROCEDURES
Animals and HSC isolation
Wild-type, TNFR1 knockout mice, TNFR2 knockout mice, TNFR-DKO mice (10–18 weeks old) (C57BL/6 strain), a generous gift of Dr. Bluethmann (Discovery Technologies, Hoffmann-La Roche, Switzerland), were obtained by propagation of homozygous pairs. The animals had free access to water and standard purified rodent diet throughout the study. All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” published by NIH. HSC were isolated by perfusion with collagenase and cultured as previously described (21).
Cell lines and culture
In addition to primary mouse HSC, we used the human HSC cell line LX2. Cells were cultured in DMEM+10% FBS, and antibiotics at 37°C in a humidified atmosphere of 95% air and 5% CO2. Cells were serum starved at 0.5% FBS before using TNF-α, PDGF-BB, IL-1α and IL-1β (Preprotech EC, UK) or LPS (from E.coli serotype 0128:B12, Sigma-Aldrich, Spain). Neutralizing antibodies against human TNFR1 and TNFR2 (R&D Systems, Minneapolis, USA) were used at a concentration of 10µg/ml. In vitro siRNA transfection was performed using commercially available siRNA (Santa Cruz Biotechnology, Germany) as previously described (21). Unless otherwise stated, all reagents were from Sigma-Aldrich.
Real time RT-PCR and primer sequences
Total RNA from HSC, mouse tissue, or LX2 cells was isolated with TRIzol reagent (Invitrogen, UK). Real-time RT-PCR was performed with iScript™ One-Step reverse transcription (RT)-PCR Kit with SYBR® Green (BioRad, Spain). The primers sequences were designed based on published sequences (Table 1).
Table 1.
Primers used in quantitative real-time RT-PCR.
| Target gene (mouse) | 5' primer | 3' primer |
|---|---|---|
| β-actin [NM_007393] | GACGGCCAGGTCATCACTAT | CGGATGTCAACGTCACACTT |
| α-SMA [NM_007392] | ACTACTGCCGAGCGTGAGAT | AAGGTAGACAGCGAAGCCAA |
| TIMP-1 [NM_001044384] | CATGGAAAGCCTCTGTGGAT | CTCAGAGTACGCCAGGGAAC |
| MMP-2 [NM_008610] | ACCTGAAGCTGGAGAACCAA | CACATCCTTCACCTGGTGTG |
| MMP-9 [NM_013599] | CAAATTCTTCTGGCGTGTGA | CGGTTGAAGCAAAGAAGGAG |
| TGF-β [NM_011577] | GTCAGACATTCGGGAAGCAG | GCGTATCAGTGGGGGTCA |
| Procollagen α1(I) [NM_007742] | GAGCGGAGAGTACTGGATCG | GTTCGGGCTGATGTACCAGT |
| TNF [NM_013693] | CTGAACTTCGGGGTGATCGGT | ACGTGGGCTACAGGCTTGTCA |
| Target gene (human) | 5' primer | 3' primer |
| β-actin [NM_001101] | GGACTTCGAGCAAGAGATGG | AGGAAGGAAGGCTGGAAGAG |
| α-SMA [NM_001613] | CCGACCGAATGCAGAAGG | ACAGAGTATTTGCGCTCCGGA |
| TIMP-1 [NM_003254] | AGTGGCACTCATTGCTTGTG | GCAGGATTCAGGCTATCTGG |
| MMP-2 [NM_004530] | ACGACCGCGACAAGAAGTAT | ATTTGTTGCCCAGGAAAGTG |
| MMP-9 [NM_004994] | GACAAGCTCTTCGGCTTCTG | CTCGCTGGTACAGGTCGAGT |
[3H] Thymidine incorporation
Proliferation was estimated as the amount of [3H]-Thymidine incorporated into TCA-precipitable material as previously described (21).
SDS-PAGE and immunoblot analysis
Total cell lysates were incubated, after transferring to nitrocellulose membranes, with rabbit anti-phospho-Akt (1:250), anti-AKT (1:200), anti-MMP-9 at (1:200) (Santa Cruz Biotechnology, Germany), anti-phospho-ERK1/2, anti-ERK, anti-phospho-JAK2, anti-JAK2 (1:2000, Cell Signaling, USA), or mouse anti-α-SMA (1:1000) and anti-β-actin (1:5000), followed by incubation with HRP-conjugated secondary antibody anti-rabbit (1:20,000) or anti-mouse (1:20,000), and developed in ECL Chemiluminescence Substrate (Pierce, Rockford, IL).
Histochemical staining
Liver tissue was fixed in 10% formalin/PBS, dehydrated in alcohols, incubated in xylene, and embedded in paraffin. 7-µm-thick tissue sections were cut and stained with Hematoxylin-Eosin according to manufacturer's protocols.
Gelatin Zymography
Medium from cultured HSC was treated with sample buffer without 2-mercaptoethanol and loaded onto SDS gel containing 0.1% gelatin. After electrophoresis the gel was washed twice with 2.5% Triton X-100 for 15 minutes and incubated overnight in developing buffer (50mmol/L Tris-HCl pH 7.4, 0.2mol/L NaCl, 10mmol/L CaCl2, 0.002% sodium azide) at 37°C. Afterwards, the gel was stained with a solution containing 0.5% Comassie Brilliant Blue, 40% methanol and 7% acetic acid, and destained. Bands were visualized using a Gel-Doc Analyzer (BioRad).
Nuclear Extracts isolation
2×106 HSC or LX2 cells were scraped in Buffer A (10mmol/L Hepes, 10mmol/L KCl, 0.1mmol/L EDTA, 0.1mmol/L EGTA, 1mmol/L DTT, 0.5 mmol/L PMSF), kept on ice for 15 minutes, and lysed by addition 1/20 (vol/vol) 10% Igepal and vortex for 10 seconds. Nuclei were pelleted (12000×g, 30sec), resuspended in Buffer C (20mmol/L Hepes, 0.4mol/L NaCl, 1mmol/L EDTA, 1mmol/L EGTA, 1mmol/L DTT, 1 mmol/L PMSF), and incubated for 15 min on ice with gentle mixing. Afterwards nuclear extract were obtained by centrifuging at 4°C, 12000×g for 5 min.
In vivo liver fibrosis following bile duct ligation
Bile duct ligation (BDL) was performed as previously described (22).
Statistical analysis
Results were routinely expressed as mean ± standard deviation with the number of individual experiments detailed in Figure legends. Statistical significance of the mean values was established by the Student’s t test.
RESULTS
Loss of TNFR1/R2 reduces pro-Collagen-α1(I) mRNA expression
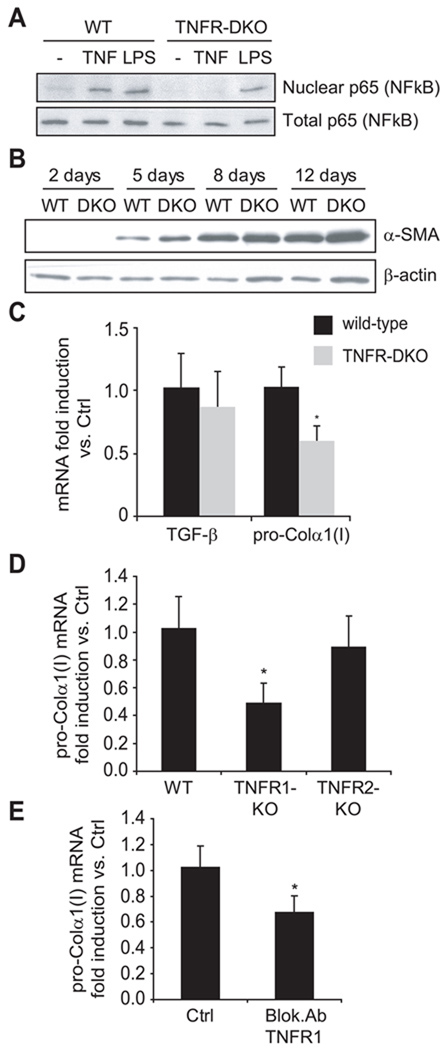
To evaluate the participation of TNF receptors on the activation of HSC, we isolated HSC from wild-type and TNFR-DKO mice and plated them on plastic with medium containing 10% FBS to allow their activation. As expected, while TNFR-DKO HSC did not respond to TNF, as shown by the lack of NF-kappaB activation after TNF challenge, represented as translocation of p65 to the nucleus, they were able to activate NF-kappaB in response to other stimuli such as LPS addition (Figure 1A). The expression of α-SMA was followed at different time-points along the activation of HSC. As shown in Figure 1B, α-SMA protein expression increased during the time of culture and its levels were similar among wild-type and TNFR-DKO HSC. Moreover, paralleling the effects on α-SMA, TGF-β mRNA levels were comparable in wild-type and TNFR-DKO HSC, after 7 days of culture. However, the pro-Collagen-α1(I) mRNA level were significantly decreased in TNFR-DKO HSC during in vitro activation (Figure 1C), and also in TNFR1-KO HSC, but not in TNFR2-KO (Figure 1D). In addition, LX2 cells incubated with neutralizing antibody against TNFR1 receptor displayed a significant decrease in pro-Collagen-α1(I) mRNA expression (Figure 1E), thus indicating that the expression of TNFR1 is necessary in HSC for optimal expression of pro-Collagen-α1(I).
Figure 1. Expression pattern of WT and TNFR-DKO HSC.
(A) p65 subunit of NF-kappaB translocation to nuclei in HSC after TNF (10ng/ml) or LPS (50ng/ml) challenge for 30 minutes. (B) Time-course of α-SMA protein expression by western blot. (C) TGF-β and pro-Collagen-α1(I) mRNA expression. Pro-Collagen-α1(I) mRNA expression in TNFR1-KO and TNFR2-KO HSC (D), or in LX2 cells (E) after incubation with blocking Ab anti-TNFR1 (10µg/mL, 24h). Data are mean±SD; in C, D and E, n≥3 and *p≤0.05 vs. WT HSC.
TNFR1 is required for PDGF-induced AKT phosphorylation and HSC proliferation
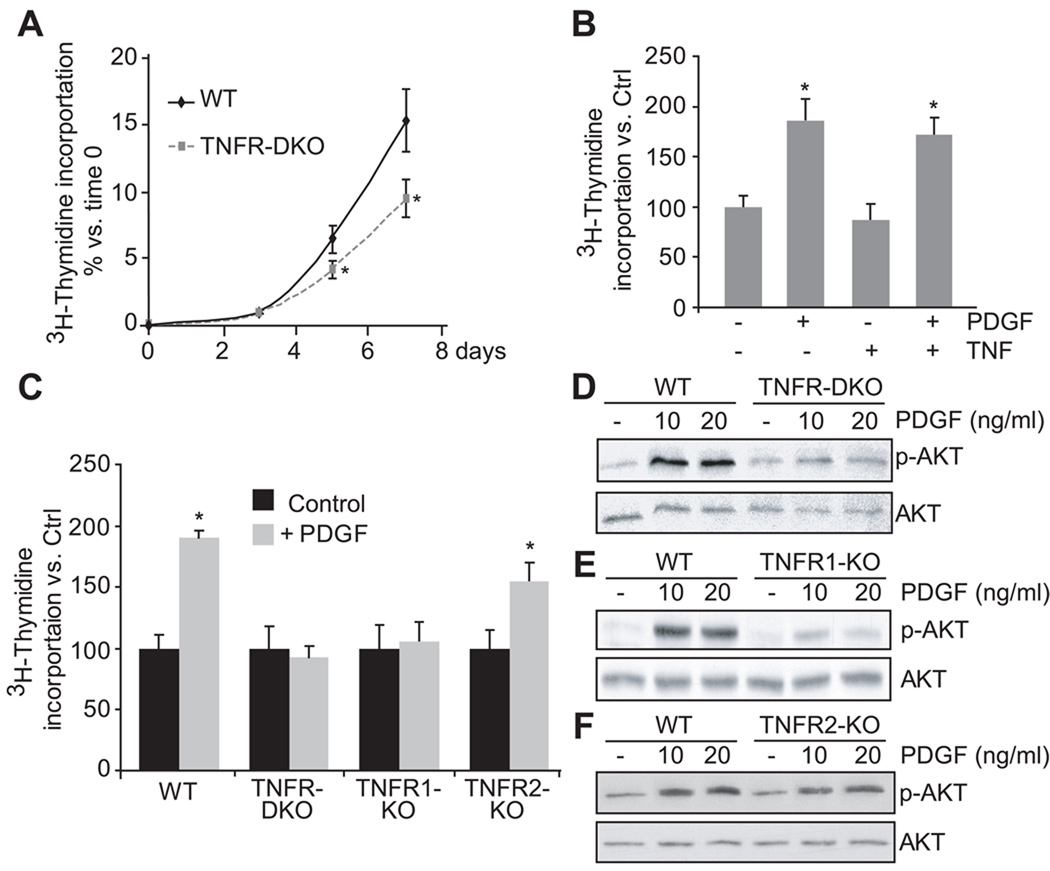
Next we assessed if lack of TNF signaling affected HSC proliferation. As shown in Figure 2A, HSC from TNFR-DKO displayed reduced proliferation rate compared to wild-type HSC during their transdifferentiation into myofibroblasts-like cells. To further evaluate the potential mechanisms involved, we first addressed whether the decreased proliferation of HSC was due to a reduced ability of TNF to stimulate proliferation. As shown in Figure 2B, TNF itself did not stimulate proliferation of HSC. Moreover, since PDGF is a potent mitogenic stimuli for HSC, we next examined if TNF potentiated PDGF signaling and stimulation of cell proliferation. As seen in Figure 2B, while PDGF stimulated wild-type HSC cell proliferation, this effect was not enhanced in the presence of TNF, thus discarding a direct role of TNF in HSC proliferation. Moreover, to examine whether TNF receptors were required for optimal PDGF signaling, we addressed the effect of PDGF in TNFR-DKO HSC. As shown, the proliferating effect of PDGF was markedly reduced in TNFR-DKO HSC (Figure 2C) due to impaired AKT phosphorylation (Figure 2D).
Figure 2. Lack of TNF receptors affects HSC proliferation.
(A) Wild-type and TNFR-DKO HSC proliferation. (B) LX2 proliferation after 24 hours of PDGF (20ng/ml) and/or TNF (50ng/ml) challenge. (C) Wild-type, TNFR-DKO, TNFR1-KO and TNFR2-KO proliferation after 24 hours of PDGF (20ng/ml) challenge. AKT phosphorylation induced by PDGF (10 or 20ng/ml) for 15 minutes in wild-type vs TNFR-DKO (D), TNFR1-KO (E), or TNFR2-KO (F) HSC. Data are mean±SD in A, C n=3 and *p≤0.05 vs. wild-type HSC; in B *p≤0.05 vs. PDGF untreated cells.
Moreover, TNFR1-KO HSC displayed a reduced phosphorylation of AKT in response to PDGF (Figure 2E), however TNFR2-KO HSC (Figure 2F) were able to phosphorylate AKT similarly to wild-type HSC, thus suggesting an intricate interplay between TNFR1 and PDGF signaling. Consistent with these observations, cell proliferation in response to PDGF was impaired in TNFR1-KO HSC but not in TNFR2-KO HSC (Figure 2C).
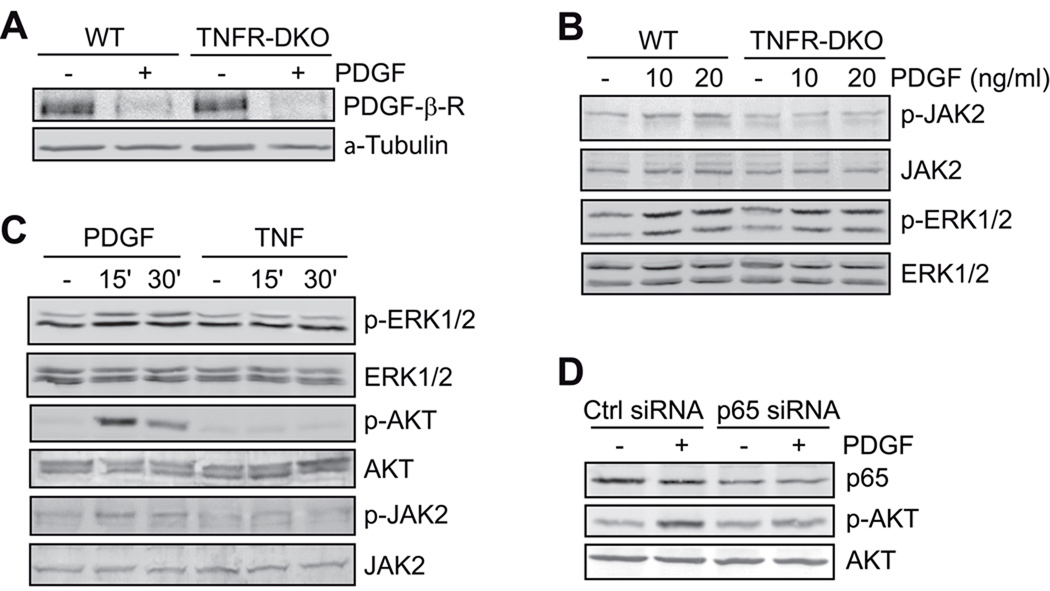
Furthermore, we addressed downstream signaling pathways involved in the proliferation of HSC induced by PDGF. First, we observed that PDGF receptor degradation stimulated by ligand binding was unimpaired in TNFR-DKO HSC (Figure 3A). Moreover, in addition to the requirement for TNFR1 for Akt phosphorylation in response to PDGF (Figure 2E), PDGF also induced the phosphorylation of ERK1/2 and JAK2 in wild type HSC or LX2 cells (Figure 3B, C). However, phosphorylation of JAK2 but not ERK1/2 was impaired in TNFR-DKO HSC (Figure 3B). Unlike ERK1/2, we did not observe p38 phosphorylation by PDGF in mouse HSC or human LX2 cells (not shown). In addition, AKT phosphorylation by PDGF was dependent on NF-kappaB activation, as p65 silencing by siRNA reduced PDGF-dependent AKT activation compared to control-siRNA transfected cells (Figure 3D).
Figure 3. PDGF-β-receptor signaling in TNF-DKO HSC.
(A) PDGF-β-receptor degradation after PDGF challenge (20ng/mL, 2 hours), and (B) JAK2 and ERK1/2 activation after PDGF challenge (15 minutes) in wild-type and TNFR-DKO HSC. (C) ERK1/2, AKT, and JAK2 activation by PDGF (20ng/ml) or TNF (50ng/ml) in LX2 cells. (D) p65 expression and AKT activation by PDGF (15 minutes) in Control and p65 siRNA transfected LX2 cells.
TNFR1 controls MMP-9 expression in HSC and human LX2 cells
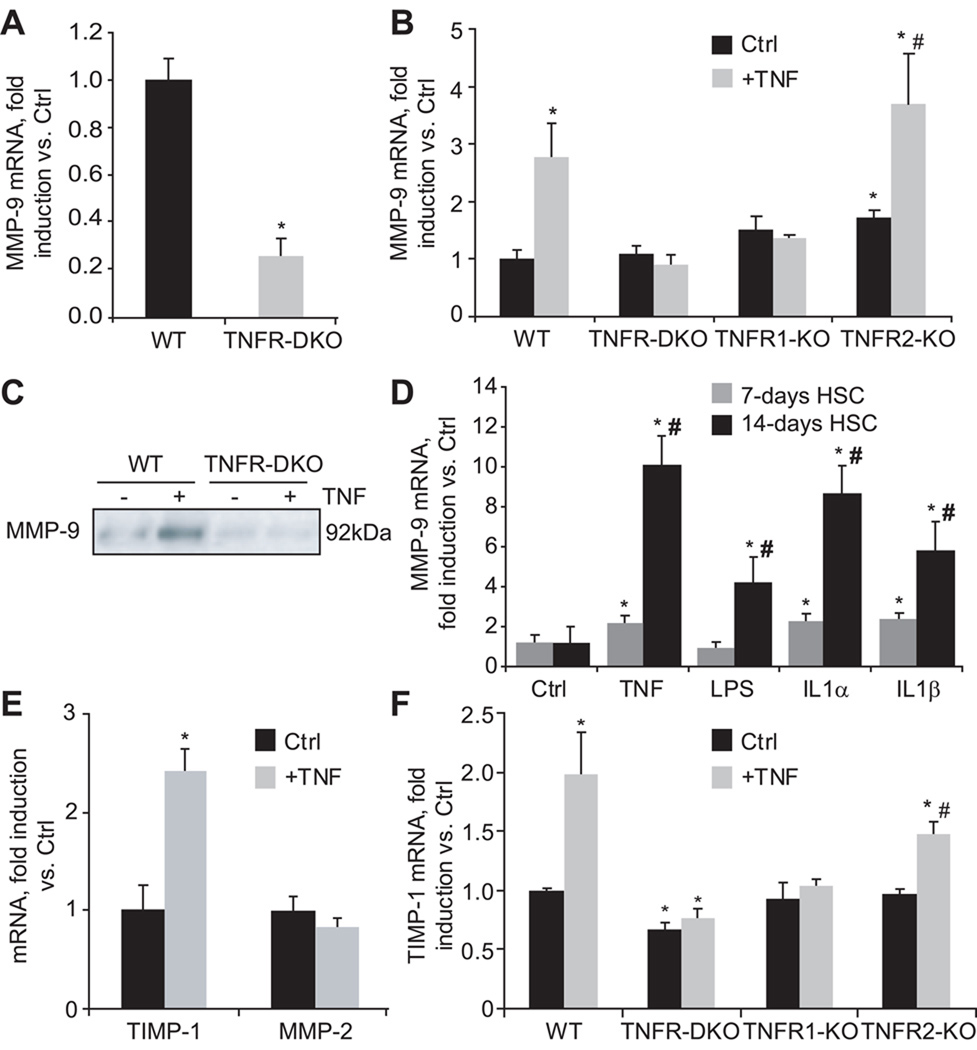
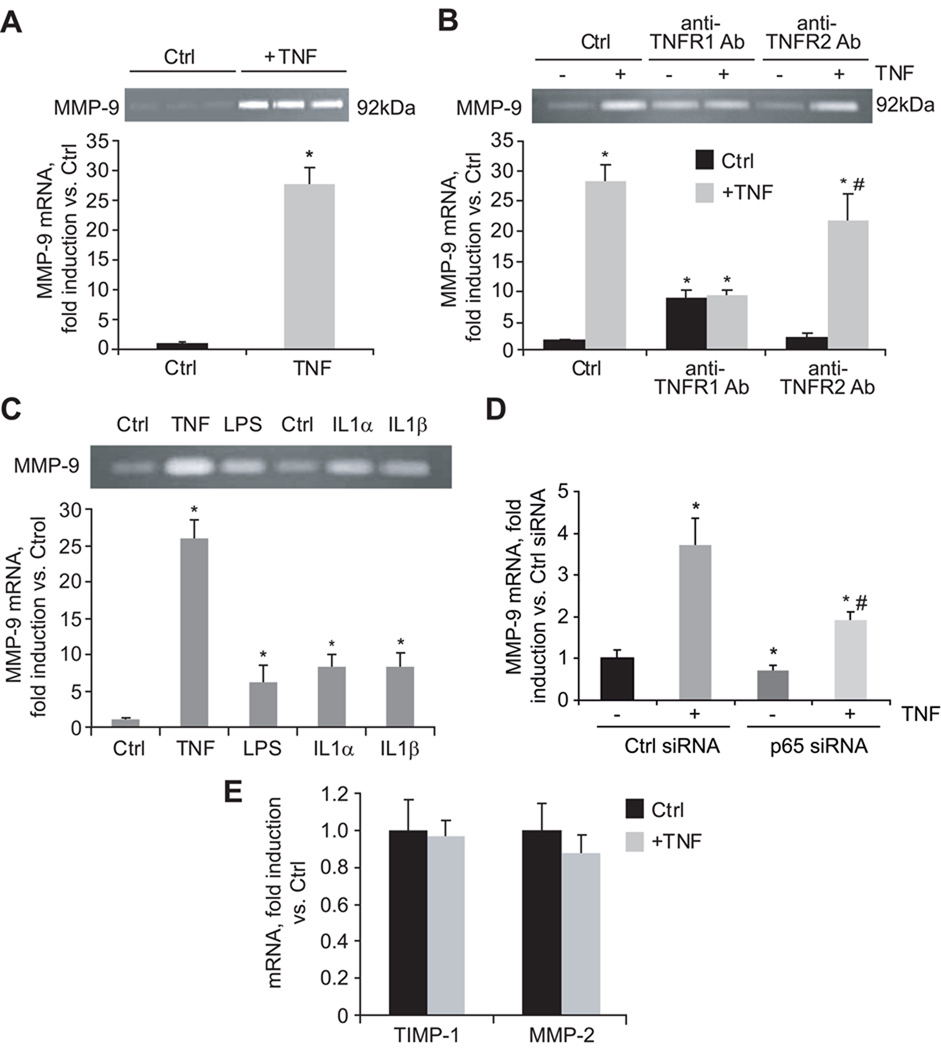
Since matrix remodeling is another critical facet of liver fibrosis and a consequence of HSC activation, we next examined the role of TNF receptors on MMP-9 expression. As shown in Figure 4A, in the presence of 10% FBS, MMP-9 mRNA expression was reduced in TNFR-DKO. To validate the importance of TNF as a putative inducer of MMP-9, HSC from wild-type and TNFR-DKO were depleted of serum up to 0.5% and incubated with TNF. This maneuver resulted in an induction of MMP-9 mRNA (Figure 4B), and protein (Figure 4C) in wild-type but not in TNFR-DKO HSC. The induction of MMP-9 was mediated by TNFR1 since TNFR2-KO HSC were able to activate MMP-9 mRNA (Figure 4B). Of note, under conditions of serum limitation (0.5% FBS) the expression of MMP-9 mRNA in wild-type HSC was similar to that of TNFR-DKO HSC, indicating that the basal induction of MMP-9 is independent of TNF but that its induction under growing conditions required TNF (Supplemental Figure 1). Moreover, the induction of MMP-9 by TNF in mouse HSC was dependent on the time of activation being higher in 14-day compared to 7-day HSC cultures and similar to the levels observed with IL-1α or IL-1β (Figure 4D). The participation of TNFR1 as the receptor responsible for MMP-9 induction was further validated in LX2 cells. As shown in Figure 5A, LX2 responded to TNF by inducing MMP-9 mRNA, and its activity could be clearly detected in the extracellular media by zymography. In addition, by using blocking antibodies against TNFR1 and TNFR2 we could confirm that TNFR1 was the receptor responsible for MMP-9 induction by TNF at the mRNA or activity level (Figure 5B). Intriguingly, MMP-9 expression by TNF in LX2 cells was higher than that caused by LPS or IL-1α/IL-1β, correlating with the nuclear translocation of p65 (not shown). Of note, neither in LX2 cells (Figure 5E) nor in wild-type HSC (Figure 4E) TNF was able to increase the expression of another important matrix collagenase, MMP-2, thus discarding the participation of TNF signaling in MMP-2 regulation. In contrast, while TNF induced TIMP-1 mRNA in wild-type HSC (Figure 4E), which required TNFR1 (Figure 4F), it failed to do so in LX2 cells (Figure 5E). Despite the divergence observed in TIMP-1 regulation, the results obtained in activated human LX2 cells emphasize the specific requirement for TNFR1-dependent signaling in the expression of matrix remodeling factors such as MMP-9 in HSC.
Figure 4. TNF controls MMP-9 expression in HSC.
(A) Basal MMP-9 mRNA expression in wild-type and TNFR-DKO HSC, and (B) after TNF challenge (50ng/ml) for 24 hours in wild-type, TNFR-DKO, TNFR1-KO, and TNFR2-KO HSC. (C) Effect of TNF (50ng/ml, 24 hours) on MMP-9 protein expression. (D) MMP-9 mRNA expression in primary 7 and 14-days old HSC incubated with equivalent concentration (50ng/ml) of TNF, LPS, IL-1α, and IL-1β for 24h. (E) MMP-2 and TIMP-1 mRNA expression in wild-type HSC after TNF challenge (50ng/ml, 24 hours). (F) TIMP-1 mRNA expression after TNF challenge (50ng/ml) for 24 hours in WT, TNFR-DKO, TNFR1-KO, and TNFR2-KO HSC. Data are mean±SD, n=4, *p≤0.05 vs. WT or untreated HSC, in B and F #p≤0.05 vs. TNFR2-KO Ctrl HSC; in D #p≤0.05 vs. 7-days old treated HSC.
Figure 5. TNF controls MMP-9 expression in LX2.
(A) Effect of TNF (50ng/ml, 24 hours) on MMP-9 mRNA expression and activity by zymography. (B) MMP-9 mRNA and activity after 24 hours challenge with TNF (50ng/ml) in LX2 cells in the presence of TNFR1 or TNFR2 blocking antibodies (pre-incubation at 10µg/ml, 30min). (C) MMP-9 mRNA expression and activity in LX2 cells incubated with equivalent concentration (50ng/ml) of TNF, LPS, IL-1α, and IL-1β for 24h. (D) MMP-9 mRNA expression in control and p65 siRNA transfected LX2 cells after TNF challenge (50ng/ml, 24h). (E) TIMP-1 and MMP-2 mRNA expression after TNF challenge (50ng/ml, 24 hours) in LX2 cells. Data are mean±SD, n=3, *p≤0.05 vs. Ctrl LX2 cells; in B, #p≤0.05 vs. anti-TNFR2-treated control LX2 cells; in D, #p≤0.05 vs. p65 siRNA-untreated LX2 cells.
Of instance, while the individual participation of IL-1 (23) or TNF (24, 25) in the induction of MMP-9 has been already described in HSC, their relative contribution to the activation of MMP-9 has not been carefully addressed, nor the comparison of their stimulating effect on MMP-9 expression between primary mouse and human HSC. To this aim, we have challenged primary mouse HSC as well as human LX2 cells in parallel with TNF, IL-1α, IL-1β, or LPS to analyze the extent of MMP-9 activation at the mRNA and protein level. As displayed in Figure 4D, TNF induced MMP-9 in 7-days old primary mouse HSC to a similar extent than IL-1, the extent of MMP-9 expression changed with activation since in 14-days old HSC MMP-9 expression by TNF and IL-1 was greatly enhanced, and at this stage cells were also responsive to LPS. In addition, TNF was a more potent inducer of MMP-9 in the human LX2 cell, as displayed also by the enhanced activity of MMP-9 in extracellular media after TNF challenge (Figure 5C). Thus, in comparison to other known MMP-9 inducers, TNF is a relevant trigger of MMP-9 expression in primary mouse and human HSC, and this induction is mediated by NF-kappaB activation since p65 silencing in LX2 cells reduced the expression of MMP-9 induced by TNF compared to control-siRNA transfected LX2 cells (Figure 5D).
TNF receptors deletion reduces early fibrogenesis in a mouse model of BDL
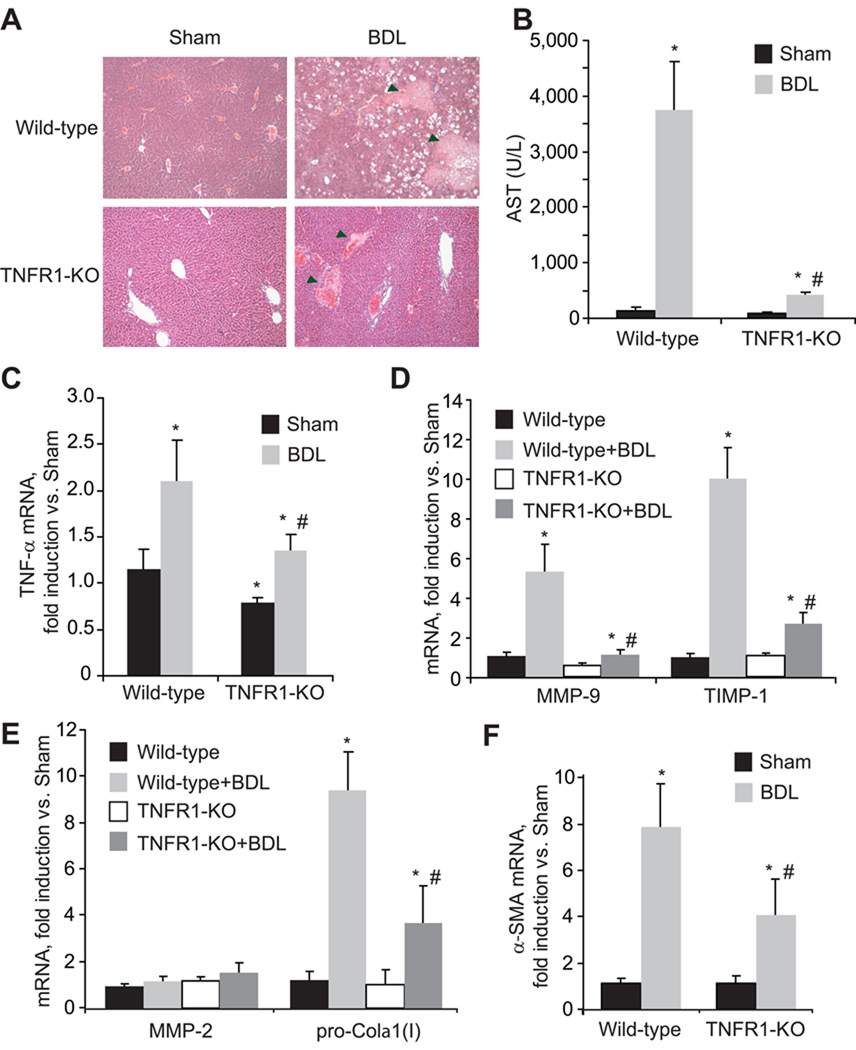
To evaluate the causal relationship between liver damage and fibrogenesis we examined in parallel the injury and fibrosis in mice with impaired TNF signaling in vivo using the bile duct ligation (BDL) model of liver fibrogenesis. TNFR1-KO mice displayed ameliorated tissue damage compared with that of the wild-type controls as indicated by the reduced volume of biliary infarcts in H&E staining and serum transaminase levels (Figure 6A and B), despite of similar bilirubin levels (8.78±1.25mg/dL in wild-type vs. 8.64±0.96mg/dL in TNFR1-KO), indicative of comparable cholestasis in both wild-type and TNFR1-KO. Interestingly, after BDL, TNFR1-KO mice displayed reduced levels of hepatic TNF mRNA (Figure 6C) compared to wild-type animals. This correlated with decreased levels of MMP-9, TIMP-1 mRNA (Figure 6D), and pro-Collagen-α1(I) mRNA (Figure 6E). In contrast, MMP-2 mRNA expression (Figure 6E) was not apparently regulated by TNF. α-SMA was also reduced in TNFR1-KO livers compared to wild-type (Figure 6F), indicating decreased HSC activation in vivo. Similar findings were observed in the TNFR-DKO mice, while TNFR2-KO mice behave similarly to wild-type animals (Supplemental Figure 2). Therefore, in vivo BDL model recapitulates the in vitro effects observed in HSC showing the dependence on TNFR1 signaling to induce changes in ECM remodeling during early fibrogenesis.
Figure 6. Reduced liver damage and fibrosis in TNFR1 KO mice after bile duct ligation.
H&E staining of liver sections (A) and serum transaminase levels (B) in wild-type and TNFR1-KO mice after BDL. TNF-α (C), MMP-9 and TIMP-1 (D), MMP-2 and pro-Colα1(I) (E), and α-SMA (F) mRNA expression in wild-type and TNFR1-KO livers after BDL. In A, Arrowheads signal bile infarcts. Data are mean±SEM, n=4–5 animals/group, *p≤0.05 vs. wild-type Sham, #p≤0.05 vs. wild-type BDL.
DISCUSSION
TNF has been implicated in the development of many chronic liver diseases and hepatic fibrosis is a hallmark of disease progression. Unlike its involvement in hepatocellular apoptosis and liver diseases, the role of TNF in liver fibrosis remains unclear, particularly whether TNF and its binding to specific TNFR1 or TNFR2 regulates HSC biology. Using genetic and pharmacological approaches, we show a profibrogenic role for TNF, specifically via binding to receptor R1. In particular, we have used primary mouse HSC from wild-type and mice deficient in TNF-receptors to analyze the role of TNF and its receptors in HSC activation and proliferation, which are critical steps in the cascade of events leading to fibrosis. Moreover, the findings were extended to human HSC in which TNF receptors were individually antagonized by specific neutralizing antibodies.
Our results indicate that although TNF does not directly participate in some fundamental traits of HSC transdifferentiation into a myofibroblast phenotype, such as increase in α-SMA or TGF-β expression; TNF, through TNFR1, has an important role in other important features such as proliferation, MMP-9 and TIMP-1 expression. A significant difference between primary mouse and human HSC was found in the participation of TNF in TIMP-1 induction. While primary mouse HSC augment TIMP-1 expression in response to TNF, we failed to observe any increase of TIMP-1 in LX2 cells under the same experimental conditions. Several conceivable possibilities could explain this differential behavior including that TIMP-1 regulation may fundamentally differ between mouse and human in HSC. Another explanation could be the fact that LX2 cells display almost negligible expression of TIMP-1, as compared to primary HSC or to its parental cell line LX1, implying that TIMP-1 expression may have been lost during its selection under low serum condition (2% FBS) (26).
A striking finding was the decrease in proliferation observed in TNFR-DKO HSC compared to wild-type HSC. Mechanistically, the decreased proliferation was mediated by a defective PI3K/AKT pathway in TNFR-DKO HSC that was reproduced in TNFR1-KO HSC but not in TNFR2-KO HSC. Indeed, both TNFR1-KO and TNFR-DKO HSC display reduced AKT phosphorylation and proliferation in response to PDGF, a potent mitogen of HSC, despite correct ligand binding and subsequent receptor degradation. These observations suggest that proteins or mediators necessary for PDGF signaling located upstream of AKT rely on NF-kappaB-dependent TNFR1 signaling indicating a cross-talk between PDGF and TNFR1 receptors. In line with our observations, previous findings in vascular smooth muscle cells indicated a similar overlapping between TNF and PDGF necessary for cell migration and proliferation (27). However, the identification of the NF-kappaB-dependent targets responsible for the reduced proliferation in response to PDGF in TNFR-DKO and TNFR1 KO HSC remains unknown and requires further work.
In contrast to TGF-β expression, we observed a decreased basal level of pro-Collagen-α1(I) in activated HSC from TNFR-DKO, and TNFR1-KO mice compared to wild-type mice, findings that were reproduced in LX2 cells using anti-TNFR1 blocking antibody. Consistent with previous studies (17, 18), TNF addition to HSC cultures did not induce pro-Collagen-α1(I) mRNA (data not shown), thus discarding a direct effect of TNF on pro-Collagen-α1(I) regulation. However, the known ability of MMP-9 to activate latent TGF-β to its active form (28), which plays an essential role in earlier stages of liver fibrogenesis when collagen production of HSC is stimulated by TGF-β (29), could explain why TNFR-DKO and TNFR1-KO HSC show decreased basal levels of pro-Collagen-α1(I) mRNA expression. In agreement, with impaired MMP-9 expression in TNFR-DKO HSC, TGF-β would be normally produced but not activated by MMP-9, thus resulting in a deficient pro-Collagen-α1(I) induction.
Unlike pro-Collagen-α1(I), interestingly, we observed a differential role of TNF receptors in the regulation of MMPs in HSC, in particular the requirement of TNFR1 in the expression of MMP-9 but not MMP-2. In relation to MMP-9, it has been described in the thioacetamide model of liver injury and fibrosis (30) that MMP-9 colocalizes predominantly to desmin-positive cells, suggesting that HSC are the source of MMP-9 cells in vivo. The importance of MMP-9 is highlighted by the observation that MMP-9-deficient mice are partially protected from liver injury and HSC activation (30). In contrast to MMP-9, although associative studies and cell culture findings suggest that MMP-2, a type IV collagenase upregulated in chronic liver diseases and considered a profibrogenic mediator, promotes hepatic fibrogenesis, no in vivo model has definitively established a pathologic role for MMP-2 in the development and progression of liver fibrosis. In fact, recent findings using MMP-2 deficient mice suggest a protective rather than pathogenic role for MMP-2 (31).
Since the above findings indicated a selective requirement for TNFR1 in specific steps of HSC activation and proliferation, we next addressed the in vivo relevance for liver fibrogenesis. The data using the BDL model of liver fibrosis, although limited in interpretation since the TNFR1-KO and TNFR-DKO and mice display both reduced liver damage and decreased matrix deposition, suggest a correlation between TNF and MMP-9, TIMP-1, and pro-Collagen-α1(I) mRNA expression. In contrast to the BDL model shown here, previous reports using the chronic administration of CCl4 reported a controversial role of TNFR1 in liver fibrosis. For instance, the lack of TNFR1 inhibited pro-Collagen-α1(I) expression and liver fibrosis following CCl4 treatment without effect on liver injury (11, 12). However, interestingly, de Meijer et al. (13) recently reported decreased liver injury and inflammation but increased collagen deposition in the CCl4 model by blocking TNF production through inhibition of its processing via TNF-alpha converting enzyme, as well as in TNFR-DKO mice.
Taken together our observations in in vitro HSC culture and in vivo point to TNF not only as an inducer of hepatocellular damage, but also as a pro-fibrogenic factor in the liver, and hence targeting TNF or its receptor TNFR1 could be of benefit to preserve hepatocellular integrity and prevent HSC proliferation and liver fibrosis.
Supplementary Material
ACKNOWLEDGEMENTS
The technical assistance of Susana Núñez is greatly appreciated. We want to thank Dr. Horst Bluethmann (Hoffmann-La Roche Ltd. Switzerland) for providing the knockout mice involved in this study. The work was carried out in part at the Esther Koplowitz Center, Barcelona.
Financial Support: The work was supported by grants PI09/00056, PI10/02114 (Instituto de Salud Carlos III), 2008-02199 and 2009-11417 (Plan Nacional de I+D), Spain; and P50-AA-11999 (Research Center for Liver and Pancreatic Diseases, NIAAA/NIH).
List of Abbreviations
- HSC
hepatic stellate cells
- TNFR
TNF-receptor
- TNFR-DKO
TNFR1/R2 double knockout
- α-SMA
alpha-smooth muscle actin
- ECM
extracellular matrix
- TIMP-1
tissue inhibitor of metalloproteinase-1
- MMP
matrix metalloproteinase
REFERENCES
- 1.Grell M, Douni E, Wajant H, Löhden M, Clauss M, Maxeiner B, et al. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 2.Grell M, Wajant H, Zimmermann G, Scheurich P. The type 1 receptor (CD120a) is the high-affinity receptor for soluble tumor necrosis factor. Proc Natl Acad Sci U S A. 1998;95:570–575. doi: 10.1073/pnas.95.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada Y, Fausto N. Deficient liver regeneration after carbon tetrachloride injury in mice lacking type 1 but not type 2 tumor necrosis factor receptor. Am J Pathol. 1998;152 1577e89. [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada Y, Webber EM, Kirillova I, Peschon JJ, Fausto N. Analysis of liver regeneration in mice lacking type 1 or type tumor necrosis factor receptor: requirement for type 1 but not type 2 receptor. Hepatology. 1998;28 doi: 10.1002/hep.510280410. 959e70. [DOI] [PubMed] [Google Scholar]
- 5.Bradham CA, Plümpe J, Manns MP, Brenner DA, Trautwein C. Mechanisms of hepatic toxicity. I. TNF-induced liver injury. Am J Physiol. 1998;275:G387–G392. doi: 10.1152/ajpgi.1998.275.3.G387. [DOI] [PubMed] [Google Scholar]
- 6.Schwabe RF, Brenner DA. Mechanisms of liver injury. I. TNF-alpha-induced liver injury: role of IKK, JNK, and ROS pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G583–G589. doi: 10.1152/ajpgi.00422.2005. [DOI] [PubMed] [Google Scholar]
- 7.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wullaert A, Heyninck K, Beyaert R. Mechanisms of crosstalk between TNF-induced NF-kappaB and JNK activation in hepatocytes. Biochem Pharmacol. 2006;72:1090–1101. doi: 10.1016/j.bcp.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Han D, Ybanez MD, Ahmadi S, Yeh K, Kaplowitz N. Redox regulation of tumor necrosis factor signaling. Antioxid Redox Signal. 2009;11:2245–2263. doi: 10.1089/ars.2009.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudo K, Yamada Y, Moriwaki H, Saito K, Seishima M. Lack of tumor necrosis factor receptor type 1 inhibits liver fibrosis induced by carbon tetrachloride in mice. Cytokine. 2005;29:236–244. doi: 10.1016/j.cyto.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Simeonova PP, Gallucci RM, Hulderman T, Wilson R, Kommineni C, Rao M, et al. The role of tumor necrosis factor-alpha in liver toxicity, inflammation, and fibrosis induced by carbon tetrachloride. Toxicol Appl Pharmacol. 2001;177:112–120. doi: 10.1006/taap.2001.9304. [DOI] [PubMed] [Google Scholar]
- 13.de Meijer VE, Sverdlov DY, Popov Y, Le HD, Meisel JA, Nosé V, et al. Broad-spectrum matrix metalloproteinase inhibition curbs inflammation and liver injury but aggravates experimental liver fibrosis in mice. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011256. e11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernández I, de la Torre P, Rey-Campos J, Garcia I, Sánchez JA, Muñoz R, et al. Collagen alpha1(I) gene contains an element responsive to tumor necrosis factor-alpha located in the 5' untranslated region of its first exon. DNA Cell Biol. 2000;19:341–352. doi: 10.1089/10445490050043317. [DOI] [PubMed] [Google Scholar]
- 15.Hernández-Muñoz I, de la Torre P, Sánchez-Alcázar JA, García I, Santiago E, Muñoz-Yagüe MT, et al. Tumor necrosis factor alpha inhibits collagen alpha 1(I) gene expression in rat hepatic stellate cells through a G protein. Gastroenterology. 1997;113:625–640. doi: 10.1053/gast.1997.v113.pm9247485. [DOI] [PubMed] [Google Scholar]
- 16.Houglum K, Buck M, Kim DJ, Chojkier M. TNF-alpha inhibits liver collagen-alpha 1(I) gene expression through a tissue-specific regulatory region. Am J Physiol. 1998;274:G840–G847. doi: 10.1152/ajpgi.1998.274.5.G840. [DOI] [PubMed] [Google Scholar]
- 17.Solis-Herruzo JA, Brenner DA, Chojkier M. Tumor necrosis factor alpha inhibits collagen gene transcription and collagen synthesis in cultured human fibroblasts. J Biol Chem. 1988;263:5841–5845. [PubMed] [Google Scholar]
- 18.Varela-Rey M, Fontán-Gabás L, Blanco P, López-Zabalza MJ, Iraburu MJ. Glutathione depletion is involved in the inhibition of procollagen alpha 1(I) mRNA levels caused by TNF-alpha on hepatic stellate cells. Cytokine. 2007;37:212–217. doi: 10.1016/j.cyto.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 19.de Leeuw AM, McCarthy SP, Geerts A, Knook DL. Purified rat liver fat-storing cells in culture divide and contain collagen. Hepatology. 1984;4:392–403. doi: 10.1002/hep.1840040307. [DOI] [PubMed] [Google Scholar]
- 20.Friedman SL, Rockey DC, McGuire RF, Maher JJ, Boyles JK, Yamasaki G. Isolated hepatic lipocytes and Kupffer cells from normal human liver: morphological and functional characteristics in primary culture. Hepatology. 1992;15:234–243. doi: 10.1002/hep.1840150211. [DOI] [PubMed] [Google Scholar]
- 21.Moles A, Tarrats N, Fernández-Checa JC, Marí M. Cathepsins B and D drive hepatic stellate cell proliferation and promote their fibrogenic potential. Hepatology. 2009;49:1297–1307. doi: 10.1002/hep.22753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moles A, Tarrats N, Morales A, Domínguez M, Bataller R, Caballería J, et al. Acidic Sphingomyelinase Controls Hepatic Stellate Cell Activation and in Vivo Liver Fibrogenesis. Am J Pathol. 2010;177:1214–1224. doi: 10.2353/ajpath.2010.091257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han YP, Zhou L, Wang J, Xiong S, Garner WL, French SW, et al. Essential role of matrix metalloproteinases in interleukin-1-induced myofibroblastic activation of hepatic stellate cell in collagen. J Biol Chem. 2004;279:4820–4828. doi: 10.1074/jbc.M310999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migita K, Maeda Y, Abiru S, Nakamura M, Komori A, Yokoyama T, et al. Immunosuppressant FK506 inhibits matrix metalloproteinase-9 induction in TNF-alpha-stimulated human hepatic stellate cells. Life Sci. 2006;78:2510–2515. doi: 10.1016/j.lfs.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Zhou L, Yan C, Gieling RG, Kida Y, Garner W, Li W, et al. Tumor necrosis factor-alpha induced expression of matrix metalloproteinase-9 through p21-activated kinase-1. BMC Immunol. 2009:10–15. doi: 10.1186/1471-2172-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peppel K, Zhang L, Orman ES, Hagen PO, Amalfitano A, Brian L, et al. Activation of vascular smooth muscle cells by TNF and PDGF: overlapping and complementary signal transduction mechanisms. Cardiovasc Res. 2005;65:674–682. doi: 10.1016/j.cardiores.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 28.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–176. [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Q, Mak KM, Lieber CS. Dilinoleoylphosphatidylcholine decreases acetaldehyde-induced TNF-alpha generation in Kupffer cells of ethanol-fed rats. Biochem Biophys Res Commun. 2002;299:459–464. doi: 10.1016/s0006-291x(02)02672-4. [DOI] [PubMed] [Google Scholar]
- 30.Gieling RG, Wallace K, Han YP. Interleukin-1 participates in the progression from liver injury to fibrosis. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1324–G1331. doi: 10.1152/ajpgi.90564.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radbill BD, Gupta R, Ramirez MC, Difeo A, Martignetti JA, Alvarez CE, et al. Loss of Matrix Metalloproteinase-2 Amplifies Murine Toxin-Induced Liver Fibrosis by Upregulating Collagen I Expression. Dig Dis Sci. 2011;56:406–416. doi: 10.1007/s10620-010-1296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.