Summary
Transcription of immediate early genes (IEGs) in neurons is exquisitely sensitive to neuronal activity, but the mechanism underlying these early transcription events is largely unknown. We demonstrate that several IEGs such as arc/arg3.1 are poised for near-instantaneous transcription by the stalling of RNA Polymerase II (Pol II) just downstream of the transcription start site in rat neurons. RNAi-depletion of Negative Elongation Factor, a mediator of Pol II stalling, reduces the Pol II occupancy of the arc promoter and compromises the rapid induction of arc and other IEGs. In contrast, reduction of Pol II stalling did not prevent transcription of IEGs that are expressed later and largely lack promoter proximal Pol II stalling. Together, our data strongly indicate that rapid induction of neuronal IEGs requires poised Pol II and suggest a role for this mechanism in a wide variety of transcription-dependent processes, including learning and memory.
Introduction
Several previous studies have shown that de novo transcription is critical for the consolidation of synaptic plasticity. Isolated dendrites that lack the soma, and hence the nucleus, fail to sustain induced long-term potentiation (LTP)1, and application of transcription blockers attenuates the later phases of LTP and long-term depression (LTD)2, 3. Interestingly, transcription inhibitors block late-LTP only if they are applied prior to or within a few minutes of stimulation, representing a critical temporal window during which they are effective3–5. Together, these observations emphasize the importance of rapid transcription in stabilizing long-term synaptic modifications.
Rapid transcription of many immediate early genes (IEGs) is triggered by neuronal activity. Products of several such IEGs can be detected within a few minutes of stimulation and are referred to as rapid IEGs in this manuscript. One of these genes is the activity-regulated cytoskeleton associated protein (arc; also called activity-regulated gene 3.1, arg3.1)6, 7. Evidence of rapid arc transcription is well documented: arc pre-mRNA (unspliced nascent transcript or hnRNA) is detectable at genomic alleles within two to five minutes of an animal’s exploration of a novel environment and newly synthesized mature arc mRNA is detected in the perikaryotic cytoplasm in as early as fifteen minutes8, 9. Similarly, elevated levels of arc mRNA have been documented in the hippocampus within minutes of high frequency stimulation or spatial learning and in the olfactory cortex after odorant exposure10–12. How is arc transcription orchestrated so rapidly? To induce genes within such a short timeframe, one model has posited that action potentials relay signals quickly through their ability to induce large influxes of calcium into the cell that can then activate gene transcription13–16. This model is in contrast to the idea that plasticity triggers synaptic protein translocations, which may not be able to reach the nucleus fast enough to meet the temporal demands of arc and other IEG transcription15, 17.
Classically, transcription is considered to be rate-limited by recruitment of RNA Polymerase II (Pol II) to form a pre-initiation complex with general transcription factors18. However, a non-canonical phenomenon has been described at a growing number of Drosophila and mammalian genes where Pol II is recruited to un-induced promoters to initiate RNA synthesis, but then ‘stalls’ after transcribing twenty to fifty nucleotides19, 20. At these genes, efficient release of Pol II into productive elongation requires a specific stimulus such as heat shock19. Referred to as ‘promoter proximal Pol II stalling’, this phenomenon has been proposed to poise genes for rapid and synchronous induction21–24. In the current study, we have tested the hypothesis that Pol II stalling is critical for near-instantaneous induction of arc and other rapid IEGs in response to neuronal activity in mammalian neurons. We propose that such mechanism would be critical for the precise timing of IEG responses with their respective nuclear and synaptic functions.
Results
TTX withdrawal induces rapid arc transcription
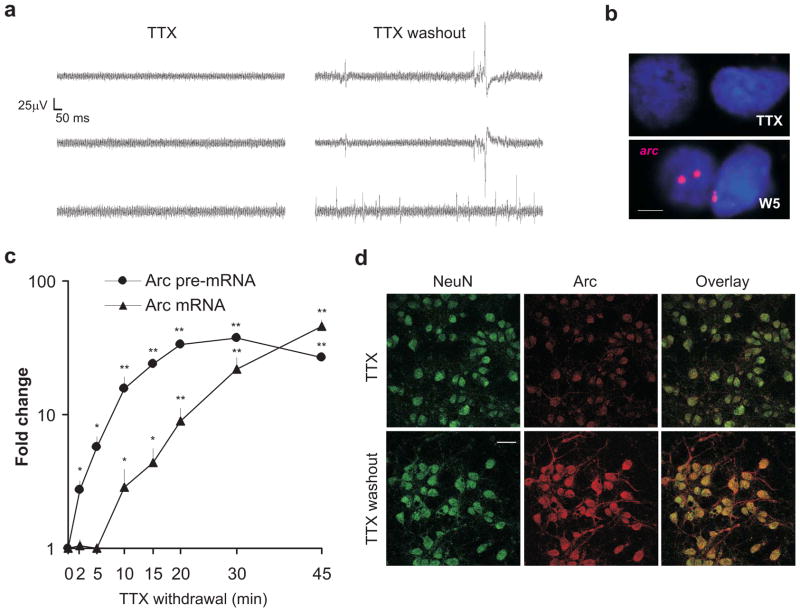
To study the mechanism underlying rapid transcription of IEGs, we chose the widely-studied arc as our model gene and used the ‘tetrodotoxin (TTX) withdrawal’ method to induce neuronal activity in neuron cultures25. This protocol utilizes prolonged treatment of neurons with TTX, a sodium channel blocker, followed by its quick washout to trigger quasi-synchronous neuronal activity (Fig. 1a). To verify rapid arc induction, we used a probe directed against the arc pre-mRNA and performed fluorescent in situ hybridization (FISH)8. Five minutes after TTX washout using TTX-free media, arc pre-mRNA was detected in discrete intranuclear foci (Fig. 1b). To quantify the level of arc pre-mRNA, we designed primers against a region spanning the first intron and second exon. Nascent arc pre-mRNA levels, as detected with these primers in one-step intron-based RT-PCR assays, increased significantly within two minutes of TTX withdrawal and continued to increase with time (Fig. 1c). No such increase was noted when neurons were mock-washed or when TTX-treated neurons were washed with TTX-containing media (n = 2 each; data not shown). We next tested for changes in mature arc mRNA levels and found that the levels increased significantly within ten minutes of TTX withdrawal and continued to increase steadily thereafter (Fig. 1c). A significant increase in arc mRNA was not detected prior to ten minutes after TTX withdrawal, which likely reflects the time required for mRNA maturation. To confirm the viability and functionality of the newly transcribed mRNA, neurons were immuno-stained for Arc protein one hour after TTX withdrawal. We found a greater preponderance of immuno-reactive Arc in the soma and dendrites of activated neurons compared to neurons left in TTX (Fig. 1d).
Figure 1. TTX withdrawal effectively induces rapid arc transcription.
(a) Recordings were performed from neurons on multi-electrode arrays in the presence of TTX (48 hours) or 1–2 minutes after TTX washout. Representative traces from different electrodes are shown for these two conditions. Note the presence of action potentials after TTX washout. (b) Representative confocal images of neurons treated with TTX for 48 hours (TTX), washed with media, and fixed after 5 minutes (W5). These were then subjected to FISH with fluorescein-labeled anti-arc riboprobes, which were detected with a Cy3-based fluorescent substrate. 48 nuclei out of 101 nuclei (W5) and 5 out of 159 nuclei (TTX) from 3 independent biological trials were positive for the arc foci. Scale bar represents 5μm. (c) Graphical representation of the level of arc pre-mRNA and mRNA at different time points after TTX washout as detected by qPCR and normalized to GAPDH. n = 3 biological trials; error bars represent S.E. of the mean. *P < 0.05 and **P < 0.01. (d) Representative fluorescent images of dissociated neurons immuno-stained with neuronal marker anti-NeuN (detected with anti-Mouse Alexa488) and anti-Arc antibody (detected with anti-rabbit Alexa633). Neurons were treated with TTX for 48 hours (TTX) and then washed with media and fixed after one hour (TTX washout). n = 3 biological trials. Average Alexa633 fluorophore intensities (representing Arc) were estimated with ImageJ software. TTX (n = 38 neurons): 42.91 ± 7.27; TTX washout (n = 46 neurons): 104.53 ± 15.98. Scale bar represents 20μm.
Taken together, these data show that TTX withdrawal induces rapid de novo transcription of arc in neurons within minutes, and this induction closely recapitulates the temporal features of rapid arc induction in vivo8, 10.
Pol II is poised in proximity of the arc transcription start site
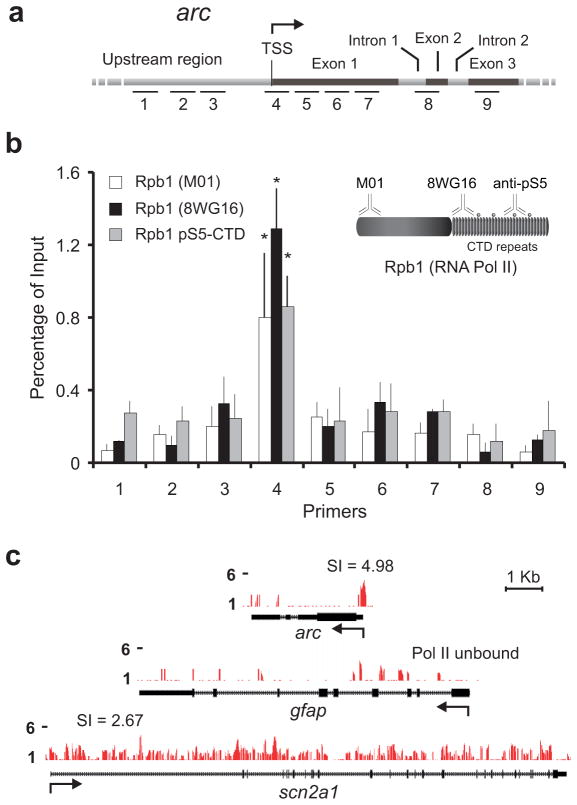
The C-terminal domain (CTD) of Rpb1, the largest subunit of eukaryotic RNA Pol II, consists of a tandemly repeated heptapeptide sequence (Y1S2P3T4S5P6S7) containing up to five potential post-translational phosphorylation sites26. To detect stalled and poised Pol II in the vicinity of the arc promoter, we conducted Chromatin Immunoprecipitation (ChIP) assays on unstimulated neurons using three different antibodies against Rpb1 (see Fig. 2b inset): first, a monoclonal antibody against the N-terminal region of Rpb1 that can detect Pol II irrespective of the phosphorylation state of the Pol II CTD (M01); second, a monoclonal antibody that detects Rpb1 with an unphosphorylated or hypophosphorylated CTD (8WG16); and third, a polyclonal antibody that detects Rpb1 with phosphorylation at the Serine 5 residue of the CTD [Rpb1 (pSer5-CTD)]. Ser5 is phosphorylated during transcription initiation and is a hallmark of polymerase stalling23. DNA obtained from immunoprecipitation with these antibodies was subjected to real-time PCR amplification using primers targeting nine different regions of the arc promoter and gene body (Fig. 2a and Supplementary Information). Primer pair 4, centered at +24 with respect to the arc transcription start site (TSS), revealed a significant enrichment of Pol II detected with all three antibodies (Fig. 2b). This enrichment was pronounced in comparison to either the upstream promoter region (primer pairs 1–3) or the gene body (primer pairs 5–9), suggesting that Pol II recruited to the arc TSS stalls in proximity of the promoter rather than being released into the gene. As a control, we also probed the TSS of glial fibrillary acidic protein (gfap), an astroglia-specific gene that remains silent in neurons. Pol II enrichment was not detected near this gene by any of our antibodies (data not shown).
Figure 2. RNA Pol II is enriched at the arc TSS.
(a) Graphical map (not to scale) to show relative position of primers used to quantify immunoprecipitated chromatin. Primer information is provided in Supplementary Materials. (b) Quantification of RNA Pol II binding to promoter regions and gene bodies of arc as determined by ChIP with three antibodies against RNA Pol II: anti-Rpb1 (M01 and 8WG16) and anti-Rpb1 pSer5 CTD. n = 3 biological trials; error bars represent S.D. of the mean. *P < 0.01. Note the enrichment near the arc TSS. Inset: Pictorial representation of Pol II antibodies and epitopes used in this study. (c) ChIP-seq signals, scaled equally on the y-axis, are displayed for arc, gfap, and scn2a1. Arrow indicates TSS for each gene and the accompanying number in arc and scn2a1 refers to the Pol II stalling indices (SI) for each gene (refer to the text). Pol II is unbound in gfap.
To substantiate Pol II enrichment further, high-throughput sequencing (ChIP-seq) was performed with M01-immunoprecipitated material to determine the genome-wide distribution of Pol II in rat neurons. The M01 antibody was used to avoid any bias for particular Rpb1-CTD phosphorylation status. Consistent with our findings using ChIP assays, this technique also revealed a discernable enrichment of Pol II near the arc TSS (Fig. 2c). No such enrichment was found near the TSS of the non-neuronal gene gfap or the activity-insensitive, constitutively expressed sodium channel gene scn2a1. Several additional observations from our ChIP-seq data are presented later in the manuscript.
Pol II is stalled irrespective of spontaneous neuronal activity
We next wondered whether spontaneous network activity was responsible for Pol II enrichment. The basal level of arc expression in cultured neurons was reduced significantly after neuronal activity was blocked with TTX, APV (an NMDA receptor antagonist), or both (Supplementary Fig. 1a). However, even when neuronal activity is blocked and arc mRNA levels are low, TSS-specific enrichment of Pol II was retained (Supplementary Fig. 1b). Enrichment in these cases was not significantly different from the enrichment in untreated neurons, demonstrating that stalling is maintained at the arc promoter irrespective of the spontaneous level of neuronal activity.
To further corroborate this finding, we determined whether any epigenetic signatures on the arc promoter were altered by spontaneous neuronal activity. Tri-methylation of histone H3 at lysine 4 (H3K4.3me) and acetylation of H3 at lysine 9 (H3K9.ac), two epigenetic signatures associated with stalled polymerase27, were detected around the arc TSS in untreated cells and, importantly, after prolonged TTX treatment (Supplementary Fig. 1c, d). ChIP with anti-H3 antibody revealed no significant alteration in the nucleosome population in TTX-treated cells with respect to untreated control cells (Supplementary Fig. 1e). These data indicate that transcription-related histone marks are maintained at the arc promoter even during low levels of network activity.
Activity mobilizes stalled Pol II into productive elongation
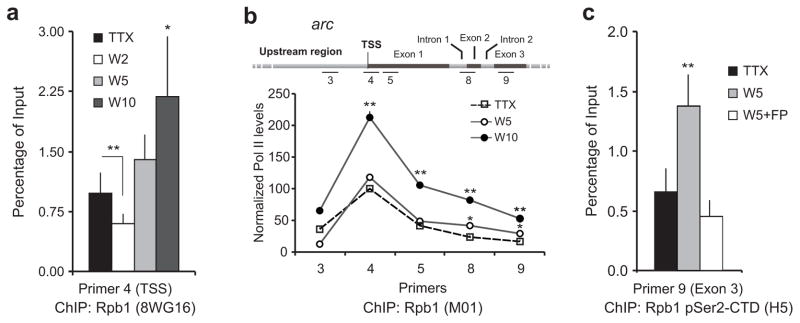
To study the immediate effects of activity on the stalled Pol II, we induced TTX-treated neurons to fire for various times and reassessed Pol II enrichment at the arc TSS. A significant drop in promoter-proximal Pol II was recorded after two minutes of activity, which probably indicates the release of stalled Pol II into the gene body (Fig. 3a). Pol II levels increased shortly thereafter, likely reflecting recruitment of additional Pol II to support multiple rounds of RNA synthesis (Fig. 3a).
Figure 3. Activity promotes Pol II escape from stalling into productive elongation.
(a) Quantification of RNA Pol II in the arc TSS as determined by ChIP with 8WG16 antibody at indicated time points after TTX withdrawal. n = 3 biological trials; error bars represent S.D. of the mean. *P < 0.05 and **P < 0.01. (b) Quantification of RNA Pol II in the arc gene body as determined by ChIP with M01 antibody at indicated time points after TTX withdrawal. Data presented as the percentage Pol II level at the arc TSS using primer pair 4 in the presence of TTX (100%). n = 3 biological trials; error bars represent S.D. of the mean. *P < 0.05 and **P < 0.01. (c) TTX washout induces enhanced presence of pSer2 Pol II (elongation competent) in arc exon 3 as estimated by ChIP with anti-pSer2 antibody five minutes after TTX withdrawal. This enhancement is blocked by FP, an inhibitor of the kinase responsible for this modification. Exon 3-specific primers (primer pair 9; see map in b) were used because RNA Pol II is predominantly phosphorylated at this epitope as it approaches termination near the 3′-end of genes43. (n = 3 biological trials; error bars represent S.D. of the mean. **P < 0.01).
To verify the escape of polymerase into the productive elongation phase of arc, we performed Pol II ChIP using primers that targeted each of the three arc exons. As expected, Pol II levels significantly increased in the distal gene body by five minutes after TTX washout (Fig. 3b). Within ten minutes of TTX washout, a significant increase was observed in all three exons (Fig. 3b). To further verify that Pol II within the gene body was fully elongation-competent, we examined the phosphorylation status of the Ser2 residue of the Pol II CTD, as phosphorylation of Ser2 facilitates the transition of Pol II to productive elongation28. Using a monoclonal antibody against Rpb1 pSer2-CTD (H5), we found that TTX washout induced an increase of transcription-competent Pol II in the arc exon 3 (Fig. 3c). This increase was sensitive to flavopiridol (FP), a potent inhibitor of the kinase responsible for the phosphorylation of Serine2 residue, the Positive Transcription Elongation Factor b (P-TEFb)29 (Fig. 3c). Consistent with known mechanisms of transcription, FP-mediated inhibition of P-TEFb led to a dramatic loss of rapid arc induction (Supplementary Fig. 2). Taken together, these data support the hypothesis that activity-induced arc transcription is facilitated by Pol II stalling and involves the rapid release of stalled Pol II downstream into the gene.
Stalling is mediated by Negative Elongation Factor (NELF)
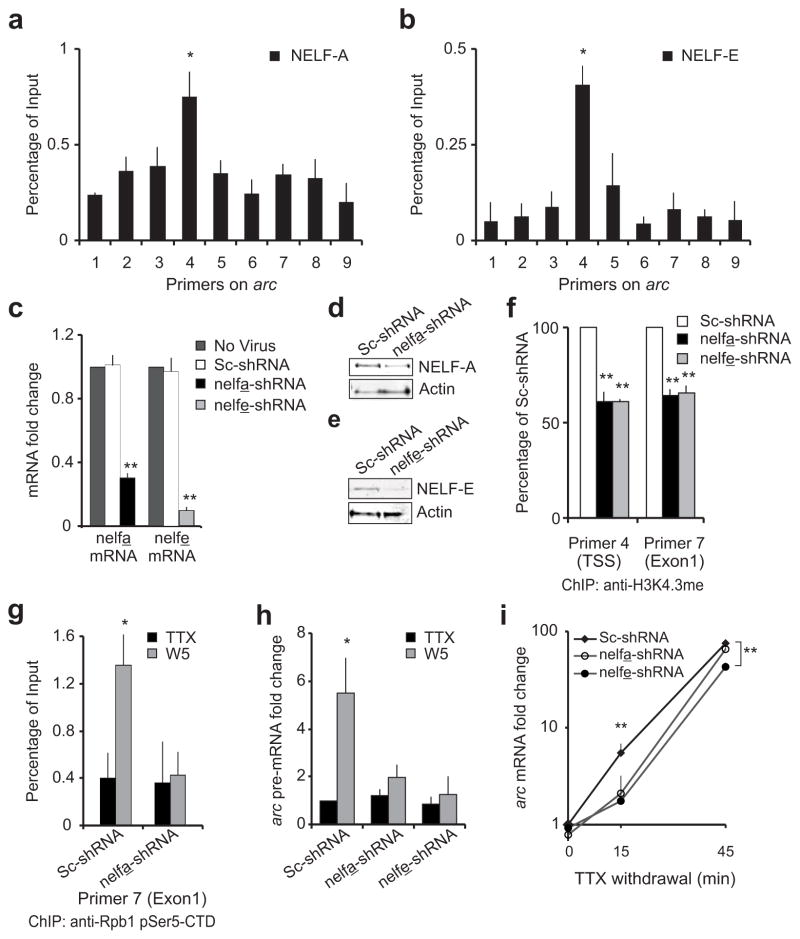
NELF, a four-subunit complex that binds to Pol II (NELF-A, NELF-B, NELF-C/D, NELF-E)30, mediates Pol II stalling in non-neuronal cells31, 32. To test for a connection between NELF and Pol II stalling at the arc TSS, we performed ChIP with antibodies against NELF-A (also known as Wolf-Hirschhorn syndrome candidate 2; WHSC2) and NELF-E (also called RNA-binding protein RD; RDBP) subunits. Both NELF subunits were enriched at the arc TSS in untreated neurons (Fig. 4a, b) and in TTX-treated cells (n = 3; data not shown), indicating the co-existence of the NELF complex in this region with stalled Pol II. Co-IP experiments confirmed their association (Supplementary Fig. 3a, c). Enrichment of NELF-A at the arc TSS was reduced significantly after TTX withdrawal (Supplementary Fig. 3b). This is consistent with previous data demonstrating dissociation of NELF during release of Pol II from stalling32, 33. Importantly, this decrease in NELF binding to Pol II was specific for the activated arc gene, as it was not observed globally (Supplementary Fig. 3a).
Figure 4. arc Pol II stalling and NELF.
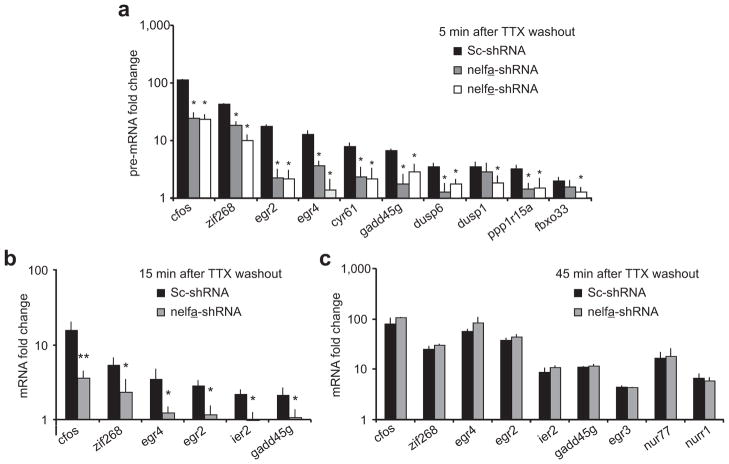
NELF enrichment near the arc TSS was estimated by ChIP with anti-NELF-A (a) and anti-NELF-E (b) antibodies. Error bars represent S.D. of the mean. (c) RT-PCR data for nelf-a and nelf-e mRNA levels in uninfected neurons and neurons expressing a scrambled sequence (Sc-shRNA) or nelf-a or nelf-e antisense shRNA (nelfa-shRNA or nelfe-shRNA). Error bars represent S.E. of the mean. (d,e) NELF-A or NELF-E levels in neurons expressing a scrambled sequence or an shRNA against the NELF subunit. Blots were immunoprobed with anti-Actin as loading control. Full length-blots are in Supplementary Fig. 8 (f) Quantification of H3K4.3me in arc by ChIP with anti-histone H3-trimethylated-lysine-4 in control neurons or neurons with depleted level of indicated NELF subunit. Error bars represent S.D. of the mean. (g) Quantification of RNA Pol II in the arc gene body as determined by ChIP with anti-pSer5 Rpb1 at W5 in neurons expressing indicated shRNAs. Error bars represent S.D. of the mean. (h) Graphical representation of arc pre-mRNA levels in neurons expressing scrambled, nelfa-shRNA or nelfe-shRNA and treated as in g. n = 5 biological trials; error bars represent S.E. of the mean. (i) Plotted are levels of arc mRNA in neurons treated as in g but harvested at either fifteen (n = 6) or forty-five minutes (n = 3) after TTX-withdrawal. Error bars represent S.E. of the mean. For all: n = 3 biological trials if not mentioned otherwise. *P < 0.05, ** P < 0.01.
To further implicate NELF in Pol II stalling in neurons, NELF-A and NELF-E were depleted using lentivirus-delivered short-hairpin (sh)RNA. After seven days of infection, neurons with nelfa-shRNA showed a seventy percent decrease in nelf-a mRNA and substantial loss of NELF-A protein compared to neurons with scrambled shRNA (Fig. 4c, d). Similarly nelfe-shRNA treatment knocked-down nelf-e mRNA by approximately ninety percent and reduced the NELF-E protein accordingly (Fig. 4c, e). Because network neuronal activity upon TTX withdrawal is critical to arc induction, we tested whether NELF depletion had any impact on the physiological health of the neurons. Depletion of NELF-A or NELF-E did not significantly interfere with excitatory synaptic function, as measured by miniature excitatory postsynaptic current (mEPSC) amplitudes and frequencies, nor were the resting membrane potentials or input resistances affected (Supplementary Fig. 4a–e).
At the arc promoter, depletion of NELF-A or NELF-E significantly reduced promoter-proximal Pol II enrichment in both untreated- and TTX-treated conditions (data not shown; n = 3 for NELF-A and n = 2 for NELF-E). This loss of Pol II with NELF depletion is consistent with genome-wide studies in Drosophila24 and indicates that Pol II stalling at the arc promoter is contingent on the inhibitory effects of NELF. Furthermore, NELF-A and NELF-E knockdown reduced tri-methylation of histone H3 lysine 4 near the arc TSS, indicating a loss of transcriptional permissiveness of the promoter (Fig. 4f).
Reduction of Pol II stalling prevents rapid arc transcription
To study the relationship between rapid arc transcription and stalled Pol II, we examined gene induction in NELF-depleted cells. As expected, we detected significantly increased levels of pSer5 Pol II in the arc gene body five minutes after TTX withdrawal in neurons treated with scrambled shRNA (Fig. 4g). We did not detect any such increase in the NELF-A depleted neurons, indicating a lack of active arc transcription five minutes after TTX washout (Fig. 4g). This is further reflected by a lack of induced arc pre-mRNA five minutes after TTX washout in NELF-A- or NELF-E-depleted neurons when compared with neurons infected with scrambled shRNA (Fig. 4h). In order to confirm the specificity of our shRNA further, we performed similar studies with an alternative shRNA (nelfa-shRNA2) that reduced NELF-A mRNA by approximately sixty-five percent and significantly compromised induction of arc at five minutes after activity (Supplementary Fig. 5a). To further implicate NELF in rapid arc induction, we generated a shRNA-insensitive-NELF-E that was cloned into the viral backbone expressing the nelfe-shRNA (Suppplementary Fig. 5b). Using this construct, which simultaneously knocked down endogenous NELF-E and expressed shRNA-insensitive-NELF-E, we achieved a fifty-five percent rescue of arc gene induction (Supplementary Fig. 5c). A full recovery was not necessarily expected as similar experiments had previously failed to completely recover the NELF complex, likely because NELF subunits are mutually regulated34. Consequently, re-establishment of one subunit after its knockdown does not appear to generate normal levels of other subunits and cannot fully revive the NELF multi-protein complex efficiently34 (Suppplementary Fig. 5d).
Finally, we quantified mature arc mRNA levels fifteen and forty-five minutes after TTX washout in NELF-A- or NELF-E-depleted neurons. At fifteen minutes past TTX washout, NELF depletion with any of the three shRNAs had a comparable significant reduction of arc induction (Fig. 4i and Supplementary Fig. 5e). By forty-five minutes after washout, transcription was able to overcome kinetic delays induced by lack of Pol II stalling in NELF-A-depleted cells, but remained significantly reduced in NELF-E-depleted cells. This disparity likely stems from the differences in silencing efficiencies of the different shRNAs. Together, these data indicate that rapid transcription of arc at very early time points is contingent upon the NELF-mediated Pol II stalling.
Identifying other IEGs
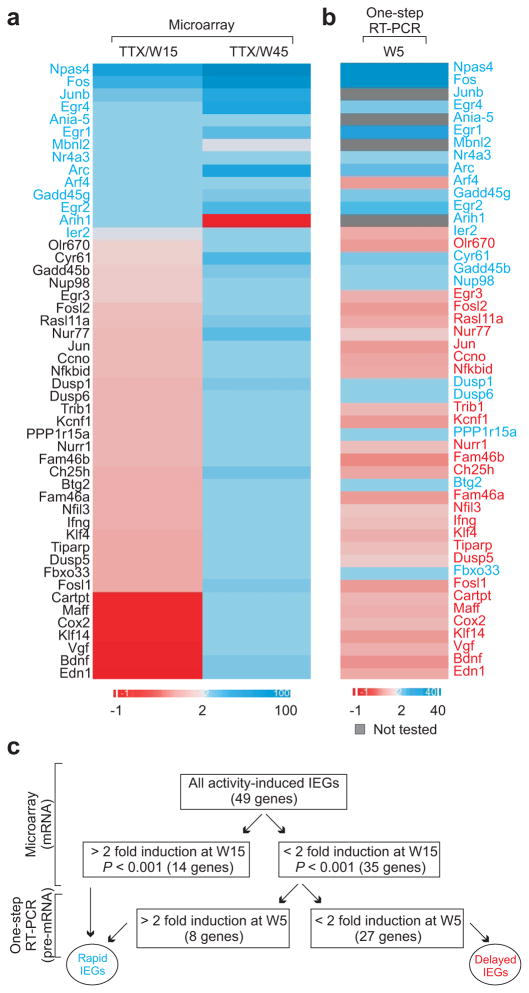
To identify other IEGs that are up-regulated by the TTX withdrawal protocol, we conducted global gene expression analysis using microarrays. Array intensities of samples collected fifteen and forty-five minutes after TTX washout were compared against their corresponding TTX-control sample. From this genome-wide screen, we identified forty-nine annotated genes that showed a two-fold or more increase in their mRNA levels within forty-five minutes after TTX washout (P < 0.01; Fig. 5a). In many instances, we validated the microarray data with quantitative RT-PCR using oligo-(dT) primers (Supplementary Table 1). Among these forty-nine genes, we could detect two-fold or more expression changes in some of them very soon after TTX withdrawal (14 genes; fifteen minutes after TTX withdrawal), whereas up-regulation of other RNAs were detected only later (forty-five minutes after TTX withdrawal). Interestingly, the expression of mbnl2 and arih1 dropped at the later time point after crossing the two-fold threshold fifteen minutes after induction, but the expression of all other rapid IEGs remained above the two-fold threshold over time (Fig. 5a).
Figure 5. Identifying other IEGs.
(a) Heat map showing relative mRNA expression of all genes 15 or 45 minutes after TTX withdrawal (derived from microarray data). (b) Heat map showing relative pre-mRNA expression of all genes 5 minutes after TTX withdrawal (derived from one-step qRT-PCR data). In both a and b, induction above two-fold is represented in shades of blue while less than two-fold induction is shown in shades of red. Genes with two-fold or more induction of mRNA at fifteen minutes or pre-mRNA transcript at five minutes were classified as rapid IEGs. (c) Schematic flowchart representing multiple techniques used to validate rapid IEGs and delayed IEGs.
Because microarrays detect mature mRNA, we next investigated whether some genes might have their rapid transcription masked by mRNA processing efficiency. Thus, we performed one-step RT-PCR assays, which use primers directed at the nascent transcripts instead of using the generalized oligo-(dT) primers and thus can detect immature transcripts that lack 3′-polyadenylation. We designed intron-based primers for intron-containing genes and against the exon of six genes lacking introns. These primers were used to assess any change in transcript levels of these genes at five minutes after TTX washout. Based on this screen, eight additional genes out of the thirty-five genes showed two-fold or more up-regulation at this time-point (Fig. 5b).
To ask whether these differences in induction kinetics were related to Pol II pausing, we grouped these genes into two classes: one, where a significant (2-fold) change of transcript was detected early (twenty-two genes), which we refer to as ‘rapid’ genes, and two, where significant up-regulation was only detected later, which we refer to as ‘delayed’ genes (twenty-seven genes; Fig. 5c and Supplementary Table 1).
Rapid IEGs have stalled Pol II
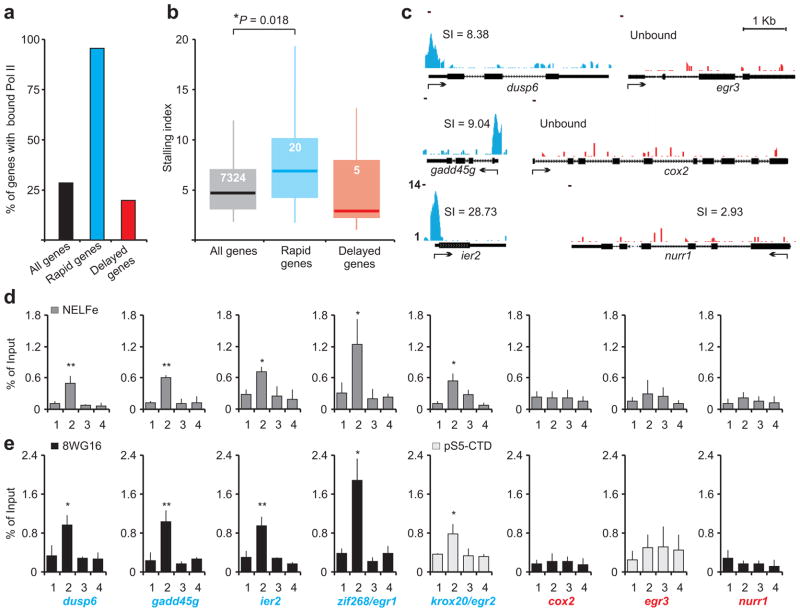
We next investigated the distribution of Pol II at rapid and delayed IEG promoters using our genome wide ChIP-seq data. To determine which promoters were occupied by significant levels of Pol II above the background, ChIP-seq reads mapping near all rat promoters (28,117 non-redundant promoter regions, +/− 200 bp) were compared with 11,426 size-matched intergenic regions. This comparison revealed that 8,015 rat promoters had significant Pol II signal and are subsequently referred to as ‘bound’. We refer to the remaining promoters as ‘unbound’. Such unbound promoters were prevalent among delayed IEGs with only five of the twenty-seven genes bound by Pol II in unstimulated neurons. This fraction is comparable to the fraction of bound genes genome wide (Fig. 6a). In contrast, twenty out of twenty-one annotated rapid IEGs were bound prior to stimulation (Fig. 6a). This indicates that rapid IEGs are primarily characterized by the presence of pre-loaded Pol II in their promoter. The promoter of the twenty-second rapid IEG (egr4) could not be assessed as it remains unsequenced (2004 Baylor3.4/rn4).
Figure 6. Pol II is enriched near all rapid IEG promoters.
(a) Pol II bound promoters were assessed from M01 ChIP-seq for all rat genes, 21 rapid IEGs, and 27 delayed IEGs. The percentage of genes with significantly greater Pol II binding than intergenic regions (noise) are represented. (n = 2 biological trials). (b) Box plot representation of the promoter proximal stalling index distribution for all rat genes (n = 7324), rapid IEGs (n = 20), and delayed IEGs (n = 5) with Pol II bound promoters. The box,and whiskers denote the 25–75th and the 5–95th percentiles respectively. The median (all genes: 4.71, rapid IEGs: 7.63 and delayed IEGs: 3.72) is represented by the horizontal bar. *P = 0.0175 (Kruskal-Wallis non-parametric test). (c) Examples of Pol II bound and unbound promoters with or without stalling. Arrow indicates TSS for each gene and the number in parentheses refers to stalling indices (SI) for Pol II bound promoters (refer to the text). (d) NELF-E enrichment in the promoter regions of rapid IEGs (blue) and delayed IEGs (red). For all genes, primer pair 1 represents upstream regions, primer pair 2 spans the TSS, and primer pairs 3 and 4 represent the gene body. n = 3 biological trials; error bars represent S.D. of the mean. (e) For comparison, detection of Pol II enrichment in same genes as in (d). n = 3; error bars represent S.D. of the mean. For d and e, *P < 0.01, **P < 0.001
Among bound genes, Pol II could either be distributed over the gene body with no enrichment at the promoter, or be enriched proximally to the promoter indicating stalling (see scn2a1 vs. arc in Fig. 2c). In an attempt to quantify stalling in bound promoters, we computed a promoter proximal stalling index for all rat genes (see Methods). The index, based on definitions described previously for identification of stalling genome-wide, is a ratio between the density of Pol II ChIP-seq ‘reads’ near the annotated TSS of a gene and the read density within the gene body24. This ratio will be low for genes that have uniform distribution of Pol II in the gene body in addition to the promoter but will be high in genes that experience slower, regulated release of stalled Pol II. The stalling indices for all bound genes are distributed between 0.23 and 64 (Supplementary Fig. 6). When we compared the stalling indices of bound rapid IEGs with all bound genes using non-parametric statistics, we found that the stalling indices of rapid IEG were significantly higher (Fig. 6b). These results support our hypothesis that rapid IEGs are characterized by promoter proximal stalling. ChIP-seq data demonstrating promoter proximal stalling in three representative rapid IEGs (dusp6, gadd45g, and ier2) are illustrated along with two unbound (egr3 and cox2) and one bound delayed IEG (nr4a2/nurr1) in Figure 6c. Unfortunately, the small sample size of bound delayed IEGs prevented us from performing any meaningful statistical comparison between them and bound genes genome-wide. Nevertheless, because Pol II binding is a pre-requisite of stalling and most delayed IEGs were unbound, we infer that delayed IEGs typically are characterized by a lack of Pol II stalling.
To further implicate promoter proximal stalling in the rapid IEG response, enrichment of NELF was also tested in promoters of several rapid IEGs with NELF-E ChIP. Enrichment of this stalling factor was detected in promoters of several rapid IEGs (Fig. 6d). Included among them were the promoter of egr1/zif268, the single rapid IEG that had a low stalling index as revealed by the M01 ChIP-seq (Supplementary Fig. 6) and krox20/egr2, the unbound rapid IEG. For reference, enrichment of Pol II was also tested in promoters of these rapid IEGs by two alternative Pol II antibodies (8WG16 and anti-pS5-CTD, Fig. 6e). Using the same primer pairs that detected NELF-E enrichment, chromatin immunoprecipitated with 8WG16 also revealed significant Pol II abundance near the TSSs of dusp6, gadd45g, ier2 and importantly, egr1/zif268. This observation is in agreement with 8WG16 ChIP-seq data from an independent study using mouse neurons35. 8WG16 however, failed to identify any stalling in krox20/egr2, the unbound rapid IEG. Nevertheless, primers spanning the annotated TSS of this unbound rapid IEG showed significant Pol II enrichment when the antibody against pSer5 epitope was used (Fig. 6e). However, neither the two antibodies against Pol II nor the antibody against NELF-E detected any enrichment of either protein near the TSS of representative delayed IEGs (egr3, cox2, or nurr1; Fig. 6d, e).
Taken together, promoter proximal Pol II stalling is associated with all twenty-one sequenced rapid IEGs as defined by the enrichment of Pol II detected by at least one Pol II antibody and NELF enrichment. In contrast, considering that more than seventy percent of delayed IEGs were unbound and all tested delayed IEGs lacked enrichment of NELF-E, we suggest that most delayed IEGs lack Pol II stalling. Lack of Pol II enrichment in all but a few delayed IEGs with high stalling indices, such as tiparp, vgf and trib1, suggests that the absence of pre-loaded Pol II at these genes could contribute to their slower activation. Enrichment of Pol II in tiparp, vgf and trib1 may support their rapid induction in response to a stimulus other than neuronal activity or TTX withdrawal.
Induction of rapid IEGs is sensitive to NELF knockdown
To study the effect of Pol II stalling on transcription of rapid IEGs, induction of several such IEGs was determined five minutes after TTX washout by assessing their pre-mRNA in neurons with normal and depleted levels of NELF-A or NELF-E. Expression of every tested rapid IEG was compromised at this time point in NELF-depleted cells (Fig. 7a). Similarly, rapid IEG mature mRNA levels at fifteen minutes after TTX washout were also reduced dramatically in NELF-A-depleted conditions (Fig. 7b). However, NELF-A RNAi had no effect on the induction of rapid IEGs or delayed IEGs at forty-five minutes after activity onset (Fig. 7c), suggesting that stalling is critical for the immediate onset of rapid IEG induction, but not the steady-state accumulation of mRNA over time. Notably, egr4, a gene with an unsequenced promoter, shows compromised immediate early transcription following depletion of NELF subunits. Thus, Pol II enrichment may be found near the egr4 TSS once the region is sequenced.
Figure 7. Pol II stalling mediates immediate transcription of several IEGs.
(a) Pre-mRNA levels of various rapid IEGs and delayed IEGs five minutes after TTX washout in neurons infected with scrambled, nelfa-shRNA or nelfe-shRNA producing lentiviruses. n = 3 biological trials; error bars represent S.E. of the mean. (b,c) NELF-A depletion blocks immediate transcription of many IEGs. mRNA levels of different IEGs in neurons infected with nelfa-shRNA- or Sc-shRNA-producing lentiviruses. Cells were harvested either fifteen (b) or forty-five (c) minutes after TTX-withdrawal. The qPCR data were normalized to GAPDH and are presented as times of the corresponding mRNA level in scrambled shRNA expressing neurons with TTX. n = 3; error bars represent S.E. of the mean. For all panels, *P < 0.05 and **P < 0.001.
Promoter proximal Pol II enrichment is detected in the brain
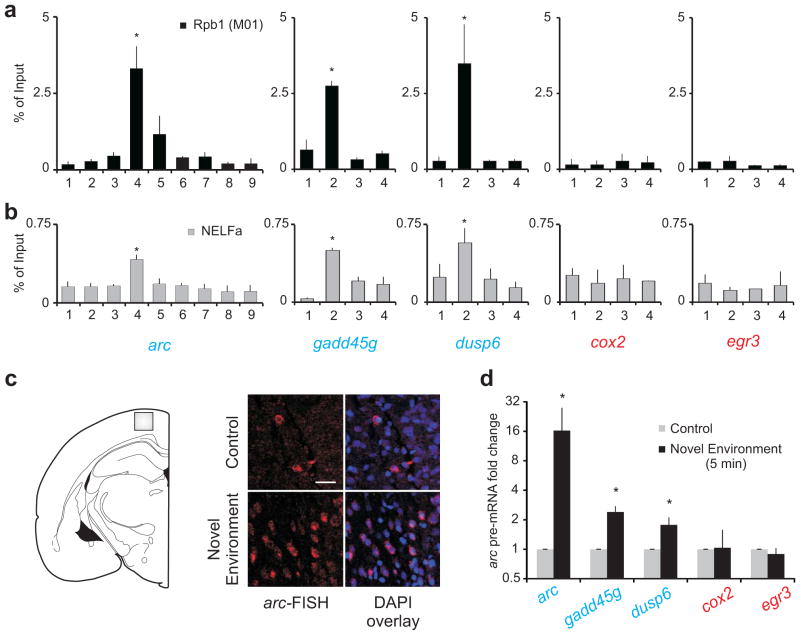
To extend our findings to brain tissue and to rule out potential artifacts related to the neuron culture preparation, we performed Pol II M01 and NELF-A ChIP using brain samples comprising the cortex and hippocampus. We found significant enrichment of Pol II and NELF-A near the TSS of arc and two other rapid IEGs (dusp6 and gadd45g) (Fig. 8a, b). Such enrichment was not noted in the delayed IEGs cox2 and egr3 (Fig. 8a, b).
Figure 8. Poised Pol II is found in vivo.
(a) Pol II enrichment in the promoter regions of rapid IEGs (blue) and delayed IEGs (red) in brain samples. The M01 antibody Pol II antibody was used. For all genes, primer pair 1 represents upstream regions, primer pair 2 spans the TSS, and primer pair 3 and 4 represents the gene body. n = 3 biological trials; error bars represent S.D. of the mean. (b) Detection of NELF-A enrichment in the promoter regions of rapid IEGs (blue) and delayed IEGs (red) using identical primers as in (a). n =3; error bars represent S.D. of the mean. *P < 0.01, **P < 0.001 (c) Representative confocal images showing transcription of arc in the cortex after a rat’s exposure to a novel environment as revealed by FISH. Scale bar represents 20μm. Images were acquired from the layer 2–3 of primary somatosensory area of the cortex (gray box in the cartoon). (n = 2) (d) Pre-mRNA levels of rapid and delayed IEGs after five minutes of exposure to a novel environment. (n = 5 for arc and cox2 and n = 3 for others); error bars represent S.D. of the mean. *P < 0.05. Y-axis is log2
To test if Pol II stalling correlates with rapid induction of genes in the brain, we exposed rats to a novel environment for five minutes. This exposure resulted in significant up-regulation of arc, dusp6 and gadd45g transcription within five minutes in the cortex (Fig. 8c, d). This is consistent with previous reports of arc upregulation in the hippocampus8. At the same time point, we did not note any significant up-regulation of either cox2 or egr3 (Fig. 8d). The same treatment induced egr3 significantly at later time points (forty-five minutes and two hours; data not shown). These data rule out the possibility of Pol II stalling being an artifact of neurons in culture and provide evidence indicating the presence of this phenomenon in the brain.
Discussion
In this study, we have made three observations supporting a role for poised Pol II in neuronal activity-induced rapid transcription: first, using genome-wide screens we demonstrate that Pol II is enriched in the proximity of the promoters of arc and all other testable rapid IEGs; second, with NELF RNAi, we demonstrate that rapid induction of arc and other rapid IEGs is dependent on Pol II stalling; and third, we show that delayed IEGs, which are expressed only later, lack Pol II stalling in most instances and remain unaffected by NELF RNAi. Thus, our evidence supports the idea that rapid IEGs are a specialized subset of genes poised for immediate response biologically mechanized by Pol II stalling. Also, because transcription is initiated prior to the onset of action potential triggers, their expression is likely rate-limited by transcription elongation.
Evidence that Pol II stalling renders a kinetic advantage to transcription of rapid IEGs comes from experiments where we have depleted components of the NELF complex and find the onset of transcription after TTX withdrawal to be significantly slower. Importantly, this effect is specific for the rapid response of transcription to activity, as NELF depletion does not interfere with the health of the neurons or impair function of the transcription machinery. This idea is supported by the gradual accumulation of arc and other rapid IEG transcripts over time in control and NELF-depleted cells. Note that NELF depletion has been found to enhance as well as suppress expression of several genes36, 37. In our experiments, NELF RNAi results in the loss of active chromatin marks on the arc promoter, attenuation of Pol II stalling, and compromised induction of all tested rapid IEGs. This includes cfos, which is up-regulated by NELF-E depletion in an immortalized non-neuronal human cell line38. This difference may be due to cell type-specific regulatory mechanisms and warrants further analysis.
Here, we propose that the presence of stalled Pol II primes an entire group of rapid IEGs for rapid transcription (Supplementary Fig. 7). Whereas the initiation step of transcription is a time-consuming process, genes with stalled polymerase already possess active chromatin marks, are pre-loaded with general transcription factors, and harbor engaged polymerase that waits in the promoter-proximal region for an activity-induced signal that likely involves calcium39. Our data suggest that neuronal activity triggers recruitment of PTEF-b to the rapid IEGs, allowing for rapid phosphorylation of the polymerase CTD and release of Pol II into these genes40. Once the initial round of RNA synthesis has occurred, additional Pol II can be rapidly recruited, making use of the already established transcription scaffold and permissive chromatin architecture. Most delayed IEGs, however, lack stalled Pol II near their promoters and are expressed with relatively slower kinetics than those of rapid IEGs. The extra time is likely to be required for transcription factor and Pol II assembly. Taken together, the presence of stalled Pol II at all rapid IEGs and its absence at most delayed IEGs suggest a non-redundant role of the Pol II stalling mechanism in rapid gene transcription (Supplementary Fig. 7). These findings are in agreement with a recent study in murine macrophages that demonstrates the occurrence of Pol II stalling at immediate early inflammatory genes, but not in genes with slower induction kinetics33.
What is the functional significance of poised Pol II in neurons? In the absence of stalling, only the immediate transcription of several rapid IEGs was impacted, suggesting that Pol II stalling would be most pertinent in regulating the precise timing and dynamics of gene responses. Well-timed responses may serve as replenishment for a dendritic mRNA pool that is rapidly exhausted during local translation41. In addition to genes, Pol II stalling may have a role in transcription of other molecules, like the recently discovered RNA transcribed from enhancers, the eRNA35. Moreover, stalling might permit a temporally coordinated response to neuronal activity that can achieve distinct roles in brain development and function. Several rapid IEGs, being transcription factors, are likely regulating successive waves of gene expression. Others, like arc, may reinforce synaptic events prior to decay of synaptic tags42. This diverse array of functions could be triggered by a single signaling pathway that targets the release of stalled Pol II21. If a single learning event triggers a pathway as such, then the poised Pol II mechanism is expected to facilitate all the downstream events leading up to consolidation of learning, for example. These are exciting possibilities that require further testing..
Methods
RNAi
Plasmids (pLKO.1-puro) carrying pre-designed anti-NELF-A or anti-NELF-E short hairpin (designed by RNAi consortium or TRC) or scrambled hairpin sequences were obtained from Sigma. Self-inactivating HIV lentivirus particles were produced by transfecting 293T cells with the vector, envelope (pMD2.G; Addgene), and packaging plasmids (psPAX2; Addgene). For infection with recombinant lentiviruses, the viral supernatant was diluted in neuronal media and cells were infected at a multiplicity of infection ranging from 2 to 5. At seven days post infection, neurons were tested for NELF-A or NELF-E mRNA knockdown. Two anti-NELF-A hairpins (sh-nelfa: CCG GCC CAA TGT TCA AGA CAT CTT ACT CGA GTA AGA TGT CTT GAA CAT TGG GTT TTT G and sh-nelfa2: CCG GGA GGA ACA GAA CCC CAA TGT TCT CGA GAA CAT TGG GGT TCT GTT CCT CTT TTT G) and one anti-NELF-E hairpin (sh-nelfe: CCG GCT GGA TTC CTT GTG CCT CAT ACT CGA GTA TGA GGC ACA AGG AAT CCA GTT TTT G) were used in this study. For electrophysiological studies, NELF-A hairpin was subsequently cloned into a GFP containing lentiviral backbone (pLKO.3G; Addgene) using SphI and EcoRI restriction enzyme sites for co-expression of GFP along with the anti-NELF-A shRNA.
Cell culture and treatment
Cultures of cortical neurons were prepared from embryonic day 18 Sprague Dawley rats44 (Institution approval: ASP 01–21). Dissociated cortical neurons were plated in Neurobasal media (Invitrogen) supplemented with 25 mM glutamate (Sigma) and 0.5 mM L-Glutamine (Sigma) and either B27 (Invitrogen) or NS21 and maintained in a similar media without the glutamate. NS21 was prepared as previously described45. Neurons were routinely used between 11–16 days in vitro. In gene induction studies, cells were treated with 1–2 μM tetrodotoxin (TTX; Calbiochem) for 48 hours. To induce transcription, cells were washed twice with excess TTX-free warm media, and then incubated in fresh media at 34°C. The withdrawal time was recorded after the second wash.
Immunocytochemistry
Cultured neurons were fixed with 4% paraformaldehyde in PBS and permeabilized with pre-chilled absolute ethanol treatment for 5 minutes in −20°C. Fixed neurons were then immuno-stained with anti-Arc antibody (1:1000; Synaptic Systems) overnight at 4°C and then with anti-Rabbit Alexa 633 dye-conjugated secondary antibody (1:2500; Molecular Probes) at room temperature for 2 hours in blocking solutions.
Gene expression analysis using quantitative PCR (qPCR)
Total RNA was isolated from cortical cell cultures using the RNeasy kit with DNase treatment (Qiagen). To measure mature mRNA (qRT-PCR), cDNA was produced using MuLV reverse transcriptase with poly-dT and random hexamer primers. Gene expression was measured in quantitative real-time PCR (qPCR). To measure pre-mRNA (one-step qRT-PCR), primers that target the arc and GAPDH introns (Supplementary Information) served for cDNA synthesis and subsequent amplification (14 cycles) using the manufacturer’s protocol in the One-Step RT-PCR kit (Qiagen). The amplified product level was quantified by qPCR using the same primers against the arc and GAPDH introns.
Expression Microarray
Gene expression analysis was conducted by the NIEHS Microarray Core using RNA obtained from three biological replicates. The Agilent Whole Rat Genome 4×44 multiplex format oligo arrays (014879, Agilent Technologies) were used following the Agilent 1-color microarray-based gene expression analysis protocol. Using 500ng of total RNA, Cy3 labeled cRNA was produced according to the manufacturer’s protocol. For each sample 1.65 μg of Cy3 labeled cRNAs were fragmented and hybridized for 17 hours in a rotating hybridization oven. Slides were washed and then scanned with an Agilent Scanner. Data was obtained using the Agilent Feature Extraction software (v9.5), using the 1-color defaults for all parameters. The Agilent Feature Extraction Software performed error modeling, adjusting for additive and multiplicative noise. The resulting data were processed using the Rosetta Resolver® system (version 7.2; Rosetta Biosoftware, Kirkland, WA).
Chromatin Immunoprecipitation (ChIP)
Cells were cross-linked with formaldehyde after treatment. In TTX samples, cells were washed with 2 μM TTX in PBS. Cross-linked cell lysates were sonicated for fifteen cycles (bursts and intervals of 30 seconds each) with the Bioruptor (Diagenode). Sonicated samples were pre-cleared for 1.5 hours with 50% A/G bead slurry (SantaCruz Biotechnology), then immunoprecipitated overnight at 4 degrees with 5 μg anti-Rpb1 (8WG16; Abcam), anti-Rpb1 pSer5 (Abcam), anti-H3 (Abcam), anti-tri methyl H3K4 (Millipore), anti-acetylated H3K9 (Millipore), anti-NELF-A or anti-NELF-E (Sigma) antibodies. The anti-NELF-A (rat) antibody was custom made (Yenzyme) with a twenty amino acid synthetic peptide (QIKLSEHTEDLPKADGQGS-amide) corresponding to 481–499 amino acids of rat NELF-A (WHSC2) as the immunogen. Immunoprecipitation with H5 (anti-Rpb1 pS2-CTD IgM; Covance) antibody was carried out overnight at 4°C and then anti-mouse IgM was added for an additional day. Antigen-antibody complexes were immunoprecipitated with 50% A/G bead slurry, washed once with low salt buffer, three times with high salt buffer for most IPs, once with LiCl buffer and finally once with Tris-EDTA. As a deviation, NELF-A imunoprecipitation was done in 200mM NaCl and two and four high salt washes were performed for the H5 and NELF-A immunocomplexes respectively. Samples were reverse cross-linked and chromatin DNA was eluted using the QIAquick Nucleotide Removal Kit (Qiagen). Eluted chromatin was quantified by qPCR. Data for ChIP with H5 was obtained by subtracting anti-mouse IgM ChIP values from the corresponding H5 values. For ChIP with brain samples, isolated minced cortices (with hippocampus) were passed through 19 and 22 gauge needles in ice-cold PBS containing 1% Formaldehyde. These cross-linked samples were then processed as above.
Chromatin Immunoprecipitation-sequencing (ChIP-seq)
ChIP was performed with Pol II M01 antibody and the immunoprecipitated DNA fragments were used to prepare ChIP-Seq libraries. These libraries were constructed using the standard protocol from Illumina with NEBNext DNA Sample Prep Reagent Set 1 and 2 (New England Biolabs; E600L; E6020L) and single-end adapters from Illumina (Genomic DNA Sample Prep Oligo Only Kit; FC-102-1003) with two modifications to the protocol. Platinum® Pfx DNA Polymerase (Invitrogen; 11708-039) was used for library amplification and Agencourt AmPure Reagent (Beckman Coulter Genomics; A29152) was used to clean up the amplification reaction. Libraries were quantitated by qPCR (Power SYBR® Green PCR Master Mix; 4368702; Life Technologies, Inc.) with following primers:
5′-CCAAGCAGAAGACGGCATACGAGCTCTTCCGATC-3′
5′-CAATGATACGGCGACCACCGAGATCTACACTCTTTCCCTAC-3′
Flow cells were prepared with Illumina’s Standard Cluster Generation Kit V 2 (FC-103-2001) on an Illumina Cluster Station and run on a Genome AnalyzerIIx within one week using 36-Cycle Sequencing Kits V4 at National Intramural Sequencing Center (NISC), NIH.
Short Read alignment and data analysis
Reads provided by NISC were aligned with Bowtie 0.12.346 to the R. norvegicus rn4 index. Biological replicates were in good agreement and were combined, resulting in a data set of 21,229,004 unique hits. ChIP-seq hit locations were centered by moving the forward and reverse strand hit location 75 nt in the appropriate direction because the average fragment size was ~150bp and placed in 25 nt bins for visualization using the UCSC genome browser. R. norvegicus Ensembl Genes 58 data were used to define TSS and gene bodies. Promoter proximal regions were defined as the physical genomic region +/− 200 nt from the annotated TSS. Gene bodies were defined as the physical region from +501 to the end of the gene. To define the significant number of reads in bound promoter proximal regions, we used the Fisher’s exact test implemented in R. We compared 28,117 promoter proximal regions to 11,426 size matched intergenic regions that are at least 5,000 nt from the start or end of an annotated gene. The p-values from the Fisher’s exact tests were corrected using the Benjamini-Hochberg method to control for false discovery, and a corrected P-value < 0.01 was deemed significant. Promoter proximal stalling index was defined as the ratio of the number of reads in a promoter proximal region to a gene body normalized by the search size. Genes with gene bodies less than 401 nt were excluded from Stalling index analyses.
arc intron-enriched riboprobes and fluorescent in situ hybridization (FISH)
The plasmid containing the arc gene sequence spanning both the introns (and the exon in between) has been previously described8 and was a kind gift from J. Guzowski. Riboprobes were generated from linearized plasmids by in vitro transcription using fluorescein-labeled dUTPs. FISH was subsequently performed with this riboprobe as detailed previously by Guzowski et al. (2006). Briefly, fixed neurons were hybridized with the riboprobe overnight at 56°C and FISH signals were then detected with anti-Fluorescein–HRP conjugate (Jackson) and amplified using cyanine-3 tyramide signal amplification kit (PerkinElmer).
Electrophysiological recordings
Whole-cell patch-clamp recordings were performed on dissociated cortical neurons co-expressing GFP and shRNA. Neurons were perfused at 1 ml/min at room temperature in ACSF (in mM): 124 NaCl, 2.5 KCl, 2 MgCl2, 2 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, 17 D-glucose, 0.5 Picrotoxin and 0.001 TTX. Patch electrodes (3–7 MΩ) were filled with (in mM): 120 K-gluconate, 10 KCl, 3 MgCl2, 0.5 EGTA, 40 HEPES, 2Na2-ATP, 0.3 Na-GTP, with pH adjusted to 7.2 by NaOH. Cells were voltage clamped at −60 mV and mEPSCs were recorded over 3 minute periods. mEPSCs were detected and analyzed using the Mini Analysis program (Version 6.0.3, Axon Instruments). For multi-electrode arrays, cortical neurons were prepared and plated onto a square grid of 64 planar electrodes (MED64; MED Sciences, Japan). Recordings were acquired with an 64-channel integrated amplifier and MED64 Conductor software (Sampling rate 20kHz, LCF 100Hz). Spontaneous activity was recorded at 34°C both at the end of the TTX treatment and immediately following washout of TTX, achieved by washing 2 times.
Co-immunoprecipitation and Western blotting
For Co-IPs, nuclear material was extracted using the Nuclear Complex Co-IP Kit (Active Motif). Extracted material was pre-cleared for one hour with A/G beads (Santa Cruz Biotechnology) and then incubated with primary antibody overnight. As negative control, normal IgG was used that showed negligible signal. The following day, A/G beads were added to the IPs for one hour. The beads were washed with IP wash buffer prepared from the CoIP Kit and were prepared for Western blotting by adding LDS Sample Buffer (NuPAGE) and boiling. To extract nuclear proteins, neurons were washed with PBS + 5 mM MgCl2 and then lysed with plasma membrane lysis buffer (10mM HEPES pH 7.5, 60 mM KCl, 1.5 mM MgCl2, 0.1% NP40, and 0.5 mM DTT). Nuclei were collected by centrifugation, resuspended in nuclear extraction buffer (20mM HEPES pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1 mM EGTA, and 20% glycerol) and disrupted by brief sonication. Proteins were resolved by gel electrophoresis and transferred to a nitrocellulose membrane using the iBlot gel transfer apparatus (Invitrogen). Immunoblots were incubated with primary antibody overnight. Blots were visualized with an Odyssey infrared scanner (Li-COR Biosciences) after immunolabeling primary antibodies with infrared fluorophore-tagged secondary antibody (Molecular Probes). Images were analyzed using the Odyssey 2.1 software.
Novel environment studies
Sixteen to twenty one day-old rat pups were handled daily for several days prior to experimentation (Institution approval: ASP 2009–0023). The novel environment was a square plexiglass platform with high walls on a temperature regulated bed. Toys were included in this environment along with cotton swabs dipped in artificial vanilla, lemon and almond extracts. The control rat was decapitated immediately upon removal from the cage in the adjacent procedure room whereas the experimental rat was allowed to explore the environment and was decapitated immediately at the designated time. The brain was split into two hemispheres and one was frozen immediately in methyl-butane pre-chilled in a dry ice and methanol slurry for FISH. The other hemisphere was utilized to obtain tissue samples for PCR. Samples and sections were taken from the primary somatosensory area of the cortex. Similar findings have been observed in the hippocampus8.
Statistics
ChIP data are reported as means and S.Ds. All remaining data are expressed as mean and S.E.Ms. Statistical comparison of datasets were performed with two-tailed Student’s t test with Bonferroni corrections (for multiple comparisons of ChIP data), Kruskal-Wallis non-parametric test (comparison of Pol II Stalling indices) and two way ANOVA (for multiple comparisons in NELF-A knockdown studies). Statistics were performed with Sigmastat 3.5 software.
Supplementary Material
Acknowledgments
We thank D. Armstrong, P. Wade and the Dudek Lab members for critical review of the manuscript, the NIEHS Viral Vector Core for lentivirus preparations, the NIEHS Microarray Core for generating, processing and analyzing microarray data, NIH Intramural Sequencing Center for ChIP-seq sample preparations and sequencing and the NIEHS Imaging Center for assistance with confocal imaging. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (Z01 ES100221 to SMD and Z01 ES101987 to KA).
Footnotes
Database accession numbers:
GEO accession number for microarray data: GSE22622
GEO accession number for ChIP-seq data: GSE22878
Author contributions:
RNS and SMD conceived and designed the study. Experiments were conducted by RNS, ERB, EMW, MZ, J-YH, JDF and KRD. Data analysis was performed by RNS, ERB, EMW and MZ. Bioinformatics analyses were performed by DCF. KA provided statistical analyses and technical and conceptual advice. RNS and SMD wrote the manuscript with inputs from EMW, ERB, DCF and KA.
References
- 1.Frey U, Krug M, Brodemann R, Reymann K, Matthies H. Long-term potentiation induced in dendrites separated from rat’s CA1 pyramidal somata does not establish a late phase. Neurosci Lett. 1989;97:135–139. doi: 10.1016/0304-3940(89)90152-3. [DOI] [PubMed] [Google Scholar]
- 2.Kauderer BS, Kandel ER. Capture of a protein synthesis-dependent component of long-term depression. Proc Natl Acad Sci USA. 2000;97:13342–13347. doi: 10.1073/pnas.97.24.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol. 1996;490:703–711. doi: 10.1113/jphysiol.1996.sp021179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- 5.Messaoudi E, Ying SW, Kanhema T, Croll SD, Bramham CR. Brain-derived neurotrophic factor triggers transcription-dependent, late phase long-term potentiation in vivo. J Neurosci. 2002;22:7453–7461. doi: 10.1523/JNEUROSCI.22-17-07453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyford GL, et al. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 7.Link W, et al. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez-Amaya V, et al. Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J Neurosci. 2005;25:1761–1768. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci. 2001;21:5089–5098. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyashita T, Kubik S, Haghighi N, Steward O, Guzowski JF. Rapid activation of plasticity-associated gene transcription in hippocampal neurons provides a mechanism for encoding of one-trial experience. J Neurosci. 2009;29:898–906. doi: 10.1523/JNEUROSCI.4588-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou Z, Buck LB. Combinatorial effects of odorant mixes in olfactory cortex. Science. 2006;311:1477–1481. doi: 10.1126/science.1124755. [DOI] [PubMed] [Google Scholar]
- 13.Saha RN, Dudek SM. Action potentials: to the nucleus and beyond. Exp Biol Med (Maywood) 2008;233:385–393. doi: 10.3181/0709-MR-241. [DOI] [PubMed] [Google Scholar]
- 14.Hardingham GE, Chawla S, Johnson CM, Bading H. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature. 1997;385:260–265. doi: 10.1038/385260a0. [DOI] [PubMed] [Google Scholar]
- 15.Adams JP, Dudek SM. Late-phase long-term potentiation: getting to the nucleus. Nat Rev Neurosci. 2005;6:737–743. doi: 10.1038/nrn1749. [DOI] [PubMed] [Google Scholar]
- 16.West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- 17.Thompson KR, et al. Synapse to nucleus signaling during long-term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron. 2004;44:997–1009. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 19.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 21.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- 24.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao VR, et al. AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nat Neurosci. 2006;9:887–895. doi: 10.1038/nn1708. [DOI] [PubMed] [Google Scholar]
- 26.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Chao SH, et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- 30.Narita T, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu CH, et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adelman K, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci USA. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun J, et al. Deregulation of cofactor of BRCA1 expression in breast cancer cells. J Cell Biochem. 2008;103:1798–1807. doi: 10.1002/jcb.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aiyar SE, Blair AL, Hopkinson DA, Bekiranov S, Li R. Regulation of clustered gene expression by cofactor of BRCA1 (COBRA1) in breast cancer cells. Oncogene. 2007;26:2543–2553. doi: 10.1038/sj.onc.1210047. [DOI] [PubMed] [Google Scholar]
- 37.Gilchrist DA, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aida M, et al. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coulon V, et al. A novel calcium signaling pathway targets the c-fos intragenic transcriptional pausing site. J Biol Chem. 1999;274:30439–30446. doi: 10.1074/jbc.274.43.30439. [DOI] [PubMed] [Google Scholar]
- 40.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 41.Giorgi C, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 42.Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 43.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 44.Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, et al. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171:239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.