Abstract
The Rbfox family of RNA binding proteins regulates alternative splicing of many important neuronal transcripts but their role in neuronal physiology is not clear1. We show here that central nervous system (CNS)-specific deletion of the Rbfox1 gene results in heightened susceptibility to spontaneous and kainic acid-induced seizures. Electrophysiological recording reveals a corresponding increase in neuronal excitability in the dentate gyrus of the knockout mice. Whole transcriptome analyses identify multiple splicing changes in the Rbfox1−/− brain with few changes in overall transcript abundance. These splicing changes alter proteins that mediate synaptic transmission and membrane excitation, some of which are implicated in human epilepsy. Thus, Rbfox1 directs a genetic program required in the prevention of neuronal hyperexcitation and seizures. The Rbfox1 knockout mice provide a new model to study the post-transcriptional regulation of synaptic function.
Alternative pre-mRNA splicing is an important mechanism for regulating gene expression in the CNS, where it both affects neuronal development and controls functions in the mature brain2,3. Developmental and tissue-specific alternative splicing is mediated by cis-acting elements in the pre-mRNA and by corresponding trans-acting protein factors that bind these elements to influence adjacent spliceosome assembly. The RNA-binding Fox (Rbfox) family of splicing factors is comprised of three members, Rbfox1 (Fox-1 or A2BP1), Rbfox2 (Fox-2 or RBM9), and Rbfox3 (Fox-3, HRNBP3 or NeuN). Rbfox proteins regulate splicing of many neuronal transcripts by binding the sequence (U)GCAUG in introns flanking alternative exons4–7. A (U)GCAUG motif that lies downstream of the alternative exon generally promotes Rbfox-dependent exon inclusion, whereas an upstream motif will usually repress exon inclusion4,6,8–12. Human RBFOX1 (A2BP1) was first identified through its interaction with Ataxin-2, the protein mutated in spinocerebellar ataxia type II13. Critical neurological functions for Rbfox1 are indicated by human mutations in the RBFOX1 gene that lead to severe disorders including mental retardation, epilepsy and autism spectrum disorder14–17.
The three mouse Rbfox paralogues show a high degree of sequence conservation, especially within the RNA binding domain, which is identical between Rbfox1 and Rbfox2 and is only slightly altered in Rbfox3 (94% amino acid identity). Rbfox1 is specifically expressed in neurons, heart and muscle6,18, while Rbfox2 is expressed in these tissues plus in other cell types, including the embryo, hematopoetic cells and embryonic stem cells (ESCs)6,19,20. Expression of Rbfox3 is limited to neurons21,22. In the mature brain, most neurons express all three Rbfox paralogues, but certain neuronal subtypes and sporadic individual cells can be seen that express only one or two of the proteins23 (Fig. 1a). All three proteins have similar N- and C-terminal domains that are diversified by alternative promoter use and alternative splicing to produce a cohort of related protein isoforms with variable localization and splicing activities5,24,25. The three proteins apparently share some target exons, including exon N30 of non-muscle myosin heavy chain II-B (NMHC-B), exon N1 of c-src and exons 9* and 33 of the L-type calcium channel Cav1.25,6,26. There are likely other exons that respond to only one Rbfox protein due to differences in their expression patterns or protein-protein interactions.
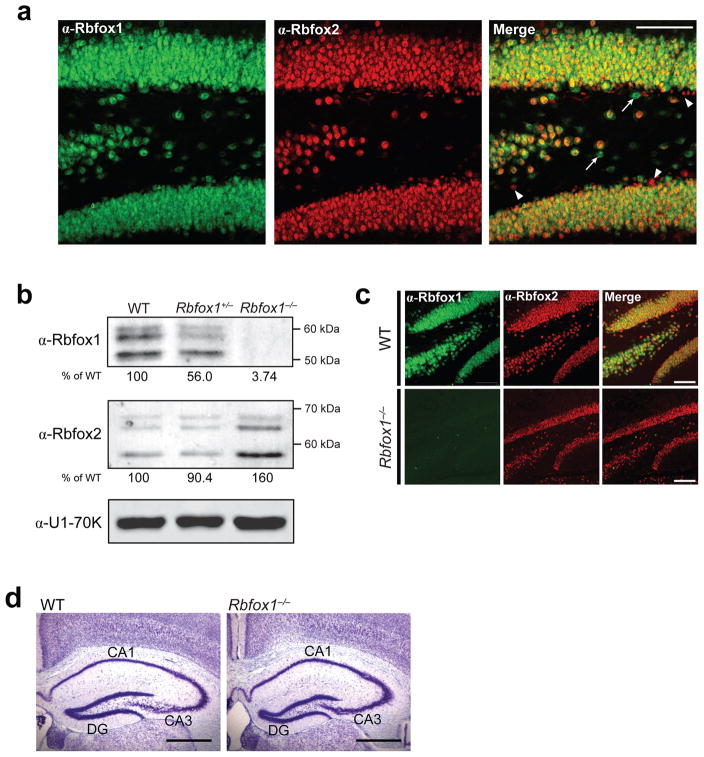
Figure 1. Rbfox1−/− brains lack Rbfox1 protein expression but possess normal morphology.
(a) Confocal immunofluorescence microscopy on coronal sections of wild-type (WT) dentate gryus probed for Rbfox1 (green) and Rbfox2 (red); overlayed images are shown in the far right panels. Arrows point to cells expressing only Rbfox1; arrowheads point to cells expressing only Rbfox2. Scale bar, 100 μm. (b) Immunoblot analysis of Rbfox1 and Rbfox2 in nuclear lysates isolated from WT, Rbfox1+/− and Rbfox1−/− brains. U1-70K was used as a loading control for total nuclear protein. Below each gel is the amount of Rbfox1 or Rbfox2 protein in each sample as a percentage of WT, normalized by U1-70K expression. (c) Confocal immunofluorescence microscopy on coronal sections of WT and Rbfox1−/− dentate gyrus probed for Rbfox1 (green) and Rbfox2 (red) expression. Overlayed images are shown in the far right panels. Scale bar, 100 μm. (d) Representative Nissl stain of WT and Rbfox1−/−hippocampus at 1 mo of age showing normal gross morphology. Scale bar, 0.5 mm. Abbreviations: CA1/CA3, pyramidal layers of the hippocampus; DG, dentate gyrus.
We recently showed that the Rbfox1 transcript itself is alternatively spliced in response to chronic depolarization of cells, resulting in the increased expression of the nuclear, splicing-active Rbfox1 isoform24. The change in nuclear concentration of Rbfox1 then alters the splicing of exons in multiple transcripts affecting neuronal excitation and calcium homeostasis, including the N-methyl D-aspartate (NMDA) receptor 1 (Grin1) and a calcium ATPase (Atp2b1). Thus, chronic stimulation controls the subcellular localization of Rbfox1, leading to an adaptive splicing response24. Together, these findings point to a unique role for Rbfox1 in regulating neuronal gene expression. However, the effect of these Rbfox1-dependent changes on neuronal physiology in the brain is not clear. To examine the effects of Rbfox-dependent splicing within functioning neuronal circuits, we generated mice with CNS-specific deletionof Rbfox1.
Mice carrying conditional Rbfox1 alleles (Rbfox1loxP/loxP; Supplementary Fig. 1) were created using standard methods and crossed with mice carrying the Cre recombinase gene driven by the rat Nestin promoter and enhancer (Nestin-Cre+/−), which express Cre in all CNS stem/neural progenitor cells27,28. The resulting heterozygous Rbfox1loxP/+/Nestin-Cre+/− mice were crossed to Rbfox1loxP/loxP mice to obtain homozygous Rbfox1loxP/loxP/Nestin-Cre+/− animals. Immunoblot analysis confirmed that Rbfox1−/− brains display loss of Rbfox1 protein (Fig. 1b). Interestingly, we observe a 60% increase in Rbfox2 protein expression in the Rbfox1−/− brain but no change in Rbfox3 expression (Fig. 1b and data not shown), suggesting a cross-regulation of Rbfox2 by Rbfox1 that balances overall Rbfox protein levels. By immunofluorescence, we confirmed that Rbfox1 expression is eliminated in the Rbfox1−/−dentate gyrus and verified that localization of the homologous splicing factors Rbfox2 and Rbfox3 are unchanged in these cells (Fig. 1c and data not shown). Rbfox1loxP/loxP/Nestin-Cre+/− mice are viable, but are slightly smaller and have reduced fertility compared to Rbfox1+/− or wild-type littermates (data not shown). Histological analysis by Nissl staining at various postnatal stages revealed that Rbfox1−/− brain gross morphology is normal, with no obvious anomalies in cellular architecture (Fig. 1d).
In observing the mutant mice, we found that they are prone to infrequent, spontaneous seizures under standard housing conditions. These seizures are accompanied by induction of the immediate early gene c-fos, an indirect marker for neuronal activity29. One hour after spontaneous seizure, c-Fos protein is highly expressed in the Rbfox1−/− amygdala and hippocampus (Fig. 2a), the primary areas affected in Mesial Temporal Lobe Epilepsy (MTLE)30. To determine if these mice also show increased susceptibility to induced seizures, we challenged them with systemic administration of kainic acid (KA), a well-described method for modeling MTLE31. The behavioral response of wild-type mice, homozygous Rbfox1loxP/loxP/Nestin-Cre+/− mice, and heterozygous Rbfox1loxP/+/Nestin-Cre+/− mice was continuously evaluated by the Racine Scale32 over a two hour period after KA administration (20 mg/kg, i.p.; Fig. 2b and data not shown). During this period, eight out of ten wild-type mice displayed relatively short tonic-clonic seizures (12.4±2.8 s). None of these mice reached stage six, defined as status epilepticus, and all survived the treatment (Fig. 2b and Supplementary Table 1). In contrast, Rbfox1loxP/loxP/Nestin-Cre+/− mice showed a dramatic response to KA, with four out of four mice reaching status epilepticus culminating in death in under 40 minutes. The mean tonic-clonic seizure duration was also significantly higher in these mice than in wild-type littermates (73.2±16.8 s; P=0.032). Interestingly, heterozygous Rbfox1loxP/+/Nestin-Cre+/− mice showed a KA sensitivity almost identical to that of homozygous mice, with four out of four dying in under 40 minutes (Supplementary Table 1). A heterozygous deletion of RBFOX1 was found in a human subject with epilepsy14; thus, increased susceptibility to seizure is caused by deletion of just one of the two copies of Rbfox1 in mice and apparently humans. Notably, Rbfox2loxP/loxP/Nestin-Cre+/− mice do not show susceptibility to spontaneous or KA-induced seizures (Fig. 2b and Supplementary Table 1), indicating that Rbfox1 and Rbfox2 are not fully redundant in their neuronal functions. The different phenotypes of the Rbfox1 and Rbfox2 knockout mice likely result, at least in part, from differences in their target exon sets. However, the infrequent neurons expressing only one protein or the other could also contribute to the observed phenotypic differences.
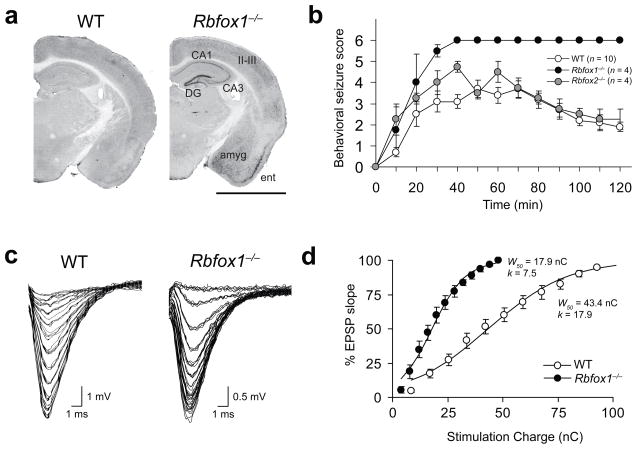
Figure 2. Rbfox1−/− brains are epileptic and hyperexcitable.
(a) c-Fos immunostaining on WT and Rbfox1−/− coronal brain sections 1 h after a spontaneous seizure in the Rbfox1loxP/loxP/Nestin-Cre+/−mouse. Relevant brain areas are indicated. Abbreviations: amyg, amygdala; CA1/CA3, pyramidal cell layers of the hippocampus; DG, dentate gyrus; ent, entorhinal cortex; II-III indicate layers of the cerebral cortex. Scale bar, 2 mm. (b) Progression of behavioral changes after systemic KA administration (20mg/kg, i.p.) in WT, Rbfox1loxP/loxP/Nestin-Cre+/−, and Rbfox2loxP/loxP/Nestin-Cre+/−mice over a 2 h observation period. Seizures were scored on the Racine Scale as described31 and data shown are mean scores. Error bars, s.e.m. (c) Representative fEPSP traces from individual electrophysiological recordings in WT and Rbfox1−/− dentate gyrus. (d) Average synaptic I/O curves in WT and Rbfox1−/− dentate fit with a Boltzmann function (solid lines). Circles are grand-averaged scores; error bars, s.e.m. W50, stimulus width that elicits 50% of the maximum response; k, slope factor. n = 3 mice, 16–20 slices per experimental group.
To explore the physiological basis of these seizure data, we investigated changes in neuronal excitability by analyzing the synaptic input/output (I/O) relationship in wild-type and Rbfox1−/− dentate gyrus. Field excitatory postsynaptic potentials (fEPSPs) were measured in the molecular layer in response to stimulation of the lateral perforant path using varying stimulus widths. We found that the stimulus intensities required to evoke fEPSPs in the Rbfox1−/− brain were lower than those required for wild-type brain, as shown by representative fEPSP raw traces (Fig. 2c) and average I/O curves (Fig. 2d). The grand average I/O relationships for individual experiments were fit with a Boltzmann function, where W50 is the stimulus width that elicits the half-maximal response and k is the slope factor. Consistent with an increase in network excitability, Rbfox1−/− slices exhibit a shift to the left in the I/O relationship and a decrease in the W50 value to 41% that of wild-type slices (Fig. 2d). Thus, the loss of Rbfox1 causes an increase in excitability of the neuronal population of the dentate gyrus.
To determine if the increase in excitability results from an increase in synapse number, we quantified spine density along granule cell dendrites of the dentate gyrus using the Golgi Cox staining method. We found a very modest decrease in spine density along Rbfox1−/− dendrites compared to wild-type dendrites (Supplementary Fig. 2a c). Similarly, the Rbfox1−/− hippocampus showed no changes in expression of the synaptic marker proteins Synapsin-I and PSD-95 (Supplementary Fig. 2d,e). Thus, altered synaptic function, rather than synapse number, is likely responsible for the increased neuronal excitability of the Rbfox1−/− brain.
To survey the changes in exon inclusion in the Rbfox1−/− brain compared to wild-type, we used Affymetrix exon-junction (MJAY) microarrays to assay transcript abundance and alternative splicing across the genome (Supplementary Table 2; full dataset is available online at Gene Expression Omnibus (GEO), at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28421). The MJAY arrays offer broad coverage but do not have probes for all exons, so to extend the analysis, we also directly assayed by RT-PCR candidate exons that are known to be regulated by Rbfox1 or to be present in genes implicated in epilepsy. Combining the RT-PCR validated array exons and directly tested candidate exons, we identified a total of 20 statistically significant splicing changes in the Rbfox1−/− brain compared to wild-type (Table 1 and Supplementary Fig. 3). This list is not comprehensive, but compared to mice lacking other splicing factors such as Nova33 or nPTB (unpublished data), Rbfox1 knockout mice show relatively few splicing changes in the brain. This may result from compensation for the loss of Rbfox1 by Rbfox2, which is upregulated in the Rbfox1−/− brain (Fig. 1b). The heterozygous Rbfox1+/− brain exhibits many of the splicing changes observed in the Rbfox1−/− brain, but these changes are generally smaller in magnitude and show large variation between biological replicates (data not shown). Many exons altered in Rbfox1−/− brain also change in the Rbfox2−/− brain, as expected from the overlap in their targets. However, some exons, including exons of the NMDA receptor 1 (Grin1), the voltage-gated potassium channel Kv4.3 (Kcnd3), and the L-type calcium channel Cav1.3α1 (Cacna1d), are more responsive to loss of Rbfox1 than Rbfox2 (Fig. 3a). Since Rbfox2loxP/loxP/Nestin-Cre+/− mice are not prone to seizures, transcripts more strongly controlled by Rbfox1 may have a crucial effect on the hyperexcitation phenotype.
Table 1.
Summary of differentially spliced exons in Rbfox1−/− brain.
| Alt event ID* | Δ PSI** (Mean ± s.e.m.) | P value† | Upstream (U)GCAUG‡ | Downstream (U)GCAUG‡ | Function*** | |
|---|---|---|---|---|---|---|
| 1 | Stx3_(46) | −33.44±3.65 | 0.00587 | (−101, −74) | (+25, +185, | SNARE complex |
| 2 | Snap25_(118_118) | −24.24±3.27 | 0.00886 | n/p_n/p | n/p_(+94, +101, | SNARE complex |
| 3 | Grin1_(63) | −23.29±2.66 | 0.00641 | n/p | (+8, +262) | Neurotransmitter receptor |
| 4 | Camta1_(31) | −22.16±1.94 | 0.00378 | n/p | (+70, +210, +252) | Calmodulin binding |
| 5 | Cacna1s_(57) | −21.46±2.61 | 0.00724 | n/p | (+13, +44, +60, +69, +165) | Ion channel |
| 6 | Cacna1d_(60) | −20.64±5.03 | 0.02726 | (−47) | n/p | Ion channel |
| 7 | Gabrg2_(24) | −13.73±2.14 | 0.01173 | n/p | (+30) | Neurotransmitter receptor |
| 8 | Scn8a_(92_92) | −12.33±0.81 | 0.00216 | n/p_n/p | n/p_(+114, +192) | Ion channel |
| 9 | Camk2g_(68) | −11.30±1.13 | 0.00496 | (−264, −65) | (+8, +65, +120, +190) | Calmodulin binding |
| 10 | Nrxn3_(27) | −7.61±1.30 | 0.01400 | n/p | n/p | Synapse assembly |
| 11 | Ablim1_(120) | −6.93±0.93 | 0.00871 | (−255) | (+123, +171) | Cytoskeletal dynamics |
| 12 | Cadps_(147) | −6.11±1.49 | 0.02745 | (−227) | (+49) | SNARE complex |
| 13 | Tpm3_(79) | +1.92±0.31 | 0.01277 | (−260) | n/p | Cytoskeletal dynamics |
| 14 | Nrcam_(57) | +5.46±0.69 | 0.00773 | (−136) | (+171) | Synapse assembly |
| 15 | Ptpro_(84) | +6.97±2.22 | 0.04400 | n/p | n/p | Tyr phosphatase |
| 16 | Cacna1d_(104_104) | +7.63±2.35 | 0.04180 | (−225, − 191)_n/p | n/p_(+93) | Ion channel |
| 17 | Ppp3ca_(30) | +8.55±1.97 | 0.02465 | (−188) | n/p | Calmodulin binding |
| 18 | Pbrm1_(156) | +13.22±2.52 | 0.01729 | (−78, −44, − 31) | (+233) | Chromatin binding |
| 19 | Stxbp5l_(72) | +13.57±4.51 | 0.04757 | (−13) | (+130) | SNARE complex |
| 20 | Kcnd3_(57) | +28.17±7.63 | 0.03233 | (−16) | (+83) | Ion channel |
The number in parenthesis after the gene ID indicates the size in nt of the alternative exon. All events listed are alternative cassette exons, except those with two exon sizes (i.e., Snap25_(118_118)), which are paired mutually exclusive exons.
ΔPSI, percent change in alternative exon inclusion for Rbfox1−/− brain compared to WT, determined by RT-PCR. For mutually exclusive exons, the value given is for the downstream exon.
RT-PCR P value was determined by paired, one-tailed Student’s t test (n = 3).
Location of (U)GCAUG binding sites in the proximal 300 nt upstream and downstream of the alternative exon are shown with distance in nt in parenthesis. n/p, not present.
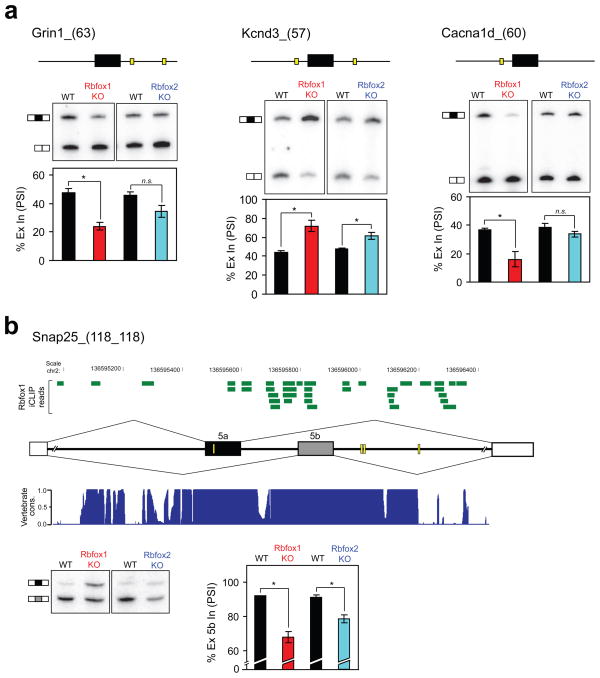
Figure 3. Rbfox1−/− brain exhibits splicing changes in transcripts affecting synaptic function and neuronal excitation.
(a) Denaturing gel electrophoresis of RT-PCR products for three Rbfox1-dependent exons. Above each gel is a schematic depicting the alternative exon (horizontal black box) and the relative location of (U)GCAUG binding sites (yellow boxes) in the flanking introns (thin horizontal lines). Shown below each gel is a graph quantifying the mean percentage of alternative exon inclusion (% Ex In, PSI) in WT (black bars), Rbfox1 knockout (KO; red bars), and Rbfox2 KO (blue bars) brain. (b) A schematic showing the Snap25 mutually exclusive exon pair, 5a (horizontal black box) and 5b (horizontal gray box), plus the intervening intron and the promixal 500 nt of the adjacent introns. Yellow boxes represent (U)GCAUG motifs. The distribution of Rbfox1 iCLIP reads (horizontal green bars) is shown above. A histogram displaying the conservation of this region among 30 vertebrate species, as determined by phastCons (http://genome.ucsc.edu), is shown below. A score of 1 indicates 100% identity among all species at that nt position. Chromosomal location in nt is shown above the iCLIP data. At the bottom, the RT-PCR assay and quantification of exons 5a and 5b splicing is shown, with the inclusion of the downstream exon plotted. For (a) and (c), error bars represent s.e.m.; n = 3. *P<0.05 and n.s. = not significant by paired, one-tailed Student’s t test. Exact P values are shown in Table 1.
Nearly all of the identified exons possess (U)GCAUG motifs in their flanking introns, and are likely to be directly regulated by Rbfox1. In total, 85% of exons (11 out of 13) showing reduced inclusion in the Rbfox1−/− brain contain a (U)GCAUG within the proximal 300 nucleotides downstream of the exon - consistent with a loss of Rbfox1 activation; 86% of exons (6 out of 7) showing increased inclusion possess a Rbfox binding site within 300 nucleotides upstream - consistent with a loss of Rbfox1 repression (Table 1). This represents a substantial enrichment of Rbfox binding motifs in the knockout-responsive exons. Of the 5166 alternative cassette exons probed by the MJAY array, only 1590 (31%) possess a (U)GCAUG motif within 300 nucleotides downstream and only 1328 (26%) possess this motif within 300 nucleotides upstream. Characterization of Rbfox1 binding sites in mouse cortex using individual-nucleotide resolution crosslinking immunoprecipitation (iCLIP)12,34 confirms that a majority of these exons exhibit adjacent Rbfox1 binding in vivo, and are thus direct targets of the protein (Figure 3b and data not shown). However, these analyses require more data before a genomewide map of Rbfox1 binding can be developed.
Notably, there were minimal changes in the overall expression of genes across the array. Besides the expected loss of Rbfox1 mRNA, only two significant changes in transcript abundance were detected in the Rbfox1−/− brain (Pop4 and AK138161, encoding a hypothetical protein expressed in mouse hypothalamus). The effect of Rbfox1 is primarily post-transcriptional. We also examined the protein expression of several well-characterized splicing factors in the Rbfox1−/− brain. Most of the proteins tested, including Ptbp1 (PTB) and Mbnl1, were unchanged in the Rbfox1−/− brain. The neuron-specific splicing factor Nova1 was moderately decreased (Supplementary Fig. 4). Rbfox1 apparently affects the post-transcriptional regulation of Nova1, with loss of Rbfox1 resulting in reduced Nova1 expression. This is interesting in light of previous findings of a significant overlap between the Nova1 and Rbfox1 protein regulatory networks35. It is also notable that Rbfox1 targets are enriched in synaptic functions, similar to Nova1. The potential cross-regulation of these factors will likely be another connection between their regulatory programs.
The splicing changes in the Rbfox1−/− brain affect multiple functional components of synaptic transmission that could clearly contribute to the epileptic phenotype (Table 1). Transcripts for ion channels such as the voltage-gated potassium channel Kv4.3 (Kcnd3) and the L-type calcium channel subunits Cav1.1α (Cacna1s) and Cav1.3α1 (Cacna1d) were significantly altered in the Rbfox1−/− brain. Transcripts encoding neurotransmitter receptors such as the gamma-aminobutyric acid (GABA)-A receptor subunit γ2 (Gabrg2) and the NMDA receptor 1 (Grin1) were also changed in the Rbfox1−/−brain. We also found splicing changes in transcripts for structural proteins of the synapse, such as neurexin 3 (Nrxn3) and for vesicle fusion proteins including syntaxin 3 (Stx3), synaptosomal-associated protein 25 (Snap25), and the calcium-dependent secretion activator (Cadps). Changes in intracellular calcium dynamics are associated with epileptogenesis, including alterations in calcium entry through voltage-gated channels or changes in calcium buffering by calcium-binding proteins36. In addition to the splicing changes in the voltage-gated channels mentioned above, we identified changes in transcripts for calcium/calmodulin-dependent protein kinase II (Camk2g), calmodulin binding transcription activator (Camta1), and calmodulin-dependent calcineurin subunit (Ppp3ca). Disruption of calcium dynamics may contribute to the epileptic phenotype observed in Rbfox1loxP/loxP/Nestin-Cre+/−mice.
Most interestingly, a number of the Rbfox1 target transcripts have been directly linked to epilepsy (Table 2). Mutations in the Gabrg2 gene have been identified in human epilepsy patients37, and changes in expression of NMDA receptor 1 (Grin1) or the voltage-gated sodium channel Nav1.6 (Scn8a) alter seizure susceptibility in mice38–40. Transgenic mouse models have revealed that changes in expression or alternative splicing of Snap25 result in seizures41,42. In particular, it was found that the splicing ratio of a pair of Snap25 mutually exclusive exons is developmentally regulated and that removal of the exon 5b predominantly expressed in adult brain results in spontaneous seizures42. We find that the splicing of this same adult-specific exon is reduced by 24% in the Rbfox1−/− brain and reduced by a much lesser extent (12%) in the Rbfox2−/− brain (Table 1 and Fig. 3b). We also identified a number of Rbfox1 iCLIP reads surrounding (U)GCAUG motifs adjacent to exon 5b (Fig. 3b), indicating that Rbfox1 directly regulates the splicing of this exon. The change in Rbfox1-dependent Snap25 splicing likely contributes to the seizure phenotype of Rbfox1 knockout mice.
Table 2.
Summary of transcripts altered in Rbfox1−/− brain with implications in epilepsy.
| Gene | Protein | Implications in epilepsy | Splicing change in Rbfox1 KO brain | Effect of splicing change on encoded protein | |
|---|---|---|---|---|---|
| 1 | Gabrg2 | GABA-A receptor γ2 | Mutations identified in patients37 | Decrease in exon 9 inclusion | Removes a protein kinase C phosphorylation site that modulates receptor function |
| 2 | Grin1 | NMDA receptor 1 | Downregulation reduces seizure susceptibility in mice38,39 | Decrease in exon 5 inclusion | Removes a portion of the extracellular domain, altering the receptor’s response to ligands |
| 3 | Scn8a | Na(v)1.6 sodium channel | Mouse mutations exhibit seizures40 | Decrease in exon 5A | Modifies transmembrane segments S3 and S4 of domain I; results in decreased expression of the adult isoform |
| 4 | Snap25 | SNAP-25 | Mouse mutations exhibit seizures41 | Decrease in exon 5b | Modifies the N-terminus; results in decreased expression of the adult isoform |
In summary, our findings demonstrate a critical role of Rbfox1 in neuronal function. We previously showed that changes in Rbfox1 activity in response to chronic stimuli can homeostatically adjust the level of splicing for its target exons. The limited response of Rbfox2 and Rbfox3 to depolarizing media (unpublished observations) and the abnormal physiology of the Rbfox1 knockout brain indicate that these proteins are not fully redundant in their functions, and that Rbfox1 plays a unique role in the cellular response to overexcitation. The Rbfox1 knockout mice provide a new model for examining how modifications of synaptic proteins determine excitation and epileptogenesis. Splicing regulation by Rbfox1 is clearly affecting neuronal excitation. However, the presence of cytoplasmic Rbfox1 protein and its binding to 3′ UTRs indicates that there will be other components to the Rbfox1−/− phenotype. Examination of the larger nuclear and cytoplasmic programs of post-transcriptional regulation by Rbfox1 should yield new insights into the control of cellular homeostasis and its misregulation in neurological disease.
MATERIALS AND METHODS
Mice
We used homologous recombination to create “floxed” Rbfox1 alleles consisting of loxP sites flanking Rbfox1 exons 11 and 12, annotated as previously described25. Southern blot hybridization probes were generated by PCR amplification (Supplementary Table 3), cloned using the TOPO TA cloning kit (Invitrogen), and labeled by PCR in the presence of α-32P-dCTP. Mouse strain 129S6/SVEvTac BAC library (RPCI-22), arrayed on high density nylon filters, was obtained from Children’s Hospital Oakland Research Institute (CHORI) and probe Rbfox1_knpr_frt was used to screen the BAC library. Positive clones were purchased from CHORI and validated by BAC end sequencing. Clone RP22-344J23 was used to construct the “floxed” Rbfox1 targeting cassette also carrying a neomycin resistance (neo) cassette flanked by Frt sites43 and the primer sequences used for recombineering are listed in Supplementary Table 3. The targeting cassette was transfected into 129S2/Sv embryonic stem cells at the University of California, San Diego (UCSD) Transgenic and Gene Targeting Core. Clones that had undergone homologous recombination were identified by Southern blot using the 5′ and 3′ probes after digesting the genomic DNA with SwaI and SpeI (Supplementary Fig. 1). Cells from positive clones were injected into a C57BL/6J blastocyst at the UCSD transgenic and gene targeting core, and one line was found to have germline transmission. Heterozygous (Rbfox1loxP/+) F1 offspring were crossed to transgenic mice expressing Flp recombinase44 to excise the neo cassette. Resulting Heterozygous (Rbfox1loxP/+) mice lacking the neo cassette were intercrossed to produce homozygous (Rbfox1loxP/loxP) mice. Homozygous mice were then crossed to transgenic Nestin-Cre+/− mice, allowing for restricted deletion of Rbfox1 in the CNS of double transgenic mice (Rbfox1loxP/loxP/Nestin-Cre+/−). Primers used for genotyping the Rbfox1+, Rbfox1loxP and Rbfox1Δ alleles are listed in Supplementary Table 3 and product sizes are given in Supplementary Fig. 1. Mice used for this study were maintained on a mixed 129S2/Sv × C57BL/6J background. Animals were housed in a 12-h light/dark cycle with food and water available ad libitum and were maintained by the University of California, Los Angeles (UCLA) Association for Assessment and Accreditation of Laboratory Animal Care accredited Division of Laboratory Medicine. All experiments were Institutional Animal Care and Use Committee approved by the UCLA Chancellor’s Animal Research Council.
Western blotting
Nuclei were isolated from wild-type, Rbfox1+/− and Rbfox1−/− brains as previously described45. Nuclei were lysed in Lysis Buffer (20 mM Hepes-KOH, pH 7.9, 300 mM NaCl, 1 mM EDTA, 0.75% NP-40) containing complete protease inhibitors (PI; Roche) for 10 min on ice. For quantification of synaptic proteins or splicing factors, hippocampi or total brains, respectively, were dissected from wild-type and Rbfox1loxP/loxP/Nestin-Cre+/− mice and homogenized by sonication in RIPA buffer containing PI. Nuclear and total protein samples were cleared, boiled in SDS loading buffer, and resolved on 10% Tris-glycine gels. Antibodies were used at the following dilutions: α-Rbfox1 1D1024, 1:2000; α-Rbfox2, 1:2000 (Bethyl); α-U1-70K, 1:5000; α-Synapsin-I, 1:2000 (Millipore); α-PSD-95, 1:2000 (Millipore); α-GAPDH, 1:10,000 (Ambion); α-PTB NT4856, 1:2500; α-nPTB IS2, 1:3000; α-Mbnl1, 1:1000 (Millipore); α-Nova-1, 1:1000 (Millipore); α-hnRNPA1, 1:1000; α-SRp20, 1:1500 (Zymed); α-β-tubulin, 1:100 (Santa Cruz); and ECL Plex Cy3-conjugated goat α-mouse and Cy5-conjugated goat α-rabbit secondary antibodies, 1:2500 (GE Healthcare).
Histology and immunohistochemistry
Rbfox1loxP/loxP/Nestin-Cre+/− and wild-type littermates were transcardially perfused with ice-cold 0.1 M phosphate buffered saline (PBS; pH 7.4), followed by ice-cold 4% paraformaldehyde in PBS (pH 7.4). Brains were cryoprotected in 30% sucrose and 40 μm free-floating sections were cut in the coronal orientation using a Microm HM505E cryostat. For Nissl histology, sections were hydrated and stained using 0.25% thionin acetate (Sigma), dehydrated through alcohols, cleared in xylenes, and mounted with DPX (Electron Microscopy Sciences). For double immunofluorescent staining, sections were blocked 30 min with blocking solution (10% normal goat serum, 0.1% Triton X-100 in PBS) and incubated with primary antibodies at 4°C overnight. Sections were washed three times and stained with secondary antibodies in PBS for 2 h. Sections were again washed three times, mounted with ProLong Gold plus DAPI reagent (Invitrogen) and imaged using a Zeiss LSM 510 Meta confocal microscope. The following primary antibodies were used: α-Rbfox1 1D10, 1:200; α-Rbfox2, 1:200 (Bethyl); α-c-Fos Ab-5, 1:1600 (Calbiochem). Alexa Fluor 488-conjugated goat α-mouse IgG and Alexa Fluor 568-conjugated goat α-rabbit IgG secondary antibodies (Molecular Probes) were used at 1:1000. For Golgi-Cox staining, wild-type and Rbfox1−/− brains were processed using the FD Rapid GolgiStain Kit (FD Neurotechnologies) according to the manufacturer’s instructions and counterstained with thionin acetate. For spine quantification, the analyzer was blind to the experimental group throughout the analysis. Dendrites of the same size in diameter from dentate gyrus granule cells were randomly selected from 4 slices per individual animal. A total of 28–29 dendritic segments from 2 animals per genotype were used for quantification. Images were collected under a 100× oil-immersion objective using the Spot RT KE camera and software system (Diagnostic Instruments). Spines were operationally defined as any protrusion less than 5 μm in length. Each spine was manually traced and dendritic length was measured using ImageJ software. The spine numbers were expressed as mean spine density per 10 μm of linear dendritic length. For all histology and immunofluorescence experiments, images shown are representative of at least 2 independent samples.
Behavioral observation of KA-induced seizures
KA (Sigma) was dissolved in sterile saline and administered i.p. at 20 mg/kg body weight. Ten wild-type mice, 4 heterozygous Rbfox1loxP/+/Nestin-Cre+/− mice, 4 homozygous Rbfox1loxP/loxP/Nestin-Cre+/−mice, and 4 homozygous Rbfox2loxP/loxP/Nestin-Cre+/− mice (2–3 mo of age) were used. In all experiments, the experimenter was blind to the genotype of the animals and each experiment used at least one wild-type littermate. Seizure severity was determined according to the Racine Scale32: stage 0: normal behavior; stage 1: mouth and facial movements; stage 2: head bobbing; stage 3: forelimb clonus; stage 4: rearing; stage 5: continuous rearing and falling (tonic-clonic seizures); stage 6: status epilepticus and/or death. For each animal, the seizure score was determined every 10 min for 2 h after KA administration. Each animal’s maximum score for each 10 min interval was used to calculate the mean seizure score (± s.e.m.) for each experimental group. Latency to first stage 5 (tonic-clonic) seizure, duration of each stage 5 seizure and time to death were measured from time of KA administration. Statistical significance was determined using one-tailed Student’s t test. The level of significance was set to P<0.05.
Electrophysiology
To obtain synaptic input/output curves, hippocampal field potential recordings were carried out as previously described46. Coronal hippocampal slices (350 μm thick) were prepared from 3 adult (2–3 mo old) wild-type and 3 Rbfox1loxP/loxP/Nestin-Cre+/− mice. Hippocampal field recordings were performed at 32–34°C under standard conditions in an interface chamber perfused with normal artificial cerebral spinal fluid (nACSF) solution containing (in mM): 126 NaCl, 2.5 KCl, 2 CaCl2, 1–2 MgCl2, 1.25 NaHPO4, 26 NaHCO3, and 10 D-glucose, pH 7.3–7.4 (bubbled with 95% O2 and 5% CO2. Field potentials were evoked by lateral perforant path stimulation at 0.05 Hz, and responses were recorded in the dentate gyrus molecular layer. Bipolar electrodes were used to deliver a constant-current stimulus. The stimulus intensity was determined by recording a threshold response at a width (W) of 60 μs with no response at 20 μs. The W of the stimulus was increased stepwise by 20 μs to create I/O curves ranging from 20 to 240 μs. Four population field EPSPs (fEPSPs) were recorded at each W, and the maximum slope of the responses (volts per second) was measured over a 0.5–1 ms window of the fEPSP rising phase. The average slope was calculated at each W used. Input/output curves were fit with a Boltzmann equation: f(W) = (MAX/(1 + exp((W - W50)/k)) + MAX), where W is stimulus width, MAX is the maximum response, k is a slope factor, and W50 is the stimulus width that elicits 50% of MAX. The fitted parameters were compared between experimental groups, and statistical significance was determined using a one-way ANOVA, followed by Bonferroni’s corrected t-tests.
Splicing microarrays
Total RNA was extracted by Trizol® reagent (Invitrogen) from whole brain of 3 wild-type 3 and Rbfox1loxP/loxP/Nestin-Cre+/− mice, all 1-mo-old males. Ribosomal RNAs were removed from samples using the RiboMinus Transcriptome Isolation Kit (Invitrogen) according to the manufacturer’s instructions. Amplified, biotinylated cDNA target was produced using the GeneChip® Whole Transcript Sense Target Labeling and Control Reagents kit (Affymetrix) according to the manufacturer’s instructions. Each sample target was hybridized overnight to a Mouse GeneSplice Array (Affymetrix PN 540092). Hybridized arrays were processed using the Affymetrix Fluidics Station 450 and scanned with an Affymetrix GeneChip scanner. Data were analyzed and each alternative event was assigned an MJAY ratio, a measure of the difference in the average ratio of inclusion to skipping for the indicated exon in the Rbfox1−/− samples compared to wild-type, as previously described47. The top 25 significant changes, ranked in descending order by the absolute value of the MJAY ratio, of alternative cassettes and mutually exclusive exons are shown in Supplementary Table 2.
RT-PCR assay
Total RNA was extracted by TrizolR reagent (Invitrogen) from whole brain of 1-mo-old wild-type, Rbfox1loxP/loxP/Nestin-Cre+/−, and Rbfox2loxP/loxP/Nestin-Cre+/− mice (n = 3 for each genotype). One μg of total RNA was reverse-transcribed with random hexamers. One-tenth of this reaction was then amplified in 24 cycles of PCR with exon-specific primers, one of which was 32P-labeled. The PCR products were resolved on 8% polyacrylamide/7.5 M urea denaturing gels. The gel was dried, exposed, and scanned in a Typhoon 9400 PhosphorImager scanner (GE Healthcare). Images were analyzed with ImageQuant TL Software. Mean exon inclusion levels were calculated for wild-type and Rbfox1−/−brains, and the percent change in inclusion (ΔPSI) was calculated by subtracting the Rbfox1−/− mean inclusion level from the wild-type mean inclusion level. Statistical significance was determined using the paired, one-tailed Student’s t test with significance set to P<0.05. RT-PCR primers are listed in Supplementary Table 3.
Rbfox1-RNA crosslinking immunoprecipitation analysis (Rbfox1 iCLIP)
The Rbfox1 iCLIP was done as previously described34 with the following modifications: cortices from 1.5-mo-old male and female wild-type C57BL/6J mice were dissected, triturated in 8 ml ice cold HBSS with 20 mM HEPES-KOH pH 7.5, and UV irradiated in a Stratalinker 1800 (Stratagene) at 750 mJ/cm2. Rbfox1 was immunoprecipitated with 1D10 antibody and washed with high salt buffer. The immobilized crosslinked RNA was fragmented as previously described12: aliquots of beads in 50 μl of 1× micrococcal nuclease buffer with 5.0 μg yeast tRNA were treated with 5 or 2,000 Kunz U/ml of micrococcal nuclease (New England Biolabs) for 5 min at 37°C. The enzyme was inhibited by addition of 0.5 ml 1× PNK buffer + EGTA. The RNA fragments were dephosphorylated for 1 h to remove the 3′-phosphorylates left by micrococcal nuclease. The RNA linker contained 3′ biotin instead of the puromycin group used previously34. Since Rbfox1 co-migrates with the IgG heavy chain, the crosslinked protein-RNA-linker products were purified on monomeric avidin beads (Fischer Scientific) before resolving on 10% NuPAGE gel (Invitrogen). The constructed Rbfox1 iCLIP library was sequenced on a Genome Analyzer IIx (Illumina) with a 76 nt read length. Mapping of sequence reads was performed against the mouse genome (version mm9/NCBI37) using Bowtie version 0.12.548 as previously described34.
Supplementary Material
Acknowledgments
This work was done in collaboration with Dr. Xiang-Dong Fu (UCSD). We thank Dr. Neal Copeland (IMCB, Singapore) for the recombineering vectors and bacterial strains used for generating the transgenic Rbfox1 mice, and John Paul Donahue for his help with the microarray analyses. Drs. Dan Geschwind, Kelsey Martin, and Tim Nielsen gave us helpful comments on the manuscript. This work was supported in part by NIH Grants R01 GM049369 to X.D.F., R37 NS30549 and R01 MH076994 to I.M., R01 GM084317 to M.A.Jr and D.L.B., and R01 GM49662 to D.L.B. D.L.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
Project conception: D.L.B., P.S. and L.T.G. Creation of transgenic mice: P.S. Phenotypic analysis, histology, immunofluorescence and RT-PCR studies: L.T.G. Behavioral seizure analyses: J.M., L.T.G. and I.M. Electrophysiology: J.M. and I.M. iCLIP study: A.D., C.H.L. Microarray studies: L.S., L.T.G. and M.A.Jr. Manuscript preparation: L.T.G. and D.L.B.
Competing interests statement: The authors declare that they have no competing financial interests.
References
- 1.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 3.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/mcb.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black DL. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 9.Huh GS, Hynes RO. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 10.Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 14.Bhalla K, et al. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- 15.Barnby G, et al. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin CL, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 17.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiehl TR, Shibata H, Vo T, Huynh DP, Pulst SM. Identification and expression of a mouse ortholog of A2BP1. Mamm Genome. 2001;12:595–601. doi: 10.1007/s00335-001-2056-4. [DOI] [PubMed] [Google Scholar]
- 19.Ponthier JL, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- 20.Yeo GW, et al. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKee AE, et al. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213×-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA. 2010;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol. 2009;29:4757–4765. doi: 10.1128/mcb.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 28.Graus-Porta D, et al. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 29.Herrera DG, Robertson HA. Activation of c-fos in the brain. Prog Neurobiol. 1996;50:83–107. doi: 10.1016/s0301-0082(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 30.Bertram EH. Temporal lobe epilepsy: where do the seizures really begin? Epilepsy Behav. 2009;14(Suppl 1):32–37. doi: 10.1016/j.yebeh.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Ari Y. Limbic seizure and brain damage produced by kainic acid: mechanisms and relevance to human temporal lobe epilepsy. Neuroscience. 1985;14:375–403. doi: 10.1016/0306-4522(85)90299-4. [DOI] [PubMed] [Google Scholar]
- 32.Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- 33.Ule J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 34.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, et al. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delorenzo RJ, Sun DA, Deshpande LS. Cellular mechanisms underlying acquired epilepsy: the calcium hypothesis of the induction and maintainance of epilepsy. Pharmacol Ther. 2005;105:229–266. doi: 10.1016/j.pharmthera.2004.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulley JC, Scheffer IE, Petrou S, Berkovic SF. Channelopathies as a genetic cause of epilepsy. Curr Opin Neurol. 2003;16:171–176. doi: 10.1097/01.wco.0000063767.15877.c7. [DOI] [PubMed] [Google Scholar]
- 38.Chapman AG, Woodburn VL, Woodruff GN, Meldrum BS. Anticonvulsant effect of reduced NMDA receptor expression in audiogenic DBA/2 mice. Epilepsy Res. 1996;26:25–35. doi: 10.1016/s0920-1211(96)00036-8. [DOI] [PubMed] [Google Scholar]
- 39.Zapata A, et al. Effects of NMDA-R1 antisense oligodeoxynucleotide administration: behavioral and radioligand binding studies. Brain Res. 1997;745:114–120. doi: 10.1016/s0006-8993(96)01134-1. [DOI] [PubMed] [Google Scholar]
- 40.Papale LA, et al. Heterozygous mutations of the voltage-gated sodium channel SCN8A are associated with spike-wave discharges and absence epilepsy in mice. Hum Mol Genet. 2009;18:1633–1641. doi: 10.1093/hmg/ddp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corradini I, Verderio C, Sala M, Wilson MC, Matteoli M. SNAP-25 in neuropsychiatric disorders. Ann N Y Acad Sci. 2009;1152:93–99. doi: 10.1111/j.1749-6632.2008.03995.×. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansson JU, et al. An ancient duplication of exon 5 in the Snap25 gene is required for complex neuronal development/function. PLoS Genet. 2008;4:e1000278. doi: 10.1371/journal.pgen.1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- 45.Grabowski PJ. Splicing-active nuclear extracts from rat brain. Methods. 2005;37:323–330. doi: 10.1016/j.ymeth.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Maguire J, Ferando I, Simonsen C, Mody I. Excitability changes related to GABAA receptor plasticity during pregnancy. J Neurosci. 2009;29:9592–9601. doi: 10.1523/jneurosci.2162-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugnet CW, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.