Abstract
Lack of safe and effective carriers for delivery of RNA therapeutics remains a barrier to its broad clinical application. We report the development of a cell tanscytosing magnetic nanovector engineered as an siRNA carrier. Iron oxide nanoparticles were modified with poly(ethylene glycol) (PEG), small interfering RNA (siRNA), and a catinionic polymer layer. Three nanovector formulations with cationic polymer coatings of poly arginine (pArg), polylysine (pLys), and polyethylimine (PEI), respectively, were prepared. The three nanovector formulations where evaluated for safety and ability to promote gene silencing in three types of cancer cells C6/GFP+, MCF7/GFP+, and TC2/GFP+, mimicing human cancers of the brain, breast, and prostate respectively. Cell viability and fluoresence quantification assays revealed that pArg-coated nanovectors were most effective in promoting gene-knockdown and least toxic of the three nanovector formulations tested. Transmission Electron Microscopy (TEM) imaging of nanovector treated cell further demonstrated that pArg-coated nanovectors enter cells through cell transcytosis, while pLys and PEI coated nanovectors enter cells endocytosis. Our findings suggest that NPs engineered to exploit the cell transcytosis intracellular trafficking pathway may offer a more safe and efficient route for siRNA delivery.
Keywords: iron oxide nanoparticle, gene therapy, transcytosis, cancer, MRI, siRNA
1. Introduction
Small interfering RNAs (siRNAs) can silence gene expression in a highly specific manner for treating genetic disorders, signifying a new approach in cancer therapy through the regulation of aberrant gene expression inherent to cancer [1]. However, the physicochemical characteristics of siRNA (i.e. high molecular weight, anionic charge, and hydrophilic character) hinder its passive diffusion across cell membranes precluding any therapeutic function [2]. Furthermore, siRNA molecules are highly vulnerable to degradation. Thus, for effective siRNA delivery, siRNA carriers are needed to protect siRNA, facilitate cellular entry, avoid endosomal compartmentalization, and promote localization in the cytoplasm where the siRNA cargo can be recognized by the RNA-induced silencing complex (RISC). Inorganic nanoparticles (NPs) designed for this application are propitious as they can be engineered for simultaneous diagnostics and therapeutics (theranostics) [2-4]. Currently, many NP core material formulations such as gold, silica, semiconductors, and metal oxides are being evaluated as siRNA carriers (nanovectors) [5-8]. Among them superparamagnetic iron oxide NPs possess superior physicochemical and biological properties ideal for in vivo magnetic resonance imaging (MRI) and drug delivery [9, 10].
The success of nanovectors relies on the apt design and integration of coatings that ensure biocompatibility and stability in a biologic milieu and proper intracellular trafficking. To date, most nanovectors developed for gene delivery applications are coated with cationic synthetic polymers (e.g. polyethylenimine (PEI) and poly amidoamines (PAMAM)) or lipids [6, 11, 12]. A common characteristic among these carriers is their high cationic charge density at physiological pH, which contributes to both the complex formation with anionic siRNA and interaction with the negatively charged cell membrane [13-15]. This interaction with cell membranes typically leads to the endocytosis of the nanovector, entrapping the nanovector within cellular endosomal vesicles [16]. Within the cellular endosomes the amino groups of cationic polymers function as proton sponges causing the swelling and eventual rupture of the endosome releasing the nanovector into the cytoplasm, a process known as endosomal escape. However, the high cationic charge density of these synthetic polymers also renders them highly cytotoxic [17].
Here we report the development of rationally designed, biocompatible, magnetic nanovectors for effective intracellular delivery of siRNA. Our strategy provides a safer alternative to the highly cationic nanovectors by utilizing the poly arginine (pArg) peptide as a coating material for NPs. pArg is a naturally occurring, biodegradable polypeptide that offers improved biocompatibility over PEI and PAMAM [18]. Furthermore, cell transcytosing proteins, which are known to avoid endosomal compartmentalization, are commonly found with arginine-rich domains, suggesting that a pArg-coated NP may provide the transcytosing ability [19, 20], and thus, offer more efficient siRNA delivery with greatly improved biocompatibility over NPs coated with synthetic polymers such as PEIs.
In this study, we synthesized magnetic nanoparticles (MNPs) coated with either pArg or PE) and functionalized with siRNA, and evaluated gene-silencing efficiency in tumor cell lines of the brain, breast, and prostate. We aimed to determine whether pArg-coated MNPs could provide efficient siRNA delivery, like with synthetic polymer (PEI) coated MNPs, but without the cytotoxicity. MNPs coated with poly lysine (pLys) were also prepared for comparison. pLys is another commonly used cationic polypeptide to complex siRNA and deliver siRNA to cytoplasm of cells via endocytosis and endosomal escape [21]. It is of great interest to understand the effect and degree of different intracellular pathways on the gene silencing efficiency for MNPs coated with different polycationic peptides.
2. Materials and Methods
2.1. Materials
All reagents were purchased from Sigma Aldrich (St. Louis, MO) unless otherwise specified.
2.2. NP-pLys, NP-pArg, and NP-PEI synthesis
Amine terminated PEG-coated iron oxide nanoparticles (NPs) with a 12-nm core diameter were synthesized according to previously established procedures [22]. After the synthesis, concentration of Fe was analyzed by inductively coupled plasma atomic emission spectroscopy (ICP-AES). The number of amine groups per NP (~70 NH2/NP) was determined by quantifying pyridine-2-thione using SPDP, according to manufacturer’s protocol. NPs were modified with cationic polymers through the formation of a thioether bond between thiolated polymers and NPs activated with iodoacetyl groups. Specifically, 10.5 mg of NP in 1 mL of thiolation buffer (0.1M sodium bicarbonate, 5mM EDTA, pH 8.0) was reacted with 10.5 mg succinimidyl iodoacetate (SIA, Molecular Biosciences, Boulder, CO) in 112.5 μl DMSO. Concurrently, 15.75 mg of poly-lysine (MW 10,000), poly-arginine (MW 10,000), or PEI (MW 10,000) were dissolved in 1.725 ml thiolation buffer and modified with 26 μl of a 10 mg/ml Traut’s reagent (2-iminothiolane, Molecular Biosciences, Boulder, CO). All of the reactions were protected from light and ensued under gentle rocking. After 2 hours, excess SIA was removed from NP-SIA using a pre-packed PD-10 desalting column (GE Healthcare, Piscataway, NJ) equilibrated with thiolation buffer. Upon purification, the NPs were split into three equal volumes, each containing 3.5 mg Fe, and added to the three polymer solutions for reaction. The reaction were protected from light and ensued for 2 hours with gentle rocking and was then stored at 4°C overnight. The reactants for each reaction were then purified to remove excess polymer using columns packed with S-200 Sephacryl resin (GE Healthcare, Piscataway, NJ) equilibrated with thiolation buffer.
2.3. siRNA preparation
siRNA sequences designed to knockdown GFP expression (5′GCAAGCUGACCCUGAAGUUCUU3′–antisense, 5′GAACUUCAGGGUCAGCUUGCUU3′–sense) were purchased from Integrated DNA Technologies, Inc (IDT, San Diego, CA). These sequences were acquired with protected-thiol modifications on the 5′ end of the sense strand and with Cy5 modification on the 5′ end of the antisense strand. siRNA sequences were received as single strands and were annealed to their complementary strand in annealing buffer (12 mM potassium chloride, 1.2 mM HEPES, 0.04 mM magnesium chloride, pH 7.5) by incubating at 95°C for five minutes, then 37°C for 1 hr, and stored at −20°C.
2.4. NP-pLys-siRNA, NP-pArg-siRNA, and NP-PEI-siRNA synthesis
NP-pLys, NP-pArg, and NP-PEI were modified with SIA at a 5:1 weight ratio of Fe:SIA. The SIA was dissolved in DMSO such that the final reaction volume was 10% DMSO. The reactions were protected from light and ensued for 2 hours with gentle rocking. Meanwhile, a 57.73 mg/ml TCEP (Tris (2-Carboxyethyl) phosphine Hydrochloride) solution was prepared. The TCEP and GFP siRNA were combined at a 1:1 volume ratio, protected from light, and allowed to react for 1 hr with gentle rocking. The resultant siRNA was purified using a 2 ml Zeba column (Thermo Fisher Scientific, Waltham, MA) equilibrated with thiolation buffer. The nanoparticles were purified using pre packed PD-10 columns equilibrated with thiolation buffer. Upon purification, the siRNA was added to each of the three types of NPs at a 20:1 Fe:siRNA weight ratio. The reactions were protected from light and allowed to proceed for 2 hrs with gentle rocking and the resultant nanoparticle:siRNA nanovectors were used without any further purification.
2.5. Proton NMR analysis of cationic polymer attachment to NP
Each NP formulation (50 μg Fe) was dissolved in 50 μL of DCl and diluted to 1 mL in D2O. Similarly, cationic polymers were dissolved in 1 mL D2O containing 50 μL of DCl. Proton NMR spectra were obtained on a Bruker AVance series spectrometer operating at 300 MHz.
2.6. Gel retardation assay
Attachment of siRNA to NPs was assessed using gel retardation assays. A 4% low melting point agarose gel was prepared with 0.05 mg/mL ethidium bromide. While maintaining a uniform concentration of siRNA, samples of nanoparticle:siRNA complexes were prepared at a weight ratio of 20:1 (Fe mass of NP: siRNA mass). Samples were either left untreated or treated with heparin (1000 units/ml, 10 mL heparin/mg siRNA), and incubated for 30 min at room temperature to block the electrostatic interaction between the nanoparticles and siRNA. siRNA binding was analyzed by gel electrophoresis at 55 V for 90 min. Images were acquired on a Gel Doc XR (Bio-Rad Laboratories, Hercules, CA).
2.7. Nanovector size and zeta potential characterization
Hydrodynamic sizes and zeta potentials of the different nanoparticle formulations were analyzed at 100 μg/mL in 20 mM HEPES buffer (pH 7.4) using a DTS Zetasizer Nano (Malvern Instruments, Worcestershire, UK).
2.8. Evaluation of nanovector magnetic properties by MRI
Nanovector formulations were diluted in PBS, and then mixed with 25 μL of 1% agarose and loaded into an agarose gel phantom block. T2 relaxation measurements were performed on a 4.7-T Bruker magnet (Bruker Medical Systems, Karlsruhe, Germany) equipped with Varian Inova spectrometer (Varian, Inc., Palo Alto, CA). A 5 cm volume coil and spin-echo imaging sequence were used to acquire T2-weight images. Images were acquired using a repetition time (TR) of 3000 ms and echo times (TE) of 13.6, 20.0, 40.0, 60.0, 90.0 and 120.0 ms. The spatial resolution parameters were: acquisition matrix of 256 × 128, field-of-view of 35 × 35 mm, section thickness of 1 mm and two averages. The T2 map was generated by NIH ImageJ (Bethesda, MD) based on the equation, SI = A exp(−TE/T2) +B, where SI is the signal intensity, TE is the echo time, A is the amplitude, and B is the offset. The R2 map was generated by taking the reciprocal of the T2 map.
2.9. Evaluation of nanovector buffering capacity
NP-pLys-siRNA, NP-pArg-siRNA, and NP-PEI-siRNA were desalted using pre-packed PD-10 columns equilibrated with DI water. 1 mg of Fe from each condition was then transferred to a 15 ml Falcon tube and brought to pH 11.0 by addition of 0.1 N NaOH. The final volume for each condition was brought up to 2 ml with pH 11.0 DI water making the Fe concentration 500 μg/ml. The titration was performed with 2 μL additions of 0.1 N HCl.
2.10. Cell culture
C6 rat glioma (ATCC, Manassas, VA) and MCF7 human adenocarcinoma (ATCC, Manassas, VA) cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA) and 1% antibiotic-antimycotic (Invitrogen, Carlsbad, CA) at 37°C and 5% CO2. Enhanced green fluorescent protein (EGFP) expressing cells were produced by stably transfecting cells with the pEGFP-N1 vector using the Effectene transfection reagent (Qiagen, Valencia, CA) following the manufacturer’s protocol. 48 hrs post-transfection, cells were sorted using a FACS Vantage and maintained in fully supplemented DMEM with 1 mg/ml G-418. TC2/GFP+ cells were kindly provided by Dr. Jennifer Wu’s Lab. TC2/GFP+ cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Atlanta Biological, Lawrenceville, GA) and 1% antibiotic-antimycotic (Invitrogen, Carlsbad, CA) at 37°C and 5% CO2
2.11. Cell transfection
The day before transfection, cells were plated at 50,000 cells per well in 24-well plates. For transfection of cells with nanovector formulations, cells were treated with nanovectors for 8 hrs under normal growth conditions. After the 8-hour incubation the media were replaced and cells incubated for an additional 48 hrs before analyses. Transfections of cells with siRNA using Dharmafect 4 (Dharmacon, Lafayette, CO) were performed according to the manufacturer’s instructions.
2.12. Cell viability and gene silencing
Potential cytotoxicity associated with the nanovector formulations was examined by the Alamar blue assay. After treatment, cells were washed with PBS three times, and incubated for 2 hrs with 10% Alamar blue (Invitrogen) in phenol red-free DMEM (supplemented with 10% FBS and 1% antibiotic–antimycotic). The percent reduction of Alamar blue was determined following the manufacturer’s protocol and used to calculate percent viability of treated samples (untreated cells represent 100% viability).
To quantify the degree of GFP gene silencing, treated cells were washed with PBS three times and lysed with 1% Triton X-100 in PBS. GFP protein expression was measured at an excitation and an emission wavelength of 488 and 520 nm, respectively. GFP fluorescence levels were normalized to the total number of viable cells, as determined by the Alamar Blue viability assay. Relative GFP expression levels were then calculated based on the reduction in GFP expression as compared to non-transfected cells.
2.13. Confocal fluorescence microscopy
50,000 treated cells were plated on each of 24 mm glass cover slips and allowed to attach for 24 hrs. Cells were then washed with PBS and fixed in 4% formaldehyde (Polysciences Inc., Warrington, PA) for 30 min. Cells were then washed three times with PBS and membrane-stained with WGA-AF647 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Cover slips were then mounted on microscope slides using Prolong Gold antifade solution (Invitrogen, Carlsbad, CA) containing DAPI for cell nuclei staining. Images were acquired on a LSM 510 Meta confocal fluorescence microscope (Carl Zeiss Inc., Peabody, MA) with the appropriate filters.
2.14. Transmission electron microscopy (TEM)
One million C6 cells were seeded in 25 cm2 flasks 24 hrs before treatment. Cells were then treated with nanovector formulations as described for gene silencing experiments. Cells were then washed three times with PBS and incubated with ice cold Karnovsky’s fixative for 24 hrs. Following the fixation, the cells were processed directly from flasks for sectioning. Cell sections were stained with osmium tetroxide, lead citrate, and uranyl acetate for TEM-contrast enhancement. Cell samples were then imaged with a Philips CM100 TEM at 100 kV with a Gatan 689 digital slow scan camera.
3. Results and Discussion
3.1. Nanovector synthesis
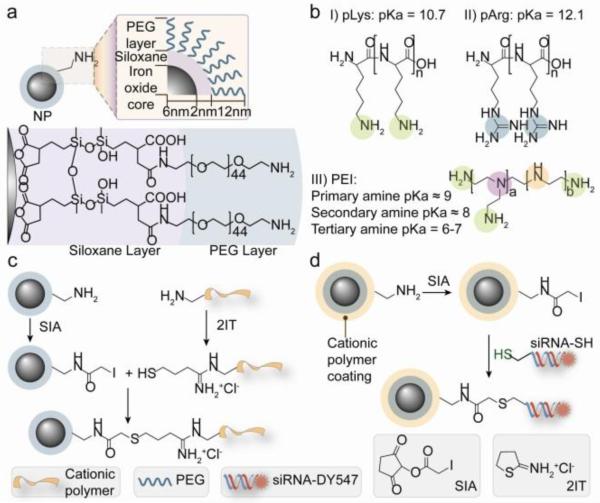
Figure 1 illustrates the fabrication scheme for the covalent attachment of cationic polymers and siRNA to the amine functionalized MNPs. The MNP consists of a 10–12 nm iron oxide core coated with siloxane to which a 44-mer of PEG-amine is anchored [22, 23] (Figure 1a). The MNP was then modified with 10-kDa cationic polymers (pLys, pArg or PEI) to produce NP-pLys, NP-pArg and NP-PEI using the conjugation scheme in Figure 1c. The chemical structures of the cationic polymer are shown in Figure 1b.
Figure 1.
Chemical scheme for synthesis of magnetic nanovectors. (a) The amidated PEG-passivated iron oxide NPs used as the base NP for construction of transfection vectors in this study. (b) Chemical structures of the cationic polymers used to functionalize the NPs. (c) Covalent attachment of cationic polymers to NPs. (d) Covalent attachment of Cy5 modified siRNA to NPs.
To assess the gene delivery efficacy, Cy5 fluorescently labeled, thiol modified siRNA (21 base-pairs, 5.7 nm length) designed to silence green fluorescence protein (GFP) transgene expression was covalently attached to the functional amine groups on the surface of NP-pLys, NP-pArg and NP-PEI using the heterobifunctional linker SIA (Figure 1d) to form NP-pLys-siRNA, NP-pArg-siRNA and NP-PEI-siRNA.
This non-labile covalent attachment of siRNA to a nanocarrier is preferable for in vivo applications because it ensures that the siRNA-carrier construct will remain intact during blood circulation [9]. Furthermore, this conjugation strategy does not compromise the knockdown efficiency of the siRNA [24]. To favor thioether bond formation over electrostatic binding of the negatively charged siRNA with cationic NP, the SIA reaction was performed in a high ionic strength buffer.
3.2. Nanovector physicochemical characterization
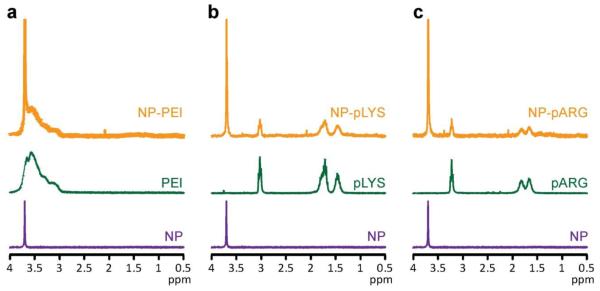
The successful immobilization of cationic polymers onto NP was verified using proton NMR (1H-NMR). Spectra from base MNPs, relevant constituent polymers, and the three different nanovector formulations confirm the successful immobilization of their expected polymer coatings (Figure 2). PEG on NP was identified at 3.7 ppm corresponding to the protons of the ethylene unit. PEI was identified at 3.75–3 ppm corresponding to the protons of its ethylene units. pLys was identified at 3, 1.75, and 1.45 ppm corresponding to the protons of the ε, δ and β, and γ carbons of the lysine side chain. pArg was identified at 3.25, 1.8, and 1.6 ppm corresponding to the protons of the δ, β, and γ carbons of the arginine side chain, respectively.
Figure 2.
Proton NMR analysis of nanovectors verifying cation polymer coating attachment. (a) Spectra of NP, PEI, and NP-PEI. (b) Spectra of NP, pLys, NP-pLys. (c) Spectra NP, pArg, NP-pArg. All samples were analyzed in D2O.
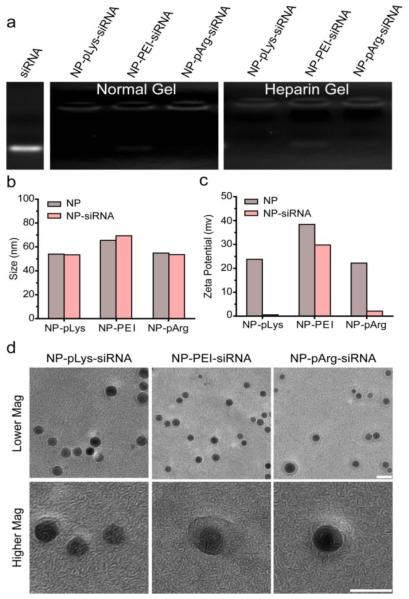
siRNA loading onto the nanovector was assessed using gel retardation assays where NP bound siRNA would not migrate down the gel. NP-pLys-siRNA, NP-PEI-siRNA and NP-pArg-siRNA were prepared at 20:1 nanoparticle:siRNA weight ratios (Fe mass of NP : siRNA mass). Without purification, the reaction products were loaded into agarose gels and unbound siRNA (bottom of gel) was separated from MNP-bound siRNA through electrophoresis. Each NP formulation showed complete binding of siRNA as no unbound siRNA was observed in the gels (Figure 3a). A similar migration profile is demonstrated in the heparin treated samples (Figure 3a), which disrupts electrostatic interactions, confirming successful covalent attachment of siRNA to MNP for all three nanovector formulations.
Figure 3.
Characterization of constructed nanovectors. (a) Gel retardation assay evaluating covalent attachment of siRNA to NPs under normal electrophoresis conditions and under heparin treatment to disrupt electrostatic interactions between cationic NPs and anionic siRNA. (b) Z-average hydrodynamic sizes of nanovectors before and after siRNA attachment. (c) Zeta potentials of nanovectors before and after siRNA attachment. (d) TEM images of nanovectors (scale bars correspond to 10 nm).
The hydrodynamic size of each nanovector formulation before and after siRNA loading was measured by dynamic light scattering (DLS) (Figure 3b). Z-average diameters of the nanovectors were 53.4 nm for NP-pLys-siRNA, 69.4 nm for NP-PEI-siRNA and 53.5 nm for NP-pArg-siRNA. No significant change in hydrodynamic size was observed after siRNA attachment to nanovectors, reflecting their stability. Notably, the hydrodynamic size falls within the acceptable size range (5 < d < 200 nm) that facilitates in vivo navigation and evasion of sequestration by macrophages [25].
TEM images of three nanovector formulations show no signs of aggregation (Figure 3d, top row). Higher magnification images in Figure 3d (bottom row) reveal a lower density shell surrounding the MNP cores, which is absent on the base MNPs (Supplementary Figure 1). This shell structure further confirms the cationic polymer coating on the MNPs.
The zeta potential of each nanovector formulation was measured at pH 7.4 in 10 mM hepes buffer by DLS. All three nanovector formulations were highly cationic prior to siRNA attachment, with zeta potentials of 22 mV for NP-pLys, 39 mV for NP-PEI and 21 mV for NP-pArg (Figure 3c). After siRNA attachment, the zeta potentials of all formulations dropped significantly confirming successful siRNA conjugation. However, NP-PEI-siRNA remained highly cationic (30 mV) while the NP-pLys-siRNA (0.5 mV), and NP-pArg-siRNA (2.5 mV) displayed near-neutral zeta potentials under physiological pH conditions. A neutral NP formulation generally has better biocompatibility and prolonged circulating time in blood than positively charged formulations [9].
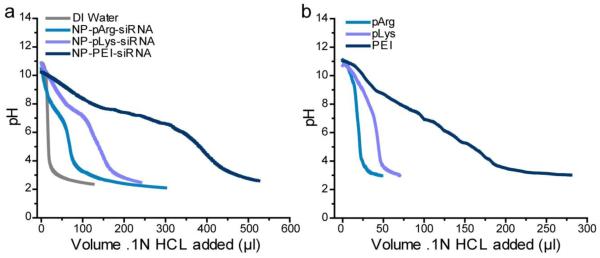
The three various nanovector formulations were similar in sizes, but had different zeta potentials. They differ in composition and abundance of surface amino groups, providing us an opportunity to compare the influence of these key parameters on siRNA delivery and cytotoxicity. The charge and composition of the cationic polymer coating regulates the buffering capacity and proton sponge behavior of the nanovector. To illustrate this, nanovectors were titrated with HCL and the change in pH was monitored. Resistance to pH change indicates an increase in proton absorption of the nanovector. The buffering capacity was seen to increase in the order of: NP-pArg-siRNA < NP-pLys-siRNA < NP-PEI-siRNA (Figure 4a). This trend mirrors the buffering capacities of the coating polymers alone (Figure 4b), indicating they were not affected by attachment to NPs or to siRNA.
Figure 4.
Titration curves evaluating the buffering capacity of each nanovector formulation (a) and its corresponding polymer (b) through addition of NaOH base.
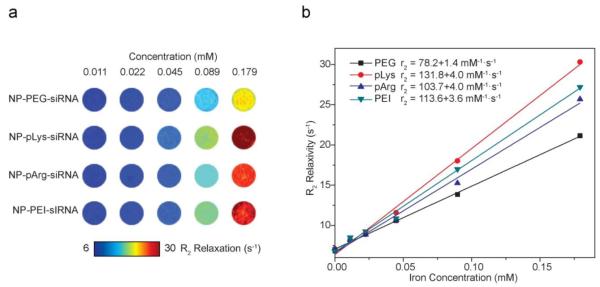
To ensure the shell structure did not compromise the magnetic properties of the nanovectors, we assessed their magnetic relaxivities on MRI. All nanovector formulations showed higher R2 relaxivities compared to the base NP (Figure 5), suggesting that the attachment of cationic polymers and siRNA to MNPs did not compromise, but rather, improved the magnetic properties. This improvement is likely due to the improved water absorption at the surface of the core MNP due to the presence of the hydrophilic cationic polymers [9].
Figure 5.
Magnetic properties of nanovectors. (a) R2 maps of gel phantoms containing nanovectors of different concentrations (mM). (b) R2 relaxivity of nanovectors as a function of Fe concentration were used to determine relaxivities, which yielded relaxivity values of 78.2 mM−1S−1 for NP-PEG-siRNA, 103.7 mM−1S−1 for NP-pArg-siRNA , 131.8 mM−1S−1 for NP-pLys-siRNA, and 113.6 mM−1S−1 for NP-PEI-siRNA.
3.3. Nanovector internalization, gene knockdown and monitoring of toxicity
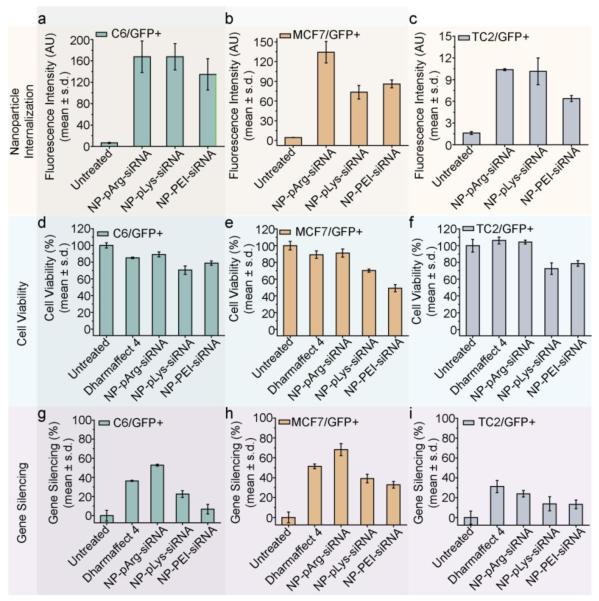
The nanovectors were evaluated in vitro for siRNA delivery and gene silencing in three cancer cell lines (C6/GFP+ brain, MCF7/GFP+ breast, and TC2/GFP+ prostate) that stably express the GFP gene. First, we evaluated the amount of siRNA delivered to cells by each nanovector formulation using the fluorescently labeled siRNA (Figure 6a-c). All three nanovectors delivered similar amounts of siRNA to C6/GFP+ cells (Figure 6a). Conversely, NP-pArg-siRNA delivered a markedly higher quantity of siRNA to MCF7/GFP+ cells than NP-pLys-siRNA and NP-PEI-siRNA (Figure 6b). In TC2/GFP+ cells, both NP-pArg-siRNA and NP-pLys-siRNA delivered more siRNA than NP-PEI-siRNA. These data demonstrate that the pArg coating was most broadly effective in facilitating the delivery of siRNA-loaded nanovectors to target cells.
Figure 6.
Evaluation of siRNA delivery, cell viability, and GFP knockdown. Relative amount of siRNA delivered to (a) C6/GFP+, (b) MCF7/GFP+ and (c) TC2/GFP+ cells by nanovectors of three different formulations. Influence of nanovector treatments on cell viability of (d) C6/GFP+, (e) MCF7/GFP+ and (f) TC2/GFP+ cells (viability was normalized to untreated cells). Efficiency of nanovector treatments on silencing GFP expression in (g) C6/GFP+, (h) MCF/GFP+ and (i) TC2/GFP+ cells (GFP Expression was normalized to untreated cells).
The influence of nanovector treatments on cell viability was evaluated using Alamar blue assays. Cells were treated with each nanovector formulation and a commercial transfection reagent Dharmafect 4 (Dharmacon Inc.) as a reference. The NP-pArg-siRNA treatment was the least toxic to C6/GFP+ cells (90% viability (%V)), with NP-pLys-siRNA (70.6 %V) and NP-PEI-siRNA (78.9 %V) most toxic (Figure 6d). A similar trend in viability was observed with MCF7/GFP+ cells (Figure 6e) where NP-pArg-siRNA treatment produced the least cytotoxicity (91.2 %V), similar to Dharmafect 4 treatment (89 %V) and much lower than NP-pLys-siRNA (70.2 %V) and NP-PEI-siRNA (49 %V) treatments. This trend was again observed in TC2/GFP+ cells (Figure 6f). Both NP-pArg-siRNA (104.3 %V) and Dharmafect 4 (92.5 %V) treatments were well tolerated, while the NP-pLys-siRNA (77.5 %V) and NP-PEI-siRNA (78.7 %V) treatments were both moderately toxic.
We then evaluated the GFP gene-silencing efficacy of each nanovector formulation (Figure 6g-i). The NP-pArg-siRNA treatment was most effective in gene silencing in C6/GFP+ cells (52.9 % GFP knockdown (%GKD)), whereas Dharmafect 4 (36.4 %GKD), NP-pLys-siRNA (22.7 %GKD) and NP-PEI-siRNA (6.8 %GKD) were all significantly less effective (Figure 6g). A similar trend was observed in MCF7/GFP+ cells (Figure 6h). NP-pArg-siRNA showed the highest potency for silencing (68.2 %GKD), whereas Dharmafect 4 (51.3 %GKD), NP-pLys-siRNA (39.1%GKD) and NP-PEI-siRNA (32.8 %GKD) treatments were all less effective. In TC2/GFP+ cells we again observed that NP-pArg-siRNA (24 %GKD) was most effective in gene silencing compared to the other two different nanovector formulations: NP-pLys-siRNA (13.9 %GKD) and NP-PEI-siRNA (13.4 %GKD) (Figure 6i). In this cell line Dharmafect 4 (31.3 %GKD) was slightly more effective than NP-pArg-siRNA.
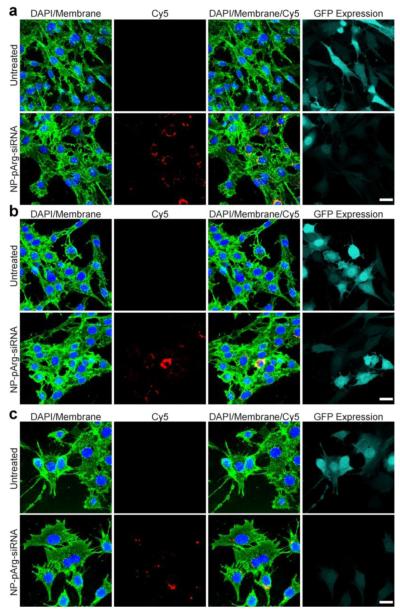
To further confirm the siRNA delivery and gene knockdown achieved by NP-pArg-siRNA to the three types of cells (C6/GFP+, MCF7/GFP+, and TC2/GFP+) confocal fluorescence microscopy was performed. Shown in Figure 7a-c are fluorescence images of untreated and NP-pArg-siRNA treated C6/GFP+ (a), MCF7/GFP+ (b), and TC2/GFP+ (c) cells. In all images cell nuclei were stained with DAPI (blue) and membranes with WGA-647 (green). Treatments with NP-pArg-siRNA were administered as described above at a concentration of 1.2 μg of siRNA/ml. As shown in all three-cell lines tested internalized siRNA (red, second column) can be visualized in cells treated with NP-pArg-siRNA. The overlay images (third column) reveal that the delivered siRNA molecules are predominantly localized in the perinuclear region of cells, the region of cell where siRNA molecules are recognized by the RISC complex [1]. This observation confirms the proper trafficking of siRNA within cells. The GFP expression analysis (light green, fourth column) showed that the NP-pArg-siRNA treatment reduced the GFP expression of cells in all cell lines compared to the untreated cells.
Figure 7.
Confocal fluorescence microscopy images evaluating siRNA internalization and subsequent GFP knockdown. Images were acquired from (a) C6/GFP+, (b) MCF7/GFP+, and (c) TC2/GFP+ cells 48 hours post treatment with NP-pArg-siRNA, with untreated cells as a reference. Cell nuclei are shown in blue, cell membranes in green, siRNA in red, and GFP expression levels in Cyan. Scale bar corresponds to 20 μm.
Based on the transfection and cell viability studies, NP-pArg-siRNA has the favorable overall properties as a theranostic agent compared to other nanovector formulations studied. This might be attributed to its neutral zeta potential and limited proton sponge effect, which minimizes the potential deleterious effects caused by non-specific interactions with anionic intracellular components (e.g. mRNA, DNA, etc.). In addition, several reports have suggested that arginine rich peptides can penetrate across cell membranes through transcytosis without being trapped in endosome vesicles [2, 16, 26]. NP-pArg-siRNA may be similarly trafficked into cells accounting for its high gene silencing efficiency despite limited proton sponge capacity.
3.4. Nanovector intracellular trafficking
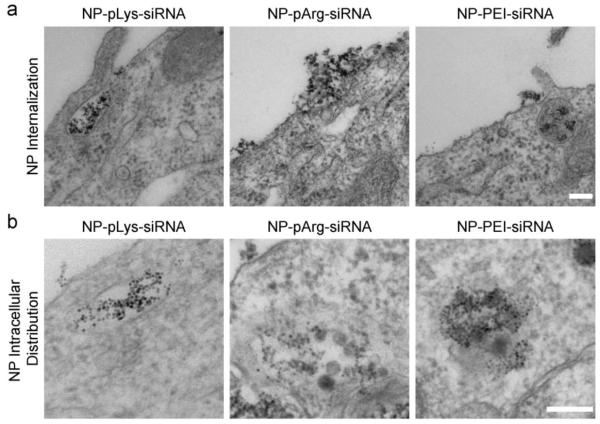
To determine the uptake mechanism of each nanovector formulation, C6/GFP+ cells treated with nanovectors were imaged with TEM (Figure 8). NP-pLys-siRNA and NP-PEI-siRNA could be observed entering the cell through endocytic pathways, whereas NP-pArg-siRNA appeared to transcytose across the cell membrane in the absence of independent endocytic engulfment (Figure 8a). Furthermore, once inside the cell NP-pLys-siRNA and NP-PEI-siRNA could be seen escaping endosomal vesicles while NP-pArg-siRNA were free in the cytoplasm with no visible endosomal vesicle (Figure 8b). These TEM images provide an explanation as to why the NP-pArg-siRNA formulation appeared efficient in inducing gene knockdown (Figure 6g-i), despite its limited buffering capacity (Figure 4). The NP-pArg-formulation does not require endosome escape properties owing to its ability to completely bypass the endocytosis pathway. Conversely, the NP-pLys-siRNA and NP-PEI-siRNA formulations must escape endosomal vesicles to localize in the cytoplasm. To our knowledge, this is the first report to demonstrate that inorganic NPs coated with pArg can be trafficked through a similar pathway as the polypeptide alone and deliver siRNA.
Figure 8.
TEM images of C6/GFP+ cells treated with three nanovector formulations. a) Internalization of nanovectors. b) Intracellular localization of nanovectors. Scale bars represent 250 nm.
4. Conclusions
Development of safe and efficient siRNA nanocarriers continues to be the major hurdle to clinical translation of siRNA technology for cancer gene therapy. Here, we have investigated the role of nanovector coating in facilitating siRNA delivery and modulating cytotoxicity. We evaluated three common cationic polymers as coating materials for MNPs for siRNA delivery to cancer lines of the breast, brain, and prostate. Our results demonstrated that nanovectors prepared with a pArg coating were most efficient and least toxic. Furthermore, we demonstrated that pArg coated NPs could penetrate across cell membranes and avoid the endocytosis pathway, precluding the need of toxic endosome lysing polymers. These findings could be utilized to engineer more safe and effective NP formulations as nanovectors for gene and drug delivery applications.
Supplementary Material
Acknowlegments
This work is supported by NIH grants R01CA134213 and R01EB006043. O. Veiseh and F. Kievit acknowledge support through NCI training grant T32CA138312. C. Fang acknowledges the support through an NSF/NCI IGERT fellowship. We acknowledge lab assistance form Surya Kotha and Andrea Nordberg, and the use of resources at the Department of Immunology’s cell analysis facility, Keck Microscopy Imaging Facility, Center for Nanotechnology, and the Diagnostic Imaging Science Center at University of Washington.
Footnotes
Appendix. Supplementary data
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–38. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Haglund E, Seale-Goldsmith MM, Leary JF. Design of multifunctional nanomedical systems. Ann Biomed Eng. 2009;37:2048–63. doi: 10.1007/s10439-009-9640-2. [DOI] [PubMed] [Google Scholar]
- [4].Hong H, Zhang Y, Cai W. In vivo imaging of RNA interference. J Nucl Med. 2010;51:169–72. doi: 10.2967/jnumed.109.066878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nature Medicine. 2007;13:372–7. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- [6].Jung J, Solanki A, Memoli KA, Kamei K, Kim H, Drahl MA, et al. Selective inhibition of human brain tumor cells through multifunctional quantum-dot-based siRNA delivery. Angew Chem Int Ed Engl. 2010;49:103–7. doi: 10.1002/anie.200905126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9:2402–6. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee JH, Lee K, Moon SH, Lee Y, Park TG, Cheon J. All-in-one target-cell-specific magnetic nanoparticles for simultaneous molecular imaging and siRNA delivery. Angew Chem Int Ed Engl. 2009;48:4174–9. doi: 10.1002/anie.200805998. [DOI] [PubMed] [Google Scholar]
- [9].Veiseh O, Gunn JW, Zhang M. Design and fabrication of magnetic nanoparticles for targeted drug delivery and imaging. Adv Drug Deliv Rev. 2010;62:284–304. doi: 10.1016/j.addr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Veiseh O, Sun C, Fang C, Bhattarai N, Gunn J, Kievit F, et al. Specific targeting of brain tumors with an optical/magnetic resonance imaging nanoprobe across the blood-brain barrier. Cancer Res. 2009;69:6200–7. doi: 10.1158/0008-5472.CAN-09-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Song WJ, Du JZ, Sun TM, Zhang PZ, Wang J. Gold nanoparticles capped with polyethyleneimine for enhanced siRNA delivery. Small. 2010;6:239–46. doi: 10.1002/smll.200901513. [DOI] [PubMed] [Google Scholar]
- [12].Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. Proton-sponge coated quantum dots for siRNA delivery and intracellular imaging. J Am Chem Soc. 2008;130:9006–12. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–84. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- [14].Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–9. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Akhtar S, Benter I. Toxicogenomics of non-viral drug delivery systems for RNAi: potential impact on siRNA-mediated gene silencing activity and specificity. Adv Drug Deliv Rev. 2007;59:164–82. doi: 10.1016/j.addr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- [16].Bareford LM, Swaan PW. Endocytic mechanisms for targeted drug delivery. Adv Drug Deliv Rev. 2007;59:748–58. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv Drug Deliv Rev. 2006;58:1523–31. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- [18].Vorhies JS, Nemunaitis JJ. Synthetic vs. natural/biodegradable polymers for delivery of shRNA-based cancer therapies. Methods Mol Biol. 2009;480:11–29. doi: 10.1007/978-1-59745-429-2_2. [DOI] [PubMed] [Google Scholar]
- [19].Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv Drug Deliv Rev. 2005;57:547–58. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- [20].Schmidt N, Mishra A, Lai GH, Wong GC. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010;584:1806–13. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- [21].Martin ME, Rice KG. Peptide-guided gene delivery. AAPS J. 2007;9:E18–29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fang C, Bhattarai N, Sun C, Zhang MQ. Functionalized Nanoparticles with Long-Term Stability in Biological Media. Small. 2009;5:1637–41. doi: 10.1002/smll.200801647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fang C, Veiseh O, Kievit F, Bhattarai N, Wang F, Stephen Z, et al. Functionalization of iron oxide magnetic nanoparticles with targeting ligands: their physicochemical properties and in vivo behavior. Nanomedicine (Lond) 2010;5:1357–69. doi: 10.2217/nnm.10.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Singh N, Agrawal A, Leung AK, Sharp PA, Bhatia SN. Effect of nanoparticle conjugation on gene silencing by RNA interference. J Am Chem Soc. 2010;132:8241–3. doi: 10.1021/ja102132e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine. 2008;3:703–17. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Howard KA. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv Drug Deliv Rev. 2009;61:710–20. doi: 10.1016/j.addr.2009.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.