Abstract
Processive reactions, such as transcription or translation, often proceed through distinct initiation and elongation phases. The processive formation of polymeric ubiquitin chains can accordingly be catalyzed by specialized initiating and elongating E2 enzymes, but the functional significance for this division of labor has remained unclear. Here, we have identified sequence motifs in several substrates of the anaphase-promoting complex (APC/C) that are required for efficient chain initiation by its E2 Ube2C. Differences in the quality and accessibility of these chain initiation motifs can determine the rate of a substrate’s degradation without affecting its affinity for the APC/C, a mechanism used by the APC/C to control the timing of substrate proteolysis during the cell cycle. Based on our results, we propose that initiation motifs and their cognate E2s allow E3 enzymes to exert precise temporal control over substrate degradation.
Keywords: ubiquitin, ubiquitin chain formation, protein degradation
Introduction
Most processive reactions, including transcription or translation, proceed through distinct initiation, elongation, and termination phases. Each of these steps requires specialized proteins, such as translation initiation or elongation factors, which are often regulated independently of each other. For example, regulation of translation initiation provides a means to rapidly change protein synthesis in response to stress (Sonenberg and Hinnebusch, 2009).
Reminiscent of translation, the formation of ubiquitin chains can be a highly processive reaction (Rape et al., 2006; Pierce et al., 2009), which is catalyzed by a cascade of E1, E2, and E3 enzymes (Ye and Rape, 2009). Dedicated factors are able to bring about initiation, i.e. the attachment of ubiquitin to a substrate lysine, or elongation, i.e. the formation of chains linked through Lys residues of ubiquitin. For example, the assembly of ubiquitin chains on PCNA requires the E3s Rad18 for initiation and Rad5 for elongation (Hoege et al., 2002). In a similar manner, chain initiation on substrates of the UFD-pathway depends on the E3 Ufd4, while elongation is catalyzed by Ufd2 (Koegl et al., 1999). Distinct E3s for initiation and elongation allow cells to produce either mono- or multiubiquitinated substrates, which results in different consequences for the modified protein (Ye and Rape, 2009).
In a variation on this theme, several E3 enzymes employ distinct E2s to catalyze chain initiation or elongation, respectively. This phenomenon was first described for the E3 Brca1-Bard1, which can utilize the E2s Ube2D, Ube2E, or Ube2W for initiation, but Ube2K or Ube2N-Uev1A for elongation (Christensen et al., 2007). In human cells, the E3 SCF appears to cooperate with Ube2D or Ube2R (Cdc34) for initiation, while it relies on Ube2R to elongate K48-linked ubiquitin chains (Petroski and Deshaies, 2005; Wu et al., 2010). A similar division of labor is observed for the human E3 APC/C, which employs Ube2C (UbcH10) for initiation, but Ube2S for the elongation of K11-linked chains (Jin et al., 2008; Williamson et al., 2009; Garnett et al., 2009; Wu et al., 2010). Both the APC/C and the SCF are able to promote chain formation with high processivity (Carroll et al., 2002; Petroski and Deshaies, 2005; Rape et al., 2006; Pierce et al., 2009), and distinct functions for mono-ubiquitinated substrates of these E3s have not been reported. This strongly suggests that specialized initiating E2s provide regulatory advantages that remain to be discovered.
The initiating E2s select Lys residues in substrates as acceptors for ubiquitin. Much of our knowledge about substrate lysine selection comes from studies of the E2 Ubc9, which transfers the ubiquitin-like modifier SUMO to hundreds of cellular proteins. Ubc9 directly binds substrate residues of the consensus SUMOylation motif, ΨKxE/D (Bernier-Villamor et al., 2002). Charged amino acids close to this motif can enhance SUMOylation (Yang et al., 2010), while phosphorylation of nearby Ser or Thr residues allows for regulation (Mohideen et al., 2009). Lysine selection by Ubc9 is, therefore, strongly influenced by degenerate sequence motifs in the substrate.
In contrast to SUMOylation, little is known about how ubiquitin-initiating E2s decide on substrate lysine residues for modification. Global analyses of ubiquitination sites did not reveal strong preferences for specific sequence environments (Xu et al., 2010), suggesting that different E2s utilize distinct strategies to determine acceptor sites for the first ubiquitin. Consistent with this hypothesis, the APC/C-specific E2 Ube2C requires residues in the APC/C-substrate securin for efficient chain initiation (Jin et al., 2008). The respective sequence in securin contained a Thr-Glu-Lys motif, and hence was labeled TEK-box, but the amino acids responsible for promoting chain initiation were not determined. In a similar manner, the SCF-specific E2 Cdc34 has been suggested to recognize residues proximal to the modified lysine of the SCF-substrate Sic1 (Sadowski et al., 2010). However, as the sequences in securin and Sic1 were not characterized in detail, the recurrence of initiation motifs in multiple substrates of the same E3, their recognition by initiating E2s, and their potential regulation remained unknown. The significance of initiation motifs and their cognate E2s for ubiquitin-dependent proteolysis is, therefore, not understood.
Here, we dissected the mechanism of ubiquitin chain initiation by the APC/C and its E2 Ube2C. We have identified a conserved initiation motif that is found in multiple APC/C-substrates. Our characterization of this motif revealed three important functions in proteolysis: initiation motifs increase the efficiency of substrate degradation to allow the APC/C to degrade its many substrates; their composition determines the rate of substrate degradation to help the APC/C coordinate cell cycle progression; and their recognition by Ube2C can be regulated to fine-tune the timing of protein degradation. Our findings, therefore, establish initiation motifs as substrate elements with important roles in protein degradation. Together with their cognate E2s, initiation motifs increase the capacity of E3s to exert precise temporal control over substrate degradation.
Results
Identification of an essential initiation motif in the APC/C-substrate geminin
We previously found that deletion of residues in the APC/C-substrate securin, its “TEK-box”, impaired chain initiation by the APC/C-specific E2 Ube2C (Jin et al., 2008). Due to the mild effects of these deletions on securin degradation, we were unable to determine functionally important residues in this motif. As a consequence, initiation motifs were not identified in other APC/C-substrates and their role in regulating degradation remained unclear.
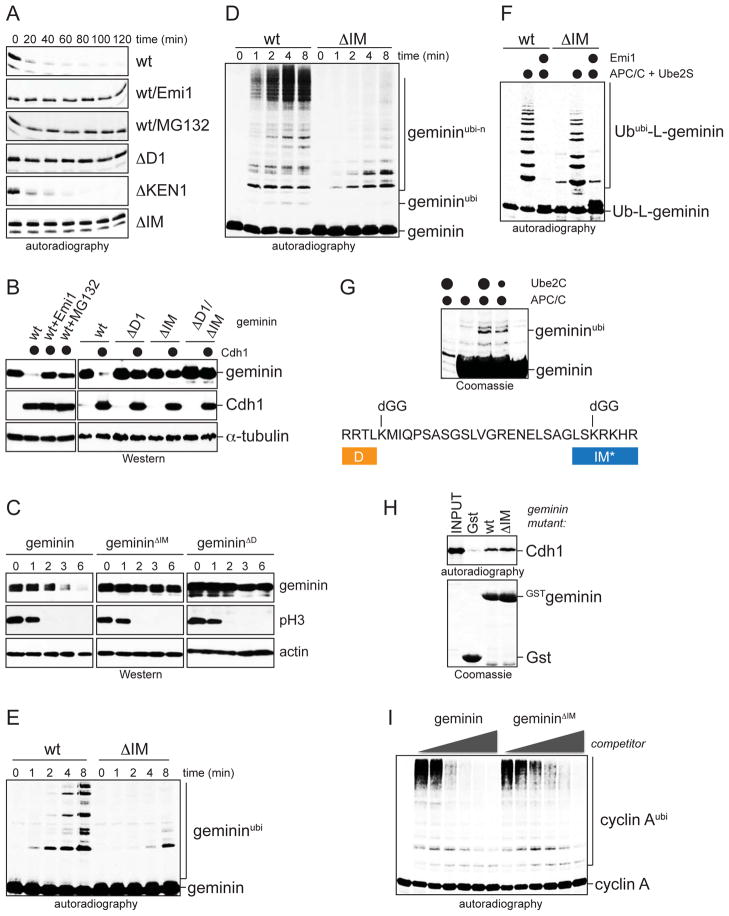
Our earlier studies had shown that the ubiquitination of the replication inhibitor geminin by the APC/C is competed by a peptide derived from the securin TEK-box, suggesting that geminin and securin use similar residues to promote initiation (Jin et al., 2008). Similar to Xenopus (McGarry and Kirschner, 1998), human geminin depends on a D-box for degradation in extracts and cells, and for ubiquitination by APC/C (Figure 1A–C; Figure S1A, B). Residues in proximity to this D-box shared similarity to the securin TEK-box (Figure S1C), and these residues (“IM” for initiation motif) were required for the APC/C-dependent ubiquitination and degradation of geminin (Figure 1A–D). As seen with stable gemininΔD (McGarry and Kirschner, 1998), injection of gemininΔIM into Xenopus embryos caused cell cycle arrest and death (Figure S1D). Thus, geminin contains a candidate initiation motif that is required for APC/C-dependent degradation and cell cycle progression.
Figure 1. Geminin requires an initiation motif for degradation.
A Degradation of 35S-geminin mutants in extracts with active APC/C was analyzed by autoradiography. B. Its initiation motif is required for geminin degradation in 293T cells after transfection with HACdh1. C. The initiation motif is required for geminin degradation in HeLa synchronized in mitosis. D. The initiation motif drives geminin ubiquitination. 35S-geminin mutants were incubated with APC/C, Ube2C, and ubiquitin, and analyzed by autoradiography. E. The initiation motif in 35S-geminin is important for chain initiation by APC/C, Ube2C, and methylubiquitin. F. The initiation motif is not required for chain elongation. Modification of 35S-Ub-L-geminin mutants by APC/C and Ube2S was analyzed by autoradiography. G. The initiation motif contains a preferential modification site (Lys50), as seen upon analysis of monoubiquitinated geminin by tandem mass spectrometry. H. The initiation motif is not essential for Gstgeminin-binding to 35S-Cdh1, as seen by autoradiography. I. The initiation motif is not required for APC/C-binding of geminin. Ubiquitination of 35S-cyclin A by APC/C and Ube2C was analyzed in the presence of increasing concentrations of Hisgeminin mutants.
Several observations suggest that the new motif in geminin specifically promotes chain initiation: First, its deletion strongly inhibited the APC/C-dependent modification of geminin Lys residues with methylubiquitin (Figure 1E). Second, a Lys residue within this motif was found to be a major initiation site for APC/C and Ube2C, as determined by mass spectrometry (Figure 1G). Third, once initiation was accomplished (Ub-L-geminin; Ub-L-gemininΔIM), the APC/C was able to elongate chains independently of whether this motif was present or not (Figure 1F; Figure S1E). Fourth, deletion of this motif did not abrogate binding of geminin to Cdh1, showing that it is not required for substrate-recruitment (Figure 1H). Fifth, geminin mutants lacking this motif inhibited the ubiquitination of other APC/C-substrates with comparable efficiency as wt-geminin, suggesting that it does not mediate APC/C-binding (Figure 1I). Together, these findings document a central and specific role for the geminin motif in promoting chain initiation and proteasomal degradation.
Initiation motifs are found in several APC/C-substrates
As deleting its initiation motif abolished geminin degradation, we used this substrate to identify key residues required for promoting initiation. We found that mutation of charged residues (RE40; KRK50-52; HR53/54) to alanine interfered with the APC/C-dependent ubiquitination and degradation of geminin (Figure 2A; Figure S2A). Assays with methylubiquitin revealed that RE40/41, KRK50-52, and HR53/54 were required for efficient chain initiation (Figure 2B). Interestingly, changing all Lys residues to arginine did not strongly affect geminin degradation or chain initiation, showing that the initiation motif has functions in addition to providing ubiquitin acceptor sites. As expected for a motif controlling the degradation of a key cell cycle regulator, functionally important, but not irrelevant, residues are highly conserved among geminin homologs from different organisms (Figure S2B).
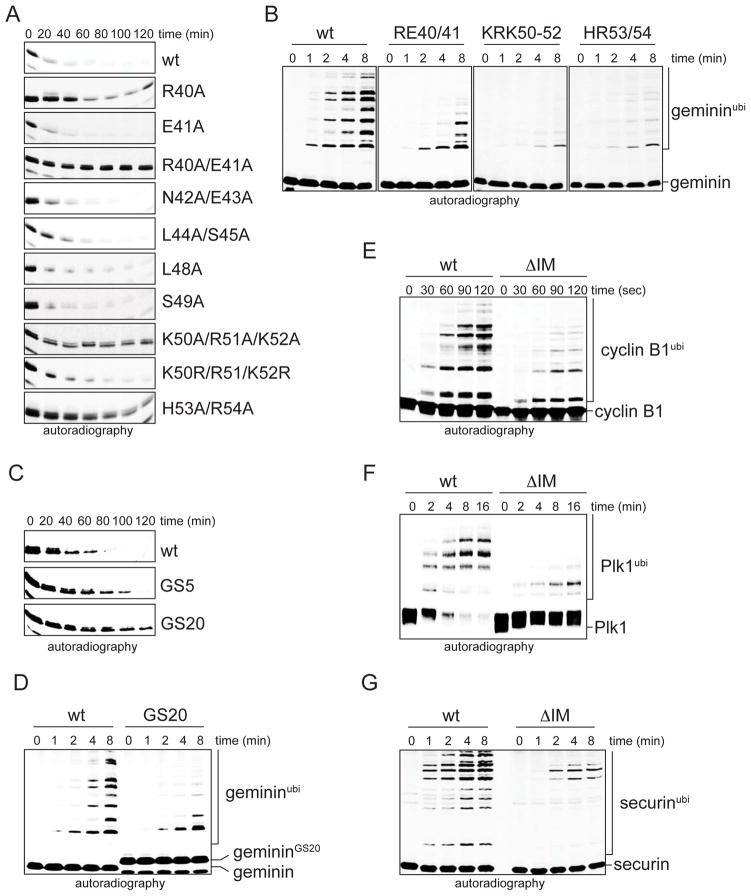
Figure 2. Initiation motifs are found in several APC/C-substrates.
A Degradation of 35S-geminin mutants in extracts with active APC/C requires positively charged residues in its initiation motif. B. Positively charged residues are required for chain initiation on 35S-geminin by APC/C, Ube2C, and methylubiquitin. C. The position of the initiation motif relative to the D-box is important for function. Degradation of 35S-geminin mutants with insertions of 5 or 20 GS-repeats between D-box and initiation motif in extracts with active APC/C was analyzed by autoradiography. D. Changing the position of the initiation motif impairs substrate Lys modification. Ubiquitination of 35S-geminin mutants by APC/C, Ube2C, and methyl-ubiquitin was analyzed by autoradiography. E. Cyclin B1 contains a chain initiation motif. Ubiquitination of 35S-cyclin B1 or an initiation motif mutant (K63KE/AAA; Δ73–78) by APC/C, Ube2C, and methylubiquitin was analyzed by autoradiography. F. Plk1 contains a chain initiation motif. Modification of 35S-Plk1 or an initiation motif mutant (Δ354–358) with methylubiquitin was analyzed as above. G. Securin contains an initiation motif. Modification of 35S-securin or an initiation motif mutant (K82QKQ/AAAA; K91K/AA) with methylubiquitin was analyzed as above.
The initiation motif in geminin is close to the D-box, its main APC/C-binding site, and the distance between the two motifs is conserved among geminin homologs (Figure S2B). This observation raised the possibility that the position of the initiation motif relative to the D-box is important for APC/C-substrate degradation. Consistent with this hypothesis, altering the distance between D-box and initiation motif through insertion of Gly/Ser-repeats impaired initiation by the APC/C, Ube2C, and methylubiquitin (Figure 2D), and stabilized geminin against proteasomal degradation (Figure 2C). The geminin initiation motif is, therefore, comprised of conserved patches of charged residues that occur in proximity to its APC/C-binding motif, the D-box.
Based on these results, we identified initiation motifs in the APC/C-substrates cyclin B1, Plk1, and securin (Figure S2C; data not shown). In securin, the motif is part of the “TEK-box”, the deletion of which provided the first evidence for a role of substrate residues in promoting initiation (Jin et al., 2008). Mutation of these motifs impaired chain initiation without strongly affecting substrate affinity to the APC/C (Figure 2E-G; Figure S2D-F). As seen before, replacing all Lys residues with arginine did not abrogate the function of the initiation motifs (Figure S2G, H). In securin, a group of Lys residues was rapidly modified despite a mutant initiation motif; we suspect that the alternative APC/C-binding of securin through its KEN-box, rather than its D-box, leads to distinct initiation (Figure S2I). Thus, initiation motifs consist of conserved patches of charged residues close to an APC/C-binding motif and are found in multiple substrates of the APC/C. The complexity of APC/C-initiation motifs is similar to the consensus SUMOylation or CDK-phosphorylation motifs that are also recognized by enzymes with a large number of cellular substrates. Thus, our observations imply a general role for initiation motifs in promoting APC/C-dependent degradation
Initiation motifs allow negative feedback regulation of the APC/C
We next set out to identify roles for initiation motifs in ubiquitin-dependent proteolysis. We hypothesized that initiation motifs could allow E3s to separate substrate binding from ubiquitination, which is required for a poorly understood aspect of APC/C-regulation: During mitosis, Ube2C, Ube2S, Cdc20, or Cdh1 bind the APC/C to trigger the ubiquitination of APC/C-substrates. However, once most substrates have been turned over, the APC/C-binding of these proteins results in their own ubiquitination and degradation, leading to APC/C-inhibition. For this feedback to work, mechanisms that independently control the binding and ubiquitination of these activators must exist.
To address this question, we focused on Ube2C. We first determined whether Ube2C contains an initiation motif that is required for its own ubiquitination, but not for APC/C-binding. Previous work had shown that the first 27 amino acids of Ube2C are pivotal for its ubiquitination by the APC/C (Rape and Kirschner, 2004), and conserved residues within this appendix have homology to the initiation motifs described above (Figure 3A). Changing positively charged residues in this motif to alanine (R6A, R17A/K18A), but not to arginine (K18R), strongly impaired the APC/C-dependent autoubiquitination of Ube2C (Figure 3B, Figure S3A). The Ala mutations also stabilized Ube2C against APC/C-dependent degradation in extracts and cells (Figure 3C; Figure S3B), while they had no (Ube2CR6A) or weak effects (Ube2CR17A/K18A) on charging by E1 (Figure S3C). Thus, Ube2C contains an initiation motif required for its degradation.
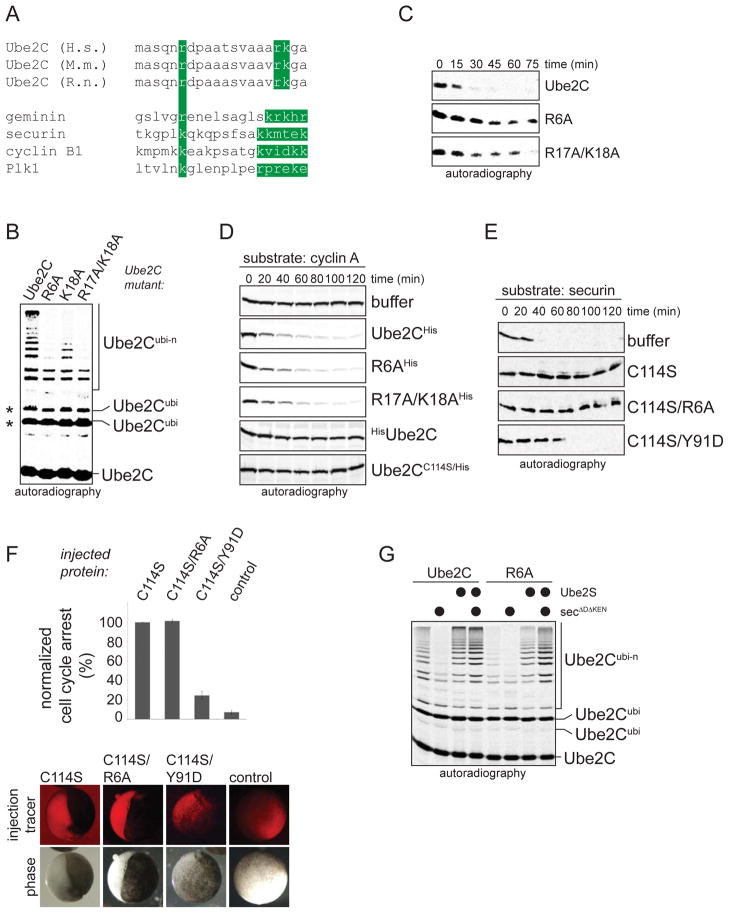
Figure 3. Initiation motifs allow negative feedback regulation of APC/C.
A The N-terminus of Ube2C contains a candidate initiation motif. B. Its initiation motif is required for ubiquitination 35S-Ube2C by APC/C, E1, and ubiquitin, as seen with autoradiography. Asterisks denote ubiquitination of K119 and K121 of Ube2C, which does not require the APC/C. C. The initiation motif is required for rapid degradation of 35S-Ube2C in extracts with active APC/C and low levels of APC/C-substrates. D. The initiation motif in Ube2C is not required for E2-activity, as seen in degradation assays using 35S-cyclin A as reporter. E. The RING-interaction loop is the main APC/C-binding site in Ube2C. Ube2CC114S, the initiation motif mutant Ube2CR6A/C114S, or RING-binding deficient Ube2CC114S/Y91D were tested for their capability to stabilize 35S-securin in extracts. F. The initiation motif is not required for APC/C-binding of Ube2C in vivo. Ube2CC114S or Ube2CR6A/C114S were injected into Xenopus tropicalis embryos at the two-cell stage, and cell cycle arrest was scored relative to Ube2CC114S. Error bars define the standard error derived from three independent experiments. G. APC/C-substrates inhibit Ube2C-ubiquitination through initiation motif competition. Recombinant securinΔDΔKEN was added to ubiquitination reactions of 35S-Ube2C by APC/C and/or Ube2S, as indicated.
To test whether its initiation motif contributes to the binding of Ube2C to the APC/C, we monitored the E2-activity of Ube2C and Ube2CR6A. We found that over a wide concentration range, Ube2CR6A catalyzed the ubiquitination of APC/C-substrates and promoted their degradation with similar efficiency as Ube2C (Figure 3D; Figure S3D, E). By contrast, mutating the RING-interaction loop, deleting an N-terminal QNP-motif (Summers et al., 2008), or appending N-terminal epitope tags reduced the activity of Ube2C towards APC/C-substrates (Figure S3D, F; data not shown). Thus, the initiation motif does not appear to mediate the binding of Ube2C to the APC/C.
To validate these findings, we introduced mutations into the catalytically inactive Ube2CC114S. If mutations impede APC/C-binding, the inhibitory effect of Ube2CC114S on degradation of APC/C-substrates in extracts or on cell cycle progression in frog embryos should be lost. Consistent with our previous observations, the initiation motif mutant Ube2CR6A/C114S stabilized APC/C-substrates as efficiently as Ube2CC114S (Figure 3E). Moreover, when injected into Xenopus embryos, Ube2CR6A/C114S produced the same cell cycle arrest as Ube2CC114S (Figure 3F). By contrast, mutation of the RING-interaction loop (Y91D) strongly diminished the capacity of Ube2CC114S to block degradation and cell cycle progression (Figures 3E, F). These findings show that Ube2C mainly interacts with the APC/C by recognizing the RING-domain of Apc11. Its initiation motif is, therefore, only required for Ube2C degradation, but not for APC/C-binding.
As a consequence, competition between initiation motifs could explain how canonical substrates stabilize Ube2C when both are bound to the APC/C. Although substrates and Ube2C are recruited to the APC/C by different means, using D-boxes or a RING-interaction loop, both require initiation motifs for chain formation. Consistent with this hypothesis, a mutant of the APC/C-substrate securin that only contains its initiation, but not its APC/C-binding motifs (securinΔDΔKEN), inhibited the ubiquitination of Ube2C by the APC/C (Figure 3G, S3G). This competitor did not impair APC/C-binding of Ube2C, as it did not block the APC/C-dependent chain elongation on Ube2C that had already undergone initiation (Figure 3G). Thus, competition by substrate initiation motifs stabilizes Ube2C that is bound to the APC/C in its role as E2; this strongly suggests that initiation motifs allow an E3, i.e. the APC/C, to regulate substrate binding and ubiquitination independently of each other.
Initiation motifs can determine the rate of substrate degradation
The APC/C degrades its substrates at different times during cell division to establish the sequence of mitotic events. The timing of APC/C-substrate degradation correlates with the processivity of ubiquitin chain formation, yet mechanisms that generate differences in the processivity remain unclear (Rape et al., 2006). As initiation can be rate-limiting for ubiquitination (Pierce et al., 2009), initiation motifs could have strong effects on the processivity of chain formation and the timing of substrate degradation.
To test this hypothesis, we replaced the initiation motif of Ube2C, which is among the last proteins to be degraded by the APC/C, with that of the early substrate geminin. Strikingly, the resulting mutant IMgem-Ube2C was ubiquitinated by the APC/C much more rapidly than wt-Ube2C, which was dependent on its own active site and RING-interaction loop (Figure 4A, B). Assays with methylubiquitin revealed that chain initiation by the APC/C occurred much more efficiently on IMgem-Ube2C compared to wt-Ube2C (Figure 4C). IMgem-Ube2C was degraded in extracts with active APC/CCdh1 much more rapidly than wt-Ube2C (Figure 4D), which was dependent on APC/CCdh1 and the proteasome (Figure S4A, B). Importantly, IMgem-Ube2C was also degraded much faster than wt-Ube2C upon APC/C-activation in cells (Figure 4E).
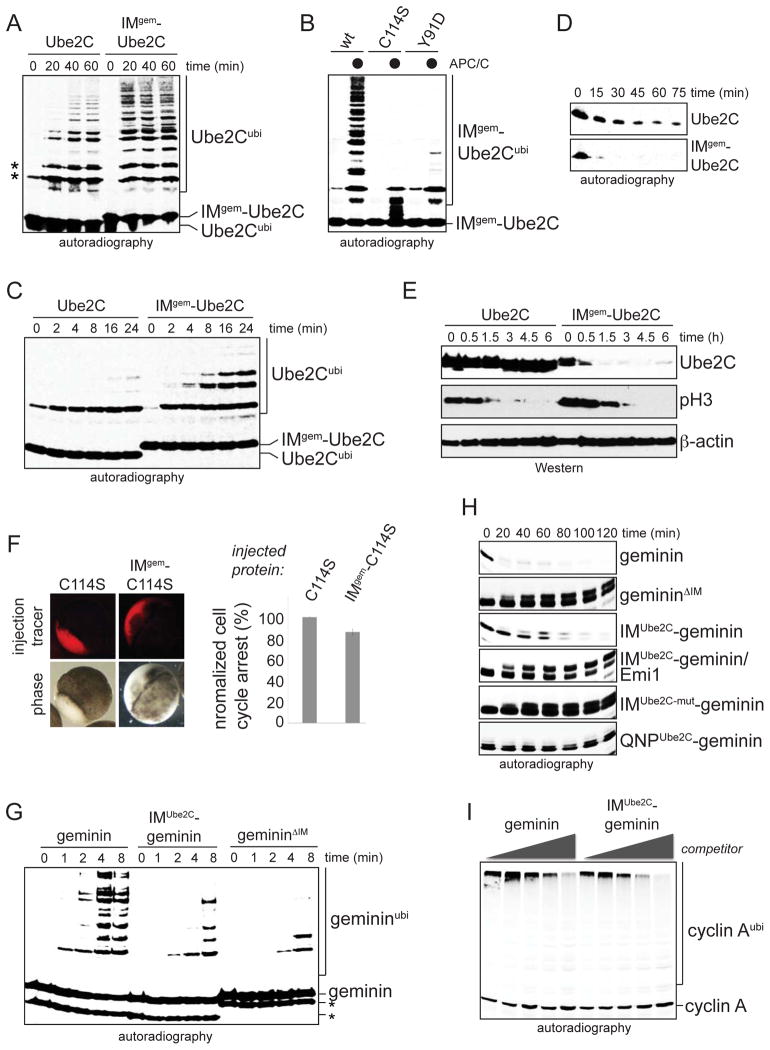
Figure 4. Initiation motifs can determine the rate of substrate degradation.
A The initiation motif determines the efficiency of Ube2C-ubiquitination. Ubiquitination of 35S-Ube2C or IMgem-Ube2C, a mutant containing the initiation motif of geminin, by APC/C was analyzed by autoradiography. The asterisk denotes the modification of Ube2C Lys-residues, which occurs without APC/C during the IVT/T. B. Ubiquitination of 35S-IMgem-Ube2C or its active site and RING-interaction mutant by the APC/C was analyzed by autoradiography. C. IMgem-Ube2C shows more efficient chain initiation than Ube2C, as seen in assays containing APC/C and methylubiquitin. The asterisk denotes APC/C-independent ubiquitination of Ube2C. D. Efficient chain initiation results in rapid degradation of 35S-IMgem-Ube2C in extracts with active APC/C. E. Efficient chain initiation results in rapid proteolysis of IMgem-Ube2CHA in synchronized HeLa cells entering anaphase. F. The initiation motif does not determine binding of Ube2C to the APC/C. Ube2CC114S or IMgem-Ube2CC114S were injected into X. tropicalis embryos at the two-cell stage, and cell cycle arrest was scored normalized to Ube2CC114S. Error bars define the standard error derived from three independent experiments. G. Less efficient initiation motifs delay geminin lysine modification. Ubiquitination of 35S-geminin, IMUbe2C-geminin, or gemininΔIM by APC/C, Ube2C, and methylubiquitin was analyzed by autoradiography. H. Less efficient initiation motifs delay proteolysis, as seen by degradation of 35S-geminin mutants in extracts with active APC/C. I. Its initiation motif does not determine the affinity of geminin to the APC/C. Increasing amounts of geminin or IMUbe2C-geminin were added as competitors to the APC/C-dependent ubiquitination of 35S-cyclin A and analyzed by autoradiography.
Altering its initiation motif did not strongly affect binding of Ube2C to the APC/C, as Ube2C and IMgem-Ube2C modified APC/C-substrates with similar efficiency (Figure S4C). The small differences between IMgem-Ube2C and Ube2C can be explained by a decrease in charging of IMgem-Ube2C by the E1, as expected from a previous report (Figure S4D; Huang et al., 2008). Moreover, the same levels of Ube2CC114S and IMgem-Ube2CC114S inhibited APC/C-substrate degradation in extracts and produced a block in cell division in embryos (Figure 4F; Figure S4E). Thus, replacing the initiation motif of Ube2C with that of geminin accelerated the ubiquitination of Ube2C without significantly affecting its binding to the APC/C or its activity as an E2.
To determine whether weak initiation motifs delay degradation, we replaced the initiation motif of geminin with that of Ube2C. The resulting mutant IMUbe2C-geminin was modified with methylubiquitin more slowly than wt-geminin (Figure 4G), indicative of impaired initiation. This caused less efficient chain formation (Figure S4F), and IMUbe2C-geminin was degraded more slowly than geminin in extracts and cells (Figure 4H, Figure S4G, H). Competition assays using recombinant geminin or IMUbe2C-geminin showed that altering its initiation motif did not change the affinity of geminin to the APC/C (Figure 4I). The initiation motif of Ube2C was functional in geminin, as IMUbe2C-geminin was modified and degraded more efficiently than gemininΔIM; moreover, IMUbe2C-geminin was stabilized by mutation of Arg6 in the Ube2C-initiation motif (Figure 4H). These swap experiments, therefore, show that the composition of initiation motifs can determine the rate of substrate degradation without affecting their affinity for the E3.
Initiation motifs allow the identification of an APC/C-substrate, PAF
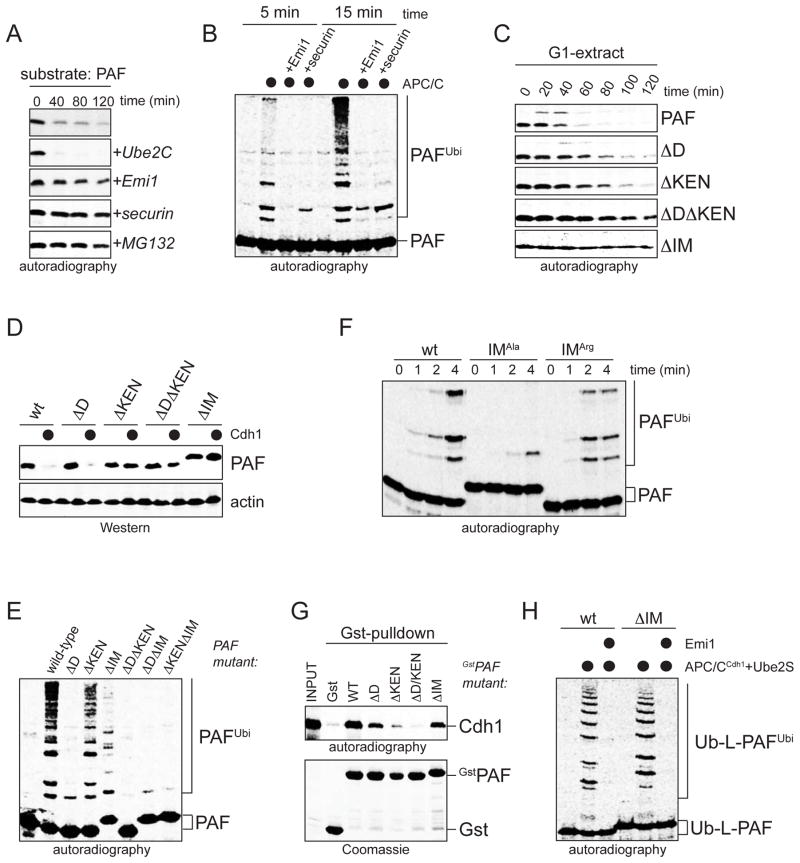
Given their role in degradation, we asked whether initiation motifs could be regulated, a control mechanism likely to occur in substrates that depend on initiation motifs for their degradation. To identify such substrates, we searched for proteins that contain D- or KEN-boxes in proximity to conserved sequences with similarity to the initiation motifs described above. A top-hit predicted by our search was PCNA-associated factor (PAF), a protein with roles in S-phase and DNA repair (Yu et al., 2001; Hosokawa et al., 2007; Turchi et al., 2009; Figure S5A). Consistent with its bioinformatic prediction, PAF was rapidly degraded in an APC/C- and proteasome-dependent manner in extracts (Figure 5A). PAF was also turned over by the APC/C in cells, as seen by Cdh1-overexpression (Figure 5D), depletion of the APC/C-inhibitor Emi1 (Figure S5B), release of nocodazole-arrested HeLa cells (Figure S5C), or serum-starvation of T24 cells (Figure S5D). Confirming its role as a substrate, PAF was ubiquitinated by purified APC/C, but not if APC/C was inhibited by Emi1 or an excess of a competing APC/C-substrate (Figure 5B). Thus, despite their degenerate nature, initiation motifs have sufficient predictive power to enable the discovery of new APC/C-substrates.
Figure 5. Initiation motifs allow identification of the APC/C-substrate PAF.
A Degradation of 35S-PAF in extracts with active APC/C was analyzed by autoradiography. B. 35S-PAF was ubiquitinated by APC/C, Ube2C, and ubiquitin, and analyzed by autoradiography. C. Degradation of 35S-PAF in extracts with active APC/C requires an initiation motif. D. The degradation of HAPAF in 293T cells with mycCdh1 requires an initiation motif. E. Efficient ubiquitination of PAF requires an initiation motif. Modification of 35S-PAF mutants by APC/C, Ube2C, and ubiquitin was analyzed by autoradiography. F. The initiation motif is required for efficient PAF lysine modification by APC/C, Ube2C, and methylubiquitin, as analyzed by autoradiography. G. The initiation motif in GstPAF is not required for binding to 35S-Cdh1. H. The initiation motif in PAF is not required for APC/C-binding or chain elongation. Ubiquitination of 35S-Ub-L-PAF mutants by APC/C, Ube2S, and ubiquitin was analyzed by autoradiography.
We next determined whether PAF-ubiquitination and degradation depend on its initiation motif. Indeed, mutation of the positively charged residues in this motif to Ala, but not to Arg, inhibited chain initiation on PAF by APC/C and methylubiquitin (Figure 5F; Figure S5E). By contrast, the initiation motif was not required for the binding of PAF to Cdh1 (Figure 5G), and ubiquitination reactions with Ub-PAF fusions showed that the initiation motif had no role in promoting Ube2S- and APC/C-dependent chain elongation (Figure 5H). Importantly, PAF-mutants in the initiation motif were not polyubiquitinated by the APC/C and therefore stabilized against APC/C-dependent degradation in extracts and cells (Figure 5C–E). Thus, the APC/C-substrate PAF requires a functional initiation motif for degradation.
Initiation motifs allow regulation of protein degradation
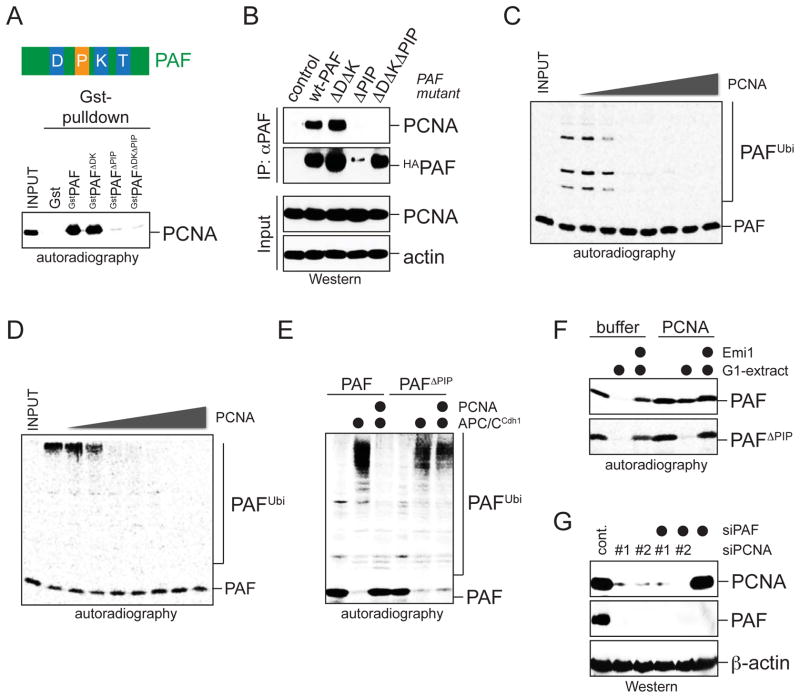
PAF binds the DNA polymerase processivity factor, PCNA, through a PIP-box located between D-box and initiation motif (Figure 6A; Yu et al., 2001). When we tested the interaction between PAF and PCNA in cells, we found that a PIP-box mutation not only ablated the binding of PAF to PCNA, but also caused a strong decrease in PAF-levels (Figure 6B). A similar effect on PAF-levels was observed upon depletion of PCNA by siRNA (Figure 6G). The low abundance of PAFΔPIP was rescued by mutation of its D- and KEN-boxes, suggesting that PCNA stabilizes PAF by antagonizing the APC/C. Supporting this notion, addition of PCNA to extracts stabilized PAF, but not PAFΔPIP, against APC/C-dependent degradation (Figure 6F).
Figure 6. PCNA inhibits chain initiation of PAF.
A PAF binds PCNA through a PIP-box. The top shows an overview of PAF-domains (D: D-box; P: PIP-box; K: KEN-box; IM: initiation motif). The bottom panel shows binding of 35S-PAF mutants to GstPCNA. B. PAF binds PCNA through a PIP-box in vivo. HAPAF mutants were precipitated from 293T lysates, and co-eluting endogenous PCNA was detected by Western blot. C. Increasing concentrations of PCNA (0.8nM-3.5μM) inhibit chain initiation on 35S-PAF by APC/C, Ube2C, methylubiquitin. D. Increasing concentrations of PCNA (0.8nM-3.5μM) inhibit the modification of 35S-PAF by APC/C, Ube2C, and ubiquitin. E. Modification of 35S-PAF or PAFΔPIP by APC/C, Ube2C, and ubiquitin was analyzed after addition of PCNA, as indicated. F. Degradation of 35S-PAF or PAFΔPIP in extracts with active APC/C was analyzed in the presence of recombinant PCNA, as indicated. G. PCNA stabilizes PAF in cells. HeLa cells were treated with siRNAs targeting PCNA or PAF, and lysates were analyzed by Western blot. The siRNAs did not result in changes in the cell cycle distribution, as analyzed by FACS (data not shown).
As the PIP-box in PAF was located between D-box and initiation motif, PCNA might interfere with initiation. Indeed, when PCNA was added to reactions containing APC/C, Ube2C, and methylubiquitin, it inhibited chain initiation on PAF in a concentration-dependent manner (Figure 6C). As a result of the reduced initiation, PCNA impaired the modification of PAF with wt-ubiquitin with an identical concentration-dependency (Figure 6D). PCNA had no effects on the APC/C-dependent ubiquitination of PAFΔPIP, and it did not inhibit the modification of APC/C-substrates that do not interact with PCNA (Figure 6E; data not shown). Thus, PCNA inhibits chain initiation on PAF in a substrate-specific manner.
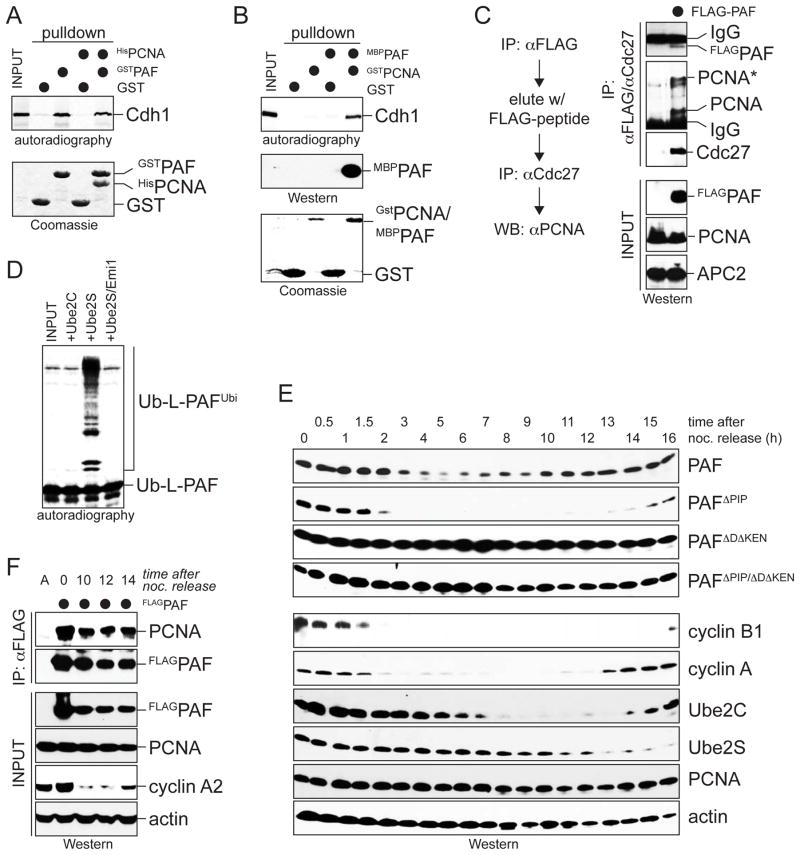
In contrast to its effect on initiation, PCNA did not inhibit binding of PAF to the APC/C. First, the PIP-box was not required for the interaction of PAF with Cdh1 or for its degradation by APC/C (Figure 6F; S6). Moreover, pulldown assays showed that GstPAF efficiently bound Cdh1, even if it was saturated with PCNA prior to incubation with Cdh1 (Figure 7A). These experiments suggest that the binding sites for Cdh1 and PCNA are far enough apart to allow PAF to bind both proteins at the same time. Indeed, PAF was able to bridge an interaction between GstPCNA and Cdh1, which provides evidence for ternary complexes between PAF, PCNA, and Cdh1 (Figure 7B). To test for this in vivo, we precipitated FLAGPAF-containing complexes from HeLa cells, eluted with peptide, re-precipitated with antibodies against the APC/C, and probed for PCNA. Confirming our in vitro studies, these experiments revealed the existence of ternary complexes between PAF, PCNA, and the APC/C in cells (Figure 7C).
Figure 7. Regulated chain initiation controls the timing of substrate degradation.
AHisPCNA does not inhibit the interaction between GstPAF and 35S-Cdh1 as analyzed by autoradiography. B. MBPPAF can form ternary complexes with GstPCNA and 35S-Cdh1, as analyzed by autoradiography. As MBPPAF and GstPCNA co-migrate, MBPPAF was detected by Western blot. C. PAF, PCNA, and APC/C form ternary complexes in vivo. FLAGPAF was precipitated from lysates of prometaphase HeLa cells. FLAG-peptide eluates were re-precipitated with antibodies recognizing the APC/C, and PCNA was detected by Western blot. The asterisk marks a modified form of PCNA. D. PCNA does not inhibit APC/C-binding or chain elongation on PAF. 35S-Ub-L-PAF was bound to GstPCNA and incubated with APC/C, ubiquitin, and Ube2C (for initiation) or Ube2S (for elongation). E. The initiation efficiency determines the timing of PAF degradation and re-accumulation. Synchronized HeLa cells stably expressing FLAGPAF, FLAGPAFΔPIP, FLAGPAFΔDΔKEN, or FLAGPAFΔPIPΔDΔKEN were released into anaphase to activate the APC/C, and levels of PAF proteins and cell cycle markers were measured by Western blot. F. PCNA interacts with PAF during late G1, when PAF, but not PAFΔPIP, is stabilized. HeLa cells expressing FLAGPAF were synchronized in prometaphase (t=0) or late G1 (10–14h after nocodazole release). PAF was precipitated on FLAG-agarose, and co-precipitating PCNA was detected by Western blot.
To test whether PCNA affects chain elongation, we studied a fusion between ubiquitin and PAF, Ub-L-PAF. We bound Ub-L-PAF to GstPCNA, and incubated these complexes with APC/C and either Ube2C for initiation or Ube2S for elongation. As seen previously, Ube2C was unable to ubiquitinate Ub-L-PAF that was stoichiometrically bound to PCNA (Figure 7D). In contrast, Ube2S efficiently elongated ubiquitin chains on PCNA-bound Ub-L-PAF in an APC/C-dependent manner. This experiment shows that PCNA does not block the binding of PAF to the APC/C nor chain elongation by Ube2S. Instead, PCNA specifically inhibits the activity of the initiating E2 Ube2C towards PAF.
The initiation efficiency controls the timing of substrate degradation
To analyze the impact of regulating chain initiation, we constructed cell lines that stably express from a constitutive promoter: FLAGPAF, which is recognized by PCNA and has impaired chain initiation; FLAGPAFΔPIP, which does not bind PCNA and has rapid initiation; and FLAGPCNAΔDΔKEN or FLAGPCNAΔDΔKEN/ΔPIP, which should be inert against the APC/C. Expression of PAF mutants at low levels did not strongly affect cell cycle progression, as seen by FACS or BrdU-staining (Figure S7A, B). Cells synchronized in prometaphase were released into fresh medium to activate APC/C, and PAF-levels were monitored until S phase, when APC/C is inhibited again. Three important observations were made (Figure 7E). First, PAF was degraded less efficiently and later than PAFΔPIP, indicating that the decreased initiation efficiency in wt-PAF delays substrate turnover. Second, PAF accumulated once Ube2C-levels had dropped below a certain threshold (Figure 7E, F), but PAFΔPIP was degraded throughout G1. Thus, reducing the initiation efficiency increases the dependency on the initiating E2. Third, PAF lacking its D- and KEN-boxes was stable independently of whether it was able to bind PCNA or not, showing that its degradation occurred through APC/C. These findings demonstrate that initiation motifs provide an effective means of controlling the timing of substrate degradation in cells.
Discussion
Here, we report that efficient chain initiation by the APC/C-specific E2 Ube2C requires motifs that are present in multiple APC/C-substrates. Variations in the composition of these initiation motifs can determine the rate of substrate degradation without affecting their affinity for the E3. Initiation motifs also provide an opportunity for regulation, which is used by APC/C to control the timing of substrate proteolysis during the cell cycle. On the basis of these results, we propose that initiation motifs and their cognate E2s increase the capacity of E3s to exert temporal control over substrate degradation.
Characteristics of initiation motifs
Studies with the ubiquitin-like modifier SUMO had shown that residues in the vicinity of a substrate lysine can affect the efficiency of modification by an E2 (Bernier-Villamor, et al. 2002; Mohiden et al., 2009). Our results indicate that chain initiation by the APC/C and Ube2C can in an analogous manner be controlled by sequence motifs in substrates that are close to or overlapping with the preferred attachment site for the first ubiquitin. As mutation of initiation motifs stabilized several APC/C-substrates, they play a key role in promoting ubiquitin-dependent degradation.
In all APC/C-substrates analyzed here, charged residues were required for the function of initiation motifs. As seen with geminin, Lys residues in the initiation motif can serve as major acceptor sites for ubiquitin. However, the initiation motifs in geminin, securin, cyclin B1, Plk1, Ube2C, and PAF were still functional if all lysines were mutated to arginine, which retains the positive charge but is unable to act as ubiquitin acceptor. We conclude that initiation motifs contain modified lysines, but also promote the interaction of the substrate with an initiating E2, similar to binding of the consensus SUMOylation motif by Ubc9 (Bernier-Villamor, et al. 2002).
With the exception of Ube2C, initiation motifs were found close to substrate D-boxes. In geminin, altering the distance between its APC/C-binding motif, the D-box, and the initiation motif impaired chain initiation and degradation, underscoring the importance of the proper location of an initiation motif. This finding is in agreement with structural analyses of the APC/C, which place the RING-domain subunit Apc11, and by inference Ube2C, in proximity to the D-box co-receptors Apc10 and Cdh1 (de Fonseca et al., 2010). This situation is also reminiscent of the SCF, which ubiquitinates β-catenin and IκBα on Lys residues in proximity to their SCFβTrCP-binding motifs (Wu et al., 2003). Thus, the presence of conserved patches of charged residues in proximity to an APC/C-binding motif are the main characteristics of initiation motifs recognized by Ube2C.
Functions of initiation motifs
The activation of the APC/C during mitosis results in the massive upregulation of K11-linked chains (Matsumoto et al., 2010), implying a large number of APC/C-substrates in human cells. As the concentration of active APC/C is comparably low, the APC/C likely acts close to saturation. Indeed, increasing the concentration of a single substrate was sufficient to bring cell division to a halt by overloading the APC/C (Marangos and Carroll, 2008). Kinetic studies with the SCF found that most encounters between this E3 and its substrates result in substrate dissociation before ubiquitination can take place (Pierce et al., 2009). Due to its large number of substrates, unproductive binding events might be detrimental for APC/C-dependent ubiquitination. Initiation motifs, however, provide an elegant means to sufficiently increase the rate of degradation to allow the APC/C to shoulder the burden of its many substrates.
Our experiments suggest that the APC/C also implements initiation motifs to fine-tune the rate of substrate degradation. Once activated, the APC/C promotes substrate turnover in a conserved sequence, ensuring that inhibitors of anaphase are degraded prior to proteins required for chromosome segregation or cytokinesis (Peters, 2006). Previous work had shown that the processivity of chain formation, but not the affinity of a substrate to the APC/C, correlates with the degradation time in cells (Rape et al., 2006). Because more rapid initiation increases the processivity of ubiquitination (Pierce et al., 2009), it should accelerate the rate of substrate degradation. As seen in initiation motif swaps using geminin and Ube2C, and in a mutational analysis of the APC/C-substrate PAF, this is the case: improving the quality or accessibility of initiation motifs accelerated the degradation of Ube2C or PAF, whereas reducing the quality of the initiation motif in geminin delayed its degradation. Altering initiation motifs had little effect on substrate affinity for the APC/C. These findings identify chain initiation as the rate-limiting step for the degradation of many APC/C-substrates, in analogy to the SCF (Pierce et al., 2009). As a result, initiation motifs play a critical role in determining the timing of substrate degradation by the APC/C.
Regulation of chain initiation
As initiation motifs differ from APC/C-binding sites, the APC/C can separate substrate binding and ubiquitination to establish negative feedback regulation: During mitosis, APC/C-activators, such as Ube2C, bind the E3 and modify its substrates without being degraded themselves. However, when Ube2C engages the APC/C after substrates have been turned over, it undergoes APC/C-dependent ubiquitination and degradation (Rape and Kirschner, 2004). We found that ubiquitination of Ube2C, but not its APC/C-binding, requires an N-terminal initiation motif. Increasing the quality of this initiation motif accelerated Ube2C-ubiquitination without affecting its association with the APC/C. Moreover, competition by substrate initiation motifs inhibited Ube2C-degradation without affecting its APC/C-binding or E2-activity. Thus, even though the initiation motif does not determine the affinity of Ube2C to the APC/C, its accessibility helps determine the timing of Ube2C-degradation.
Regulating the initiation efficiency also controls the timing of degradation and re-accumulation of canonical substrates, such as PAF. PAF is strongly stabilized by its binding to PCNA, which selectively inhibits chain initiation, yet has no effects on APC/C-binding or chain elongation. As the PIP-box in PAF is located between D-box and initiation motif, PCNA might sterically block access of Ube2C to the PAF-initiation motif. The stabilization of PAF by PCNA might play a role during late G1 or upon DNA damage, when PAF has been suggested to cooperate with PCNA, yet APC/C is active (Hosokawa et al, 2007). It is also possible that PAF bridges an interaction between PCNA and APC/C, thereby recruiting APC/C to sites of ubiquitination on DNA; the degradation of PAF upon dissociation of PCNA might then provide negative feedback.
These findings provide an unappreciated function for PCNA in cell cycle control. Initially identified as a processivity factor for DNA polymerases, PCNA had recently been shown to be an important regulator of protein degradation during S phase. By binding to an extended PIP-box, PCNA increases the affinity of the CDK-inhibitor p21 for the E3 Cul4Cdt2, thereby triggering p21 degradation (Havens and Walter, 2009). By contrast, PCNA stabilizes PAF, a protein required for S phase progression. Although more work is required, this may indicate that PCNA acts as a switchboard to degrade or stabilize cell cycle regulators, depending on their function in DNA replication or repair.
In conclusion, we show that initiation motifs allow the APC/C to precisely control the timing of substrate degradation. Our observations extend the similarities between the formation of ubiquitin chains and other processive reactions, such as transcription or translation, which can be divided into distinct initiation, elongation, and termination steps. Whether we can learn more by comparing the mechanisms of ubiquitin chain formation and those long-studied processive reactions will be an avenue of future research.
Methods
A detailed description of the methods can be found in the supplementary information accompanying this manuscript.
Plasmids and reagents
Table S1 lists all constructs, antibodies, and siRNAs. pCS2 or pCS2-ZZ/TEV were used for IVT/T and expression in cells, pET28, pGEX, or pMAL for purification, and pCMV for generation of stable lines.
Degradation assays
Degradation assays were performed as described (Rape et al., 2006). Ube2C and IMTEK-Ube2C were synthesized as ZZ/TEV-fusions, purified over IgG-sepharose and eluted by TEV-cleavage, before addition to extracts.
In vivo degradation and siRNA experiments
293T cells were transfected with substrate plasmid, mycCdh1, and Emi1 using TransIT-293. 24hr post transfection, lysates were analyzed by Western blot. Release experiments were performed in HeLa cells transfected with substrate (TransIT-LT1), arrested by thymidine/nocodazole, and replated after shake-off in fresh medium. For analysis of PAF degradation, HeLa cells stably expressing FLAGPAF mutants were used.
siRNA experiments were performed in HeLa cells transfected with siRNA against the 3′-UTR of PAF or PCNA using HiPerfect. Phenotypes were identical for different siRNAs per gene, and strong effects on cell cycle progression were excluded by FACS.
Ubiquitination assays
Ubiquitination assays were performed as described (Rape et al., 2006). Ub-L-geminin and Ub-L-PAF were synthesized in reticulocyte lysate containing 175μM ubiK29R to inhibit the UFD-pathway. Ub-L-Geminin, Ub-L-PAF, PAF, and Ube2C were synthesized as ZZ/TEV-fusions, purified on IgG Sepharose, and eluted by TEV cleavage. GstPCNA/Ub-L-PAF complexes were purified by incubating Ub-L-PAF with GstPCNA coupled to Glutathione Sepharose 4B.
Detection of ternary complexes between APC/C, PAF, and PCNA in cells
HeLa cells stably expressing FLAGPAF or control cells were synchronized in prometaphase. Lysates were incubated with anti-FLAG M2 affinity resin. After washing, bound proteins were eluted with peptide. Pooled eluates were incubated with αCdc27- antibodies and Protein G agarose. After washing, proteins were eluted with SDS sample buffer and analyzed by Western blot.
Xenopus tropicalis embryo injections
Embryos collected from mating X. tropicalis couples were dejellied for 5min in 3% cysteine, pH 8.0, and transferred to 3% ficoll in 1/9x Modified Ringer solution. One cell of a two-cell stage embryo was injected with 3ng or 6ng of protein (final conc. ~1μM at the two cell stage) premixed with rhodamine-tubulin for lineage tracing. Injected embryos were evaluated by uorescence, and scored for stage and phenotype.
Determination of ubiquitination sites on geminin
Recombinant geminin was incubated with APC/C, E1, Ube2C and methylubiquitin. Monoubiquitinated geminin was excised from SDS-gels, trypsinized, and subjected to tandem mass spectrometry, as detailed in the supplementary information.
Supplementary Material
Acknowledgments
We thank the Rape lab for helpful discussions; the Heald lab for rhodamine-tubulin; and Julia Schaletzky for reading the manuscript and many suggestions. Mass spectrometry instrumentation was supported by NIH (1S10RR022393). AW is a CRCC fellow. This work was supported by an NIH grant (5R01GM83064-3), a March of Dimes grant, and an NIH New Innovator Award to MR. MR is a Pew fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–56. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat Cell Biol. 2002;4:880–7. doi: 10.1038/ncb871. [DOI] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–8. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2010 Nov 24; doi: 10.1038/nature09625. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–9. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens CG, Walter JC. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell. 2009;35:93–104. doi: 10.1016/j.molcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, et al. Oncogenic role of KIAA0101 interacting with proliferating cell nuclear antigen in pancreatic cancer. Cancer Res. 2007;67:2568–76. doi: 10.1158/0008-5472.CAN-06-4356. [DOI] [PubMed] [Google Scholar]
- Huang DT, Zhuang M, Ayrault O, Schulman BA. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat Struct Mol Biol. 2008;15:280–7. doi: 10.1038/nsmb.1387. [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Phillip I, Rape M. Mechanism of ubiquitin chain formation by the human Anaphase-Promoting Complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–44. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Marangos P, Carroll J. Securin regulates entry into M-phase by modulating the stability of cyclin B. Nat Cell Biol. 2008;10:445–51. doi: 10.1038/ncb1707. [DOI] [PubMed] [Google Scholar]
- Matsumoto ML, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11-linkage specific antibody. Mol Cell. 2010;39:477–84. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–53. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Mohideen F, Capili AD, Bilimoria PM, Yamada T, Bonni A, Lima CD. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat Struct Mol Biol. 2009;16:945–52. doi: 10.1038/nsmb.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–20. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–9. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Sadowski M, Suryadinata R, Lai X, Heierhorst J, Sarcevic B. Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34. Mol Cell Biol. 2010;30:2316–29. doi: 10.1128/MCB.01094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2010;36:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31:544–56. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi L, Fareh M, Aberdam E, Kitajima S, Simpson F, Wicking C, Aberdam D, Virolle T. ATF3 and p15PAF are novel gatekeepers of genomic integrity upon UV stress. Cell Death Differ. 2009;16:728–37. doi: 10.1038/cdd.2009.2. [DOI] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci USA. 2009;106:18213–8. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–56. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107:1355–60. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol Cell. 2010;37:784–96. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2010;25:5083–93. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Huang B, Shen M, Lau C, Chan E, Michel J, Xiong Y, Payan DG, Luo Y. p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene. 2001;20:484–9. doi: 10.1038/sj.onc.1204113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.