Abstract
Peroxiredoxin 6 (Prdx6) is the prototype and the only mammalian 1-Cys member of the Prdx family. Major differences from 2-Cys Prdxs include the use of glutathione (GSH) instead of thioredoxin as the physiological reductant, heterodimerization with πGSH S-transferase as part of the catalytic cycle, and the ability either to reduce the oxidized sn-2 fatty acyl group of phospholipids (peroxidase activity) or to hydrolyze the sn-2 ester (alkyl) bond of phospholipids (phospholipase A2 [PLA2] activity). The bifunctional protein has separate active sites for peroxidase (C47, R132, H39) and PLA2 (S32, D140, H26) activities. These activities are dependent on binding of the protein to phospholipids at acidic pH and to oxidized phospholipids at cytosolic pH. Prdx6 can be phosphorylated by MAP kinases at T177, which markedly increases its PLA2 activity and broadens its pH-activity spectrum. Prdx6 is primarily cytosolic but also is targeted to acidic organelles (lysosomes, lamellar bodies) by a specific targeting sequence (amino acids 31–40). Oxidant stress and keratinocyte growth factor are potent regulators of Prdx6 gene expression. Prdx6 has important roles in both antioxidant defense based on its ability to reduce peroxidized membrane phospholipids and in phospholipid homeostasis based on its ability to generate lysophospholipid substrate for the remodeling pathway of phospholipid synthesis. Antioxid. Redox Signal. 15, 831–844.
Introduction
Peroxiredoxin 6 (Prdx6) was the sixth (and final) mammalian member of the Prdx family to be described. Prdx6 shares structural and functional properties with other members of this family but has important characteristics that make it unique among the Prdxs. Like the other Prdxs, Prdx6 is a nonseleno peroxidase. However, unlike the other members of the family, Prdx6 has a single conserved cysteine residue and has been called a 1-Cys Prdx, in contrast to the other members which are classified as 2-Cys (Prdx 1–4) or atypical 2-Cys (Prdx5). Because of its single active-site Cys residue, the catalytic cycle for Prdx6 differs from the 2-Cys family members. A second important distinguishing characteristic of Prdx6 is that thioredoxin does not participate in the catalytic cycle. Indeed, the Prdxs were previously called thioredoxin peroxidases, but the family name was changed to Prdxs after the discovery of Prdx6 (34). Although some are yet to be convinced, glutathione (GSH) appears to be the physiological reductant for Prdx6. A third important characteristic that distinguishes Prdx6 is its ability to bind and reduce phospholipid hydroperoxides. This enzymatic activity has an important role in antioxidant defense. Finally, Prdx6, unlike the other Prdxs, is a bifunctional enzyme with phospholipase A2 (PLA2) activity in addition to its peroxidase function.
The protein that is now called Prdx6 was first isolated from the ciliary body of the bovine eye (86). The protein was characterized as unique by its N-terminal amino acid sequence and was called a nonselenium GSH peroxidase. Subsequently, the cDNA (called ORF6) was identified by random cloning (64) and the deduced protein sequence was recognized as a member of the Prdx family (4). Shortly thereafter, the protein was isolated from rat olfactory mucosa, where it was described as a GSH peroxidase (70), and from rat lung, where its PLA2 activity was identified (38). Subsequently, recombinant protein generated in Escherichia coli was shown to have both peroxidase and PLA2 activities (6, 34). Before the consensus on nomenclature, the protein had been called nonselenium GSH peroxidase (86), aiPLA2 (38), 1-Cys Prdx (34), antioxidant protein 2 (AOP2) (75), Clara cell protein 26 (CC26), and p29 (47), whereas the gene in addition to ORF6 (64) has been called LTW4 (30) and keratinocyte growth factor (KGF)-regulated gene 1 (24).
The designation Prdx6 is based on the protein found in mammalian cells. However, homologous 1-Cys proteins are widely distributed throughout all kingdoms. These proteins, based on identity of the catalytic center, have been described in archaea, bacteria, parasites, yeast, insects, mollusks, amphibians, birds, and other orders although differences exist with respect to activity and structure compared with the mammalian enzymes (66). As examples, yeast 1-Cys Prdx is thioredoxin dependent, unlike the mammalian enzymes, whereas nonmammalian species show considerable heterogeneity in the amino acid sequence corresponding to the 1-Cys Prdx lipase motif (66); whether any of these alternate motifs support PLA2 activity is yet to be determined. Several reviews of Prdx6 have been published previously (53, 66, 73, 84). This review will focus on the mammalian enzyme.
Prdx6 Structure
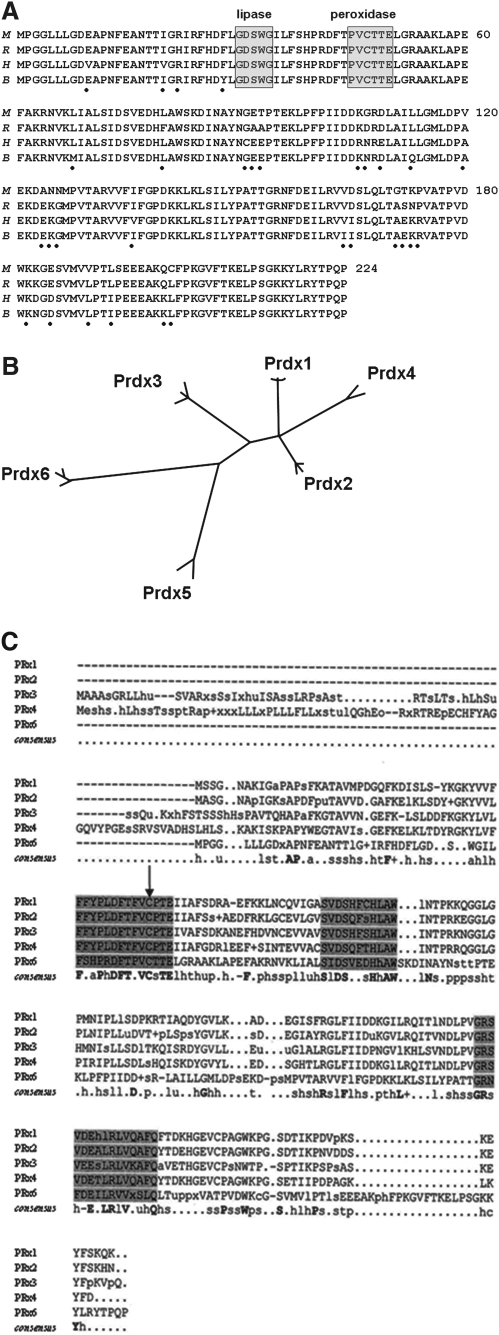
The structural properties of Prdx6 have been elucidated through the deduced amino acid sequence, gel electrophoresis, circular dichroism measurements, and protein crystallization. The primary sequence consists of 224 amino acids and indicates a molecular mass of 25,100 Da (for the human enzyme), although the protein generally migrates at 26–29 kDa on SDS/PAGE (38). The pI of the native protein is ∼5.1 and varies with oxidation or phosphorylation state of the protein (78, 102). Comparison of mammalian (human, rat, mouse, and bovine) Prdx6 proteins indicates 87% identity of the amino acid sequences (Fig. 1A). Further, population-based sequencing of the human gene has indicated a relatively low frequency of polymorphisms, with almost none in the coding region (Jason Christie, personal communication). This high degree of conservation suggests an important role for Prdx6 in the metabolism of mammalian cells. Like the other members of the Prdx family, Prdx6 has a conserved Cys residue near the N-terminus (C47); the rat and bovine proteins contain no additional Cys residues, whereas one additional Cys is present in the human (C91) and mouse (C201) proteins.
FIG. 1.
The primary structure of peroxiredoxin 6 (Prdx6). (A) Deduced Prdx6 amino acid sequence for the mouse (M), rat (R), human (H), and bovine (B) proteins. The dots beneath the sequence indicate amino acids that are not fully conserved among the four species. The lipase and peroxidase motifs are indicated. Modified from Kim et al. (37) and Fisher et al. (21). (B) Phylogenetic tree for mammalian Prdxs constructed using ClustalW (1.7). Modified from Knoops et al. (41). (C) Consensus sequences aligned using ClustalW (1.7). The consensus sequences for the human, mouse, and rat homologs for Prdxs 1, 2, 3, 4, and 6 were generated using the consensus C program. The catalytic Cys residue is indicated by the arrow. The shading indicates regions of relatively high homology. Modified from Phelan (73).
For the most part, the protein has limited amino acid sequence homology to other Prdxs, and Clustal analysis indicates divergences of Prdx6 (and Prdx5) from the more conserved Prdx1–4 (Fig. 1B). However, there are several stretches where residues show greater conservation (Fig. 1C). This is especially evident around the catalytic C47 residue but also at 2 other stretches unrelated to the sites for enzymatic activity. The sequence surrounding the active site of the oxidase activity in Prdx6 is PVCTTE, analogous to the surrounding sequence in Prdx 1–4, FVCPTE. In addition, the protein has a sequence at positions 30–34 that has been termed a lipase motif (GXSXG) and is present in a diverse group of lipolytic enzymes (10), but is not present in the 2-Cys Prdxs. The S32 in this motif is crucial for the PLA2 activity of Prdx6 (6). Circular dichroism analysis of Prdx6 shows approximately equal contributions of alpha helix, β sheets and turns, and random regions to its secondary structure (54, 55).
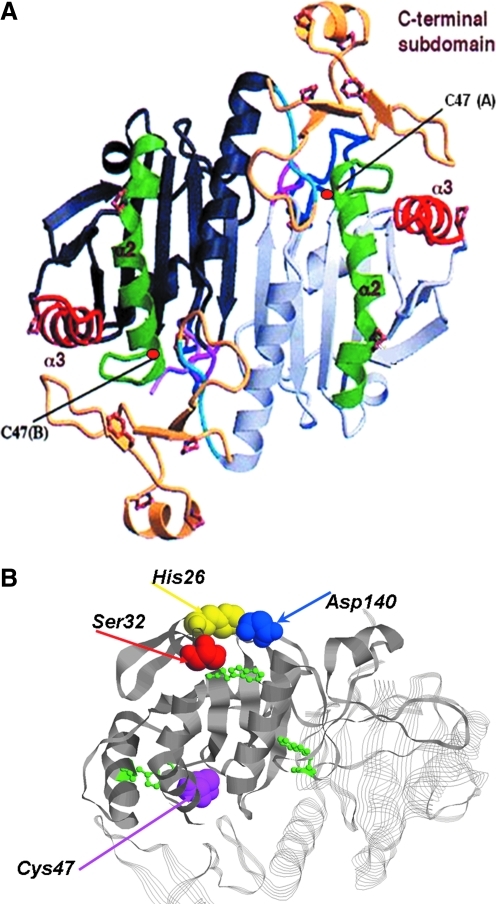
Human Prdx6 protein has been crystallized in the oxidized state and analyzed at 2 Å resolution (7) (Fig. 2A). It was necessary to mutate the nonconserved C91 residue to prevent disulfide formation in order to obtain crystals. The protein crystallized as a homodimer but not as a disulfide as the C47 residues in the individual monomers are at a considerable distance apart. Unlike the 2-Cys Prdxs where the catalytic Cys residues are on the surface and available to form disulfides (homodimers) as well as higher forms, the conserved Cys in Prdx6 is buried at the base of a narrow pocket (7) (Fig. 2B). This location renders it unable to dimerize through disulfide formation in the native configuration, but homodimers (and multimers) can arise through hydrophobic interactions. Disulfide formation may occur with denatured proteins (72) and heterodimerization also occurs normally as part of the catalytic cycle (see below). Like the other Prdxs and many other redox-active proteins, Prdx6 contains a typical thioredoxin fold consisting of ∼80 amino acids arranged in four antiparallel β sheets sandwiched between two alpha helices (7) (Fig. 2A). This region is essential for the peroxidatic function of the enzyme. The protein also contains a surface expressed catalytic triad (S-D-H) (Fig. 2B) that is important for phospholipid binding and enzymatic activities as described below.
FIG. 2.
Tertiary structure of Prdx6. (A) Crystal structure of mutated (C91S) human Prdx6 that was oxidized by air exposure. The protein crystallized as a homodimer. Notable features are the thioredoxin fold consisting of a four-stranded β sheet with two flanking α helices. The c-terminal domain comprises amino acid residues 175–224 and consists of 3 β strands and an α helix attached to the larger internal domain by a short loop. The catalytic C47 in each monomer is indicated. Modified from Choi et al. (7). (B) Ribbon diagram showing the relative position of the active site for peroxidase activity (C47) and the catalytic triad for phospholipase A2 (PLA2) activity (S32, H26, D140). The SDH catalytic triad is on the protein surface, whereas C47 is at the base of a narrow pocket. Also indicated (in green) are the positions of the three Trp residues that have been used to analyze substrate binding (54, 55). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Tissue Distribution
Initial immunohistochemical studies in mammals demonstrated the presence of Prdx6 in lungs but were unable to detect the protein in most other organs (37). However, this appears to have been due to a relatively low reactivity of the available antibodies. Subsequent study with more reactive antibodies and by measurement of mRNA expression have demonstrated wide-spread distribution in essentially all organs and essentially all cell types (53). Consistent with the initial observations, the expression level of Prdx6 mRNA and protein is greatest in the lung with high levels of expression also seen brain, testis, kidney, and liver (25, 37, 49, 50, 60). A previous report suggesting highest expression levels for the mRNA in mouse liver may have been skewed by the use of β-actin in normalization for comparison of different tissues (94); the liver content of β-actin is relatively low so that normalization would result in an inaccurately high value.
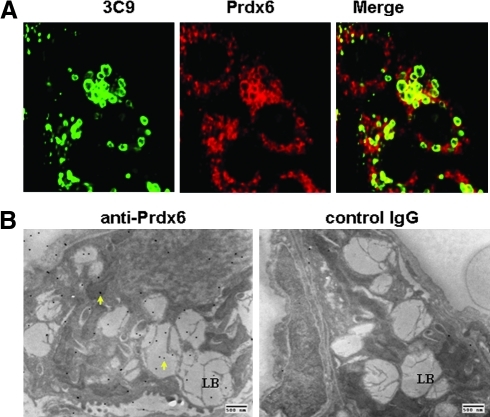
Within organs, expression of Prdx is greatest in epithelium such as apical regions of respiratory epithelium and skin epidermis (68). Within the lung, Prdx6 is expressed at relatively high levels within the type II alveolar epithelial cells and bronchiolar Clara cells; a lower level of expression is seen in alveolar macrophages and microvascular endothelium (37, 39, 78). Prdx6 expression within cells is cytosolic. The protein also is present in lysosomes in lung epithelial cells and alveolar macrophages as well as in lung lamellar bodies, the surfactant storage and secretory organelle (1, 89, 103) (Fig. 3A, B). The latter is thought to represent a modified secretory lysosome and, like lysosomes, maintains an acidic internal pH. Prdx6 has not been demonstrated in other organelles under normal conditions, but it does translocate to hepatocyte mitochondria when the liver is subjected to experimental ischemia-reperfusion injury (13). Expression of Prdx6 in lysosomes and organelles of other tissues and possible translocation of the protein with other forms of oxidant stress has not been reported.
FIG. 3.
Subcellular localization of Prdx6 in rat lung alveolar epithelial type II cells. (A) Immunofluorescence using antibodies to 3C9, a marker for lung lamellar body membranes, and Prdx6. The merged image shows colocalization for the two proteins, indicating the presence of Prdx6 in lung lamellar bodies. (B) Ultrastructural localization of Prdx6 using electron microscopic immunohistochemistry with 20 nm gold-coupled antibody. Arrows indicate gold grains of Prdx6 in lamellar bodies (LB) with a lesser concentration of grains in other organelles. The right panel is a control in the absence of primary antibody. Modified from Wu et al. (103).
We have recently explored the mechanism for delivery of Prdx6 to organelles in lung cell lines. Using a protein truncation approach, we demonstrated that targeting of Prdx6 from the site of its synthesis (endoplasmic reticulum) to the lysosomal/lamellar body compartments depends on the sequence between amino acids 31 and 40 in the N-terminal region of Prdx6 (89). Detailed analysis demonstrated that S32, G34, and at least one or more additional residues between amino acids 35 and 40 are essential for targeting. Targeting does not depend on binding of Prdx6 to phospholipids (89) but rather depends on binding to 14-3-3ɛ as a molecular chaperone (88).
Regulation of Expression
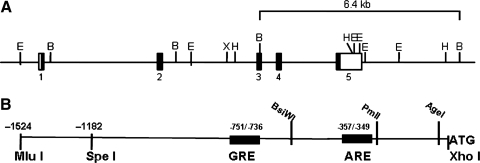
The Prdx6 gene from the mouse comprises five exons and four introns spanning ∼11 kb (Fig. 4A) and is located on chromosome 1 (75). The gene has been mapped to a 0.66 cM interval between D1Mit 159 and D1Mit 398 (74), The cDNA for various mammalian species (mouse, rat, bovine, and human) has been reported in the range of 1.4–1.7 kb, varying with the length of the 3′ untranslated region (21, 38, 62, 73). Analysis of the gene promoter region has shown an antioxidant response element (ARE) (8), a putative glucocorticoid response element (GRE) (Fig. 4B), and potential binding sites for SP1, sterol response element binding protein (SREBP), upstream stimulatory factor (USF), alcohol dehydrogenase regulatory protein 1 (ADR1), heat shock factor, Pit1, cJun, cMyc and several other transcription factors (49, 75). SREBP and upstream stimulatory factor have been linked to regulation of genes involved in lipid metabolism (75), consistent with one of the important roles of Prdx6. Several alternate murine Prdx6 transcripts have been described and appear to be related to 2 intronless genes (49, 75). Several of these transcripts are specific to liver (90). Another transcript related to one of the intronless genes has been shown in testes but is not present in lung (60, 90). None of these alternate transcripts has been shown to result in protein expression.
FIG. 4.
Regulation of Prdx6 expression. (A) The structure of the Prdx6 gene indicating five exons and intervening introns. The black boxes represent the protein coding regions in the exons and the open boxes represent noncoding regions. The sequence from exon 1 to exon 5 represents 10.3 kb. Restriction sites are indicated: B, BAMH1; E, EcoR1; H, Hind3; X, Xho1. (B) Translational regulatory elements and restriction recognition sites upstream of the Prdx6 translational start site (ATG). The figure indicates the position of an antioxidant (electrophilic) response element (ARE) (8) and glutocorticoid response element (GRE) (unpublished results). The numbers indicated are relative to the site for start of transcription. Modified from Mo et al. (60).
Oxidant stress is a potent inducer of Prdx6 expression. Exposure to hyperoxia as a model of oxidant stress resulted in an approximate doubling of Prdx6 expression in rat and mouse lungs and alveolar type II cells (35). Induction by oxidant exposure (H2O2, paraquat) also was shown in lung epithelial and hepatocyte cell lines and was blocked by the presence of antioxidants (35, 90). Using classical methods to investigate promoter activity, we have shown in a lung epithelial cell line that oxidant stress stimulates Prdx6 gene expression by a transcriptional mechanism involving its ARE (8). The ARE consensus sequence is located between positions −357 and −349 upstream from the translational start site in the Prdx6 gene (Fig. 4B). [Note that the original publication (8) incorrectly referred to this as the transcriptional start site.] Transcription is activated by binding of the transcription factor Nrf2 to the ARE, whereas transcription is inhibited by the binding of Nrf3 (8). This mechanism for transcriptional regulation of Prdx6 is similar to that described for many antioxidant enzymes (32).
Prdx6 expression also is responsive to hormonal regulation. This protein is expressed at low levels in mouse embryos and in fetal rat lungs and kidneys, but shows a marked increase immediately after birth (25, 36, 49). Treatment of neonatal rats with a corticosteroid (dexamethasone) resulted in a modest induction of expression and a similar effect was seen with human fetal lung cells during in vitro culture. Dexamethasone also induces Prdx6 expression in adult lung cells and regulates transcription through its interaction with a GRE in the promoter region of the Prdx6 gene (Fig. 4B) (unpublished studies from our laboratory). This response to corticosteroids may have physiological relevance related to the role of Prdx6 in lung surfactant metabolism (36). Transcription of Prdx6 message in dermal epithelium is induced by treatment with KGF, a response of presumed importance related to wound healing (24). Our preliminary studies (unpublished) indicate that the ARE and Nrf2 are required for the effect of KGF on Prdx6 expression.
Enzymatic Activities
Assays with recombinant protein have demonstrated that Prdx6 is a bifunctional protein with both GSH peroxidase and PLA2 activities (6). Site directed mutation analysis has clearly shown that these activities require two distinct active sites, namely, a Cys 47-dependent peroxidase activity and a Ser32-dependent PLA2 activity (6). These results have been confirmed by the use of specific inhibitors (6, 21). Mercaptosuccinate is a thiol-active agent that inhibited the peroxidase activity of Prdx6, whereas the PLA2 activity was unaffected. Both MJ33, a phospholipid substrate intermediate analog, and serine protease inhibitors inhibited the PLA2 activity of Prdx6 but had no effect on peroxidase activity. The two catalytic centers (C47 and S32) according to the crystal structure are ∼25 Å distant so that a direct interaction between them is unlikely (53).
GSH peroxidase
As described above, the original isolates of Prdx6 from bovine ciliary body and subsequently from rat olfactory mucosa showed a protein that demonstrated GSH peroxidase activity but did not contain the seleno-cysteine moiety that is characteristic of GSH peroxidases (70, 86). Defining this protein as a nonseleno-GSH peroxidase created some confusion with the GSH S-transferases (GST), which also are nonseleno enzymes, although Prdx6 does not have transferase activity (38, 86). The confusion was compounded when initial in vitro studies of recombinant protein generated in E. coli showed peroxidase activity in the presence of reductants such as dithiothreitol but no significant activity with GSH (34). Clearly, dithiothreitol is not a physiological reductant and thioredoxin, the reductant that is active in the catalytic cycle for the other Prdxs, was not effective as a reductant for Prdx6 (21, 34). Subsequent studies showed that some, but not all, preparations of recombinant protein could utilize GSH as a reductant of oxidized Prdx6 (6, 17), but this has been variable (52, 72). The unpredictability of this response has been attributed to the vagaries associated with the in vitro generation of recombinant protein although similar vagaries of response to GSH have been noted for isolated native protein (70, 71).
Analysis of the crystal structure of the protein sheds some light on the nature of the problem regarding reduction of oxidized Prdx6, although the reasons for the disparities still are not fully resolved. C47 is the only conserved Cys in Prdx6 and analysis of C47 mutants has clearly demonstrated that this residue is essential for its peroxidase activity. According to the crystal structure, the C47 residue is at the base of a relatively narrow pocket (Fig. 2B). Thus, depending on folding, the pocket could admit or exclude access of the tripeptide GSH to the catalytic site. This formulation raises the possibility that one or another cofactor could regulate Prdx6-folding and thereby regulate access of the physiological reductant. In vitro studies have indicated that Prdx6 can interact with the π isoform of GST, resulting in reduction of oxidized Prdx6 by GSH and the regeneration of active enzyme (52, 81, 82). Ascorbate (61), dihydrolipoic acid (72), or cyclophilin (48) might also reduce the oxidized form of Prdx6. Reduction of Prdx6 by ascorbate in vitro was dependent on the presence of the H39 residue to create a positively charged environment as well as the presence of a stable sulfenic acid intermediate (61). GSH, based on its high intracellular concentration (5–10 mM in most cells), would appear to be most likely as the primary physiologic reductant.
Catalytic cycle
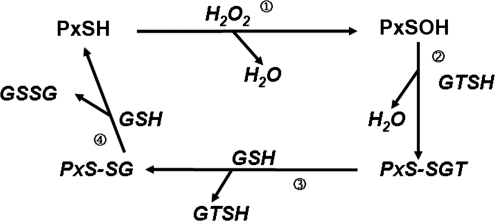
The catalytic cycle presented here is based on the observations that both πGST and GSH are required for continuous substrate reduction (52, 81, 82). The initial step in Prdx6 enzymatic function is similar to that observed with the other Prdxs, namely, oxidation of the catalytic Cys to the sulfenic acid (−SOH) (61, 72) (Fig. 5). Based on the crystal structure, it has been proposed that C47 in Prdx6 is hydrogen bonded to H39 and electrostatically activated by R132; thus, C47-H39-R132 form a catalytic triad for peroxidase activity (7, 53). With the 2-Cys Prdxs, the Cys sulfenic acid reacts with a resolving Cys in the C-terminus of the homodimeric protein to form a disulfide. As an analogous mechanism, the sulfenic acid of Prdx6 forms a heterodimeric disulfide through its interaction with πGST (Fig. 5). With 2-Cys Prdxs, the disulfide is reduced by thioredoxin to regenerate the active enzyme, whereas with Prdx6, this function is served by GSH. The Prdx6: πGST heterodimer interacts with GSH resulting in glutathionylation of Prdx6; the latter is then reduced with another GSH, regenerating Prdx6 while oxidizing GSH to GSSG. Thus, the catalytic cycles are analogous between Prdx6 and the other Prdxs, although the identity of the resolving protein and the physiological reductant differ. The sulfenic cys (S-OH) of Prdx6 can be hyperoxidized to the sulfinic and sulfonic states, although the susceptibility to hyperoxidation in vitro is less than for 2-Cys Prdxs and requires supraphysiological concentrations of H2O2 (500 μM) (72). Unlike the 2-Cys Prdxs, hyperoxidized Prdx6 cannot be reduced by sulfiredoxin and its hyperoxidation after H2O2 treatment of Hela cells is not reversed during a subsequent 6 h period (101).
FIG. 5.
The catalytic cycle for Prdx6 and role of GSH transferase (GT). Prdx6 (PxSH) is shown indicating the active-site sulfhydryl on C47. πGSH S-transferase (GTSH) also is shown with its active sulfhydryl group indicated (by coincidence, also at C47). In its antioxidant role, Prdx6 interacts with an oxidant (H2O2) to generate the sulfenic acid (reaction 1), which then interacts with the SH of πGST to generate the Prdx6:πGST heterodimer (reaction 2). The GSH bound to the πGST glutathionylates Prdx6, liberating πGST (reaction 3). Finally, a second GSH reduces the –SSG bond and regenerates the active (reduced) enzyme (reaction 4). Modified from Ralat et al. (81).
Substrate specificity
Similar to the other Prdxs, Prdx6 reduces H2O2 and other short chain hydroperoxides. The turnover number (k2) with these substrates is ∼106 M−1 s−1 (21). This compares with a turnover number that is approximately an order of magnitude greater for cytosolic GSH peroxidase (GPx1). The maximal activity for the peroxidase activity of recombinant Prdx6 is ∼5 μmol/min/mg protein (52, 81). Prdx6 also can reduce phospholipid hydroperoxides as demonstrated with oxidized phospholipids in solution or incorporated into liposomes (21, 55). Thus, by analogy, this enzyme has the ability to reduce peroxidized membrane phospholipids. As mentioned above, this ability to reduce phospholipid hydroperoxides is not shared by GPx1 or the other Prdxs. The rate constants for reduction of oxidized phosphatidylcholines by Prdx6 are similar to those obtained with H2O2 as substrate and also are similar for reduction of phosphatidylcholine with either oxidized arachidonyl or linoleoyl in the sn-2 position (21). The possible reduction of oxidized complex lipids other than phosphatidylcholine (such as cholesterol) by Prdx6 has not been tested.
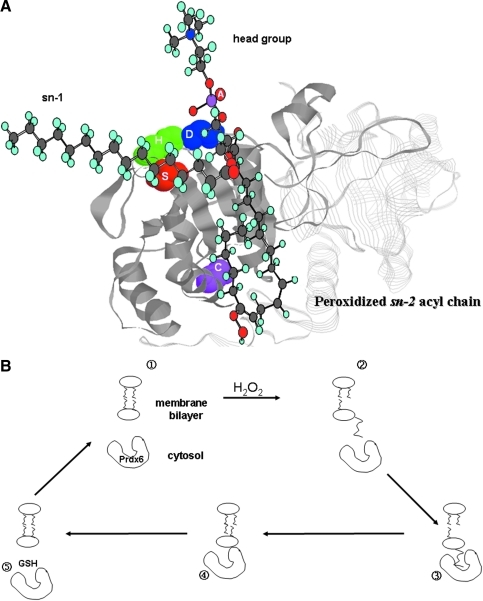
The key factor in the ability of Prdx6 to reduce peroxides of phospholipids is the binding of Prdx6 to the oxidized lipid substrate. Mutagenesis studies based on the crystal structure have indicated that the binding site for protein to phospholipid substrate is on the β turn represented by the amino acid residues H26, S32, and W33 (54, 55) (Fig. 2B). These residues are on the protein surface in proximity to the surface opening for the pocket containing the catalytic C47 residue. We propose that binding of the head group (e.g., phosphorylcholine) of the peroxidized phospholipid to this protein docking site results in insertion of the peroxidized sn-2 fatty acid into the hydrophobic pocket, generating proximity to and subsequent interaction with C47 (Fig. 6A). Of note, Prdx6 at cytosolic pH (pH 7.0) does not bind to normal membranes (reduced phospholipids), whereas the enzyme binds readily to peroxidized phospholipids (55). Thus, we have proposed that cytosolic Prdx6 could bind to and reduce peroxidized membrane phospholipids followed by its dissociation from the membrane and return to the cytosolic compartment (Fig. 6B). We presume that reduction of the sulfenic Cys in Prdx6 occurs within the cytoplasm.
FIG. 6.
Binding of phospholipids to Prdx6. (A) Theoretical analysis of the binding of an oxidized phospholipid to Prdx6. The phospholipid head group binds at the protein surface in the vicinity of the PLA2 catalytic triad (SDH), whereas the oxidized sn-2 oxidized fatty acyl group inserts into the hydrophobic pocket where the oxidized double bond is in proximity to the catalytic Cys47. (B) Scheme for reduction of oxidized membrane phospholipids by cytosolic Prdx6. The resting state shows phospholipids of the membrane bilayer and Prdx6 contained in the cytosol (panel 1). Oxidative stress (H2O) generates a phospholipid hydroperoxide, increasing its aqueous solubility resulting in flotation of the peroxidized sn-2 acyl chain (panel 2). Cytosolic Prdx6 binds to the oxidized phospholipid (panel 3) resulting in reduction of the phospholipid hydroperoxide (panel 4) and dissociation of Prdx6 from the membrane (panel 5). Reduction of Prdx6 with GSH restores the resting state. At this time, it is not clear whether Prdx6 is reduced while attached to the membrane or in the cytosol subsequent to its dissociation. Modified from Manevich et al. (55). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
PLA2 activity
The PLA2 activity of Prdx6 was discovered through follow-up of studies using a novel inhibitor of phospholipid metabolism in the intact lung. This agent, MJ33, is a transition state phospholipid analog that functions as a competitive inhibitor for some PLA2s; within the lung, it appears to specifically inhibit a calcium-independent activity that is maximal at pH 4 (17, 19). This pH versus activity spectrum is compatible with a lysosomal localization. Using these characteristics, a protein with PLA2 activity (called aiPLA2 for acidic, Ca2+-independent PLA2) was isolated from rat lung and, after molecular sequencing, was demonstrated to be identical to the deduced sequence of Prdx6 (37, 38). The PLA2 activity of Prdx6 has been demonstrated with native and recombinant protein (1, 6, 38, 103) as well as with mammalian cells that were transfected with a construct to generate Prdx6 (29).
PLA2s represent a relatively large family of enzymes that can be categorized as: (i) secreted PLA2s (e.g., pancreatic PLA2) that function extracellularly and are strongly calcium dependent; (ii) intracellular PLA2 (cytosolic or cPLA2) that shows specificity for sn-2 arachidonate and does not require calcium for the catalytic mechanism, although low levels of Ca2+ enhance binding; (iii) various other intracellular PLA2s that are Ca2+-independent (iPLA2); and (iv) lysosomal PLA2s that function at acidic pH (63). Prdx6 was the first lysosomal PLA2 to be described although another has been identified subsequently (28). All PLA2 enzymes function to hydrolyze an acyl or alkyl linkage at the sn-2 position of phospholipids. The secreted enzymes utilize a His-Asp dyad as the catalytic center, whereas the other members of the family are serine based, either as a Ser-Asp dyad or Ser-Asp-His catalytic triad (23). This triad is similar to that found in many proteases as well as lipolytic enzymes (66). The use of serine protease inhibitors identified aiPLA2 as a serine-based enzyme (1, 38). Mutation analysis indicated that Ser32 is essential for activity along with His26 and Asp140, which constitute the catalytic triad (6, 54). As pointed out above, S32 and H26 are crucial as surface amino acids involved in Prdx6 binding to phospholipids in addition to their role in catalysis (54, 55). With regard to enzymatic characteristics, aiPLA2 shows preference for phosphatidylcholine with lesser activity toward phosphatidylethanolamine and other phospholipids (1, 38). Activity is not altered by the particular acyl substituent at the sn-2 position of phosphatidylcholine but an alkyl substituent results in 50% less activity (1, 38).
Why two enzymatic activities in one protein?
Through what evolutionary quirk does this protein come to have two seemingly disparate activities (GSH peroxidase, PLA2)? A clue lies in the ability of Prdx6 to reduce phospholipid hydroperoxides consequent to phospholipid binding as described above. Based on mutagenesis studies and the crystal structure, we have proposed that attachment of the phospholipid to the protein occurs initially through the phospholipid head group in the vicinity of the PLA2 active site (S32, H26) with insertion of the sn-2 fatty acyl group into the pocket containing the catalytic C47 (54, 55) (Fig. 6A). Thus, the protein is in a position to carry out either reduction of the peroxidized fatty acyl group (peroxidase activity) via C47 within the pocket or hydrolysis of the ester bond (PLA2 activity) via S32, D140, and H26 at the protein surface. In this formulation, binding between the protein and substrate represents the essential feature defining the enzymatic properties. We have demonstrated that the native protein readily binds to oxidized phospholipids but binds to reduced (native) phospholipids only at acidic pH (54, 55). So, Prdx6 can remain free in the cytoplasm until membrane phospholipids are oxidized at which point binding of the protein (and subsequent reduction of the oxidized membrane components) occurs (Fig. 6B). The situation is different within the acidic organelles (lysosomes or lamellar bodies) where Prdx6 can bind and hydrolyze native phospholipids as necessary for phospholipid catabolism. These binding properties account for the differing pH profiles for the two activities of the protein. That is, hydrolysis of phospholipids (PLA2 activity) by native (nonphosphorylated) Prdx6 is maximal at pH 4, whereas reduction of oxidized phospholipid (peroxidase activity) occurs at pH 7.
Phosphorylation of Prdx6 and regulation of PLA2 activity
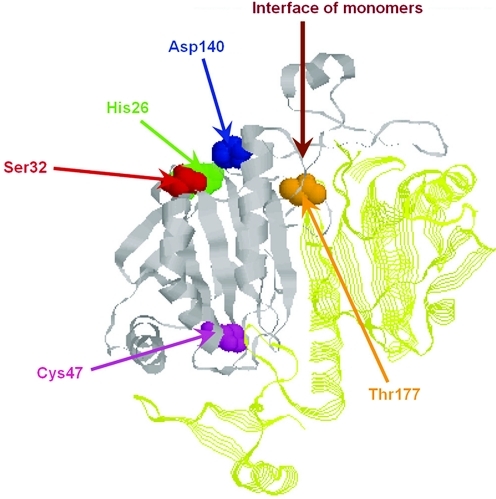
Treatment of lung cells with a phorbol ester (PMA) was found to increase phospholipid turnover and also to increase aiPLA2 activity (102). Subsequent study indicated that treatment with PMA resulted in MAP kinase-mediated phosphorylation of Prdx6. The use of inhibitors with intact cells and in vitro studies using individual enzymes suggested that either Erk or p38 MAP kinase was effective at phosphorylating the protein. Putative docking sites for the MAP kinases on Prdx6 have been described (102). By mass spectroscopic analysis, phosphorylation was at T177, and this effect (phosphorylation) was lost by mutation of the T177 residue (102) (Fig. 7). Phosphorylation resulted in a marked increase of PLA2 activity; for the rat protein, activity at pH 4 was increased more than 10-fold from 100 to 1300 nmol/min/mg protein (102). Since only 45% of the protein was phosphorylated, the maximum activity might be ∼3 μmol/min/mg protein, only slightly less than the maximal peroxidase activity (∼5 μmol/min/mg protein). Further, phosphorylation broadens the activity versus pH spectrum so that PLA2 activity at cytosolic and acidic pH values is similar (102). This effect was corroborated by demonstrating that, unlike the nonphosphorylated protein, phosphorylated Prdx6 binds to phospholipid vesicles (liposomes) at pH 7 (55). The mechanism for the enhancement of activity by phosphorylation at T177 is unclear, as this residue, based on the crystal structure, is buried within the molecule without an obvious relationship to the sites for phospholipid binding or PLA2 catalysis (Fig. 7). The physiological significance of the increase in cytosolic PLA2 activity resulting from phosphorylation also is not yet apparent although our preliminary studies (unpublished results) suggest a signaling function related to the generation of fatty acids or lysoPC. Phosphorylation has no effect on the peroxidase activity of Prdx6 (102).
FIG. 7.
Site for MAP kinase-mediated phosphorylation of Prdx6. The phosphorylation site is T177. This site, which is distant from the PLA2 catalytic triad and the peroxidatic Cys47, is located within the protein globule relatively close to the interface of Prdx6 monomers. The buried nature of the site suggests that change in configuration is required for phosphorylation. Reprinted from Wu et al. (102). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
aiPLA2 activity also can be regulated by its interaction with the protein known as lung surfactant protein A (SP-A) (18, 103). This member of the collectin family of proteins is stored within the lung lamellar bodies and secreted as a component of the pulmonary surfactant. SP-A is a multimeric protein comprising six trimers with a nominal total molecular mass of ∼ 680 kDa. SP-A and Prdx6 have been shown to interact in vitro by a variety of methods with a stoichiometry of one Prdx6 per SP-A trimer (103). This interaction results in a concentration-dependent decrease in PLA2 activity. Conversely, inhibition or knock-out of SP-A expression results in increased cellular aPLA2 activity (18, 31). Immunocytochemical studies of intact cells have demonstrated colocalization of SP-A and Prdx6 in lung lamellar bodies (103). Thus, SP-A has the requisite properties to be a physiological regulator of aiPLA2 activity; for example, by limiting PLA2 activity of Prdx6 in lamellar bodies or in the extracellular (alveolar) space.
Physiological Roles of Prdx6
In keeping with the two major enzymatic activities of the protein, Prdx6 has two major physiological roles. These can be characterized as related to antioxidant defense and to phospholipid turnover. A suspected role in the pathogenesis of atherosclerosis has not been confirmed in mouse studies and is considered to be unlikely (95). Because of the major interest of our lab, the bulk of the studies related to the physiological role of the protein has concerned its role in lung physiology. These roles have been investigated primarily using the Prdx6 null mouse or the PLA2 inhibitor, MJ33.
The Prdx6 gene targeted mouse
The study of the physiological role of Prdx6 has been greatly facilitated by the use of a gene-targeted mouse model resulting in deletion of the protein and its associated function. Prdx6 null mice have been generated in two different laboratories by targeting different sites in the 5 exon gene; our laboratory targeted exon 3, whereas another laboratory targeted exons 1 and 2 (60, 94). Mice generated by either strategy display a similar phenotype although overlapping studies have been reported for only a relatively few conditions. Mice targeted at exon 3 were on a mixed background when originally studied (96–98), but these have now been fully backcrossed (>99.9%) to the C57/Bl6 background, whereas exon 1/2-targeted mice are on the 129/SvJ background. Prdx6 gene-targeted mice show abnormalities of response to oxidant stress and altered lung phospholipid metabolism (as described below), but otherwise develop and reproduce normally and show no other deviations from the wild type. A transgenic Prdx6 overexpressing mouse has been generated by injecting a genomic clone containing the entire Prdx6 gene into C57/Bl6 oocytes; these mice under control conditions also show no obvious deviations from the wild-type phenotype (76).
Antioxidant defense
Based on its activity as a scavenger of H2O2 and other hydroperoxides, it is natural to postulate a role for Prdx6 in antioxidant defense. Studies using various cell lines showed that overexpression of Prdx6 increases resistance to experimental oxidative stress (12, 14, 56, 96), whereas a decrease of Prdx6 expression after treatment with antisense oligonucleotides results in oxidant sensitivity (69). This latter effect could be rescued by treatment of cells with an adenoviral vector directing Prdx6 expression. Subsequent studies using either primary lung epithelial cells or peritoneal macrophages derived from Prdx6 null mice confirmed that the absence of Prdx6 increased their oxidant sensitivity as compared with wild type (94, 96). There were no significant changes in expression of other enzymes that could explain this effect.
The results with cell models have been confirmed by studies with intact mice. Mice that overexpress Prdx6, either through administration of an adenoviral vector directing Prdx6 expression or by transgenic techniques, were more resistant to the oxidant stress generated by exposure to oxygen at elevated partial pressures (hyperoxia) (99, 100). By contrast, Prdx6 null mice showed a significant increase in sensitivity to the toxic effects of hyperoxia and to administration of the redox cycling drug, paraquat (94, 97, 98). The manifestations of increased oxidant sensitivity were increased mortality and increased tissue damage in lungs and liver of the exposed mice. Prdx6 null mice also showed increased injury in the isolated heart (65) and the liver (13) subjected to the oxidative stress of ischemia followed by reperfusion. Thus, these studies utilizing isolated cells, intact organs, and mice demonstrate unequivocally that Prdx6 functions in vivo as an antioxidant enzyme.
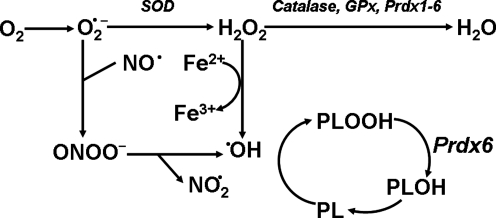
One of the unresolved issues concerning the antioxidant role of Prdx6 relates to the multiple other enzymes that also are able to scavenge H2O2, leading to the question why any one enzyme could be critical. These other enzymes include catalase, the family of GSH peroxidases, and other Prdxs. Cytosolic GPx (GPx1) has been regarded as a major enzyme for reduction of hydroperoxides and antioxidant protection (9). However, our recent comparative studies of oxidant stress in lungs and lung cells (hyperoxia, tert-butyl hydroperoxide, H2O2, and paraquat) indicate significantly greater susceptibility to injury with knock-out of Prdx6 as compared with knock-out of GPx1 (51). These results suggest that the presence of Prdx6 has a significantly greater protective effect than the presence of GPx1. We postulate that the unique role of Prdx6 is based on its ability to reduce phospholipid hydroperoxides. Initiation of lipid peroxidation can induce an oxidant-generated chain reaction resulting in progressive damage to cell plasma and organellar membranes resulting in altered volume regulation and other physiological derangements eventuating in cell death. Thus, the chemical reduction of peroxidized phospholipids is essential for cell survival. To date, only two enzymes have been shown with the ability to reduce cellular phospholipid hydroperoxides. Phospholipid hydroperoxide glutathione peroxidase (GPx4), the first enzyme described with this activity, is highly expressed in testis, where it appears to also have a structural role, but is expressed at much lower levels in other organs such as the lung, where antioxidant defense is crucial (2, 93). More recently, Prdx6 also has been shown to reduce phospholipid hydroperoxides (21). Lung homogenates from Prdx6 null mice have lost 95% of their phospholipid hydroperoxide reduction activity compared with wild type, indicating that Prdx6 is the major source of this activity in the lung (97). Evidence for significantly increased lipid peroxidation associated with oxidative stress in the lungs of Prdx6 null mice provides support for the hypothesis that Prdx6 normally functions to reduce phospholipid hydroperoxides (97, 98). This activity of Prdx6 provides a basis for the observed important role of Prdx6 in lung antioxidant defense (Fig. 8). As noted above, the ubiquitous GPx1 does not show this activity (21, 59). The reduction of phospholipid hydroperoxides generates a hydroxyphospholipid that must then be further reduced (by unspecified enzymes) to regenerate the native compound (Fig. 8). An alternate mechanism for restoring the reduced form of peroxidized membrane phospholipids involves the sequential activity of a PLA2 and an acyl transferase (85). It has been estimated that the direct reduction of peroxidized phospholipids is 104 times more efficient than the alternate mechanism (106).
FIG. 8.
Scheme for intracellular generation of reactive oxygen species (ROS) and the role of Prdx6. O2•− is dismutated to H2O2, which can generate •OH in the presence of Fe2+. •OH is a powerful oxidant that can peroxidize cell membrane phospholipids (PL). Thus, cell resistance to oxidant stress and repair depends in large part on removal of H2O2 and reduction of phospholipid hydroperoxides (there is no specific scavenger for •OH). H2O2 can be removed by multiple enzymes, including catalase, GSH peroxidases, and all Prdxs. Phospholipid hydroperoxides (PLOOH) are reduced in lung cells by Prdx6; GPx4 represents an alternate enzyme for reduction in some cells (not shown). The product of Prdx6 activity, the hydroxy phospholipid (PLOH), is further reduced by unspecified enzymes to regenerate the native phospholipid. Modified from Manevich and Fisher (53).
Lung surfactant metabolism
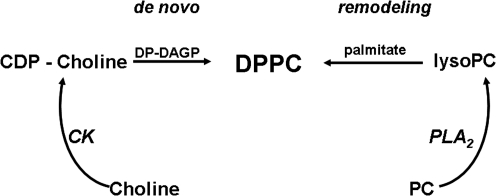
Lung surfactant is a phospholipid–protein complex secreted by the lung alveolar epithelial cells that functions to lower surface tension at the air–liquid interface in the lung alveolar space and thereby enhances lung stability. The principle surface active agent is dipalmitoylphosphatidylcholine (DPPC), which functions in concert with several hydrophobic surfactant proteins. Lung surfactant is synthesized in the endoplasmic reticulum of alveolar epithelial type II cells, transferred to modified secretory lysosomes (lamellar bodies) for storage, and is subsequently exocytosed. Lung DPPC is actively recycled by the epithelial cells with an extracellular half-time of ∼6–10 h. The material to be recycled is endocytosed by a receptor-mediated process, targeted to intermediate organelles where it may be resecreted or degraded, and components utilized for resynthesis of DPPC. Lung DPPC is synthesized by both the de novo and by the remodeling pathways. In the former pathway, cytidine 5-diphosphocholine interacts with diacylglycerol containing two palmitoyl residues to generate DPPC; for the latter pathway, phosphatidylcholine with an unsaturated fatty acid in the sn-2 position is hydrolyzed by PLA2 followed by its reacylation with a palmitoyl moiety to generate the disaturated phospholipid (Fig. 9).
FIG. 9.
Pathways for synthesis of dipalmitoylphosphatidylcholine (DPPC) by lung alveolar epithelial cells. The de novo pathway proceeds by the phosphorylation of choline (choline kinase [CK]) and then generation of cytidine 5-diphosphocholine (CDP-choline), which combines with 1,2-palmitoyl diacylglycerol phosphate (DP-DAGP) to form DPPC. The remodeling pathway requires a PLA2 acting on phosphatidylcholine (PC) to generate a lysoPC, which then forms DPPC by reacylation in the sn-2 position with palmitate. Prdx6 has been shown to be the major PLA2 involved in the remodeling pathway for surfactant biosynthesis in lung alveolar epithelial cells.
Lungs treated with the aiPLA2 inhibitor MJ33 or isolated from Prdx6 null mice show a significant reduction in the degradation of DPPC after its reuptake from the alveolar extracellular space (15, 20), whereas overexpression results in increased DPPC metabolism (22). Further, inhibition of aiPLA2 activity by treatment with MJ33 or by gene targeting of Prdx6 results in a marked decrease in the ability of lung epithelial cells to incorporate palmitate into DPPC by the remodeling pathway of phospholipid synthesis (15, 16). This effect is compatible with a role for Prdx6 in the generation of lysoPC as the substrate for the reacylation pathway for DPPC synthesis. Prdx6 null mice also show a progressive age related increase in the content of phospholipids in both the lung parenchyma as well as its alveolar space (20). This progressive increase suggests a lifelong deficit in the ability of these lungs to normally metabolize phospholipid. To date, possible abnormalities in phospholipid metabolism in organs other than the lung have not been studied.
Prdx6 in the Pathobiology of Disease
Oxidant stress is being increasingly recognized as an important mechanism in disease pathogenesis. Although it is expected that altered expression of Prdx6 might be seen with conditions that are clearly associated with oxidant stress, proteomic screens have frequently demonstrated that altered Prdx6 expression occurs in diseases where the involvement of oxidant stress is less clear. High expression of Prdx6 has been noted in degenerative afflictions of the central nervous system, including Alzheimer's (77), Parkinson's (79), Pick's (42), and Crutzfeld-Jakob diseases (43), schizophrenia (57), and an experimental mouse model of amyotrophic lateral sclerosis (91). As a potential mechanism for some of these associations, binding of Prdx6 to the protein saitohin may be linked to tau splicing and subsequent neurodegeneration (26). Elevated Prdx6 also occurs in various neoplastic diseases, including malignant mesothelioma (40), bronchogenic squamous cell carcinoma (50), and cancer of the gingivo-buccal area (87), bladder (80), and breast (5). It has been proposed that the peroxidase activity of Prdx6 promotes cancer growth, whereas the PLA2 activity facilitates invasiveness (29). Elevated Prdx6 has been shown at the leading edges of wounds in humans as well as in animal models and has been postulated to be of importance in wound healing (45, 46, 62). On the other side of the coin, abnormally low levels of Prdx6 have been described in cataracts of the lens (27), whereas overexpression of Prdx6 in mice protects against the development of lens opacity (44). Altered Prdx6 expression also has been reported in animal and cellular models of alcoholic liver disease (67, 83), cellular premature senescence (11), and various drug treatments (melatonin, luteolin, paroxetine, ochratoxin A, and capsaicin) (3, 58, 92, 104, 105). Whether any of these associations will provide definitive insights into pathogenesis or will provide targets for therapy remains to be determined. Finally, expression of Prdx6 in bovine thoracic muscles was found to correlate with tenderness of the meat (33), so think of Prdx6 at that next meal with an especially tender steak.
Summary
Prdx6, the only mammalian 1-Cys member of the Prdx family, has several unique properties that distinguish it from other Prdxs. First, its catalytic mechanism for peroxidase activity is not based on thioredoxin but rather utilizes GSH (or possibly other agents) as the physiologic reductant. Second, Prdx6 is able to reduce phospholipid hydroperoxides in addition to short chain hydroperoxides. Third, Prdx6 has PLA2 activity that is distinct from its peroxidatic function. These properties provide the basis for the physiologic role of Prdx6 as a key enzyme for both antioxidant defense and phospholipid homeostasis.
Abbreviations Used
- ARE
antioxidant response element
- DPPC
dipalmitoylphosphatidylcholine
- KGF
keratinocyte growth factor
- GPx
glutathione peroxidase
- GRE
glucocorticoid response element
- GSH
glutathione
- GST
GSH S-transferase
- PLA2
phospholipase A2
- Prdx6
peroxiredoxin 6
- SP-A
surfactant protein A
Acknowledgments
The author thanks the many coworkers who have contributed to the investigations of Prdx6 during the past 15 years, and in particular thanks to Mahendra Jain, Ye-Shih Ho, Roberta Colman, Sheldon Feinstein, Yefim Manevich, David Speicher, and Chandra Dodia. The author also thanks Susan Turbitt and Victoria Brown for their help in preparing this article and Sheldon Feinstein for critical review. Support for research related to Prdx6 has been provided by the NHLBI through HL19737 and HL79063.
References
- 1.Akiba S. Dodia C. Chen X. Fisher AB. Characterization of acidic Ca(2+)-independent phospholipase A2 of bovine lung. Comp Biochem Physiol B Biochem Mol Biol. 1998;120:393–404. doi: 10.1016/s0305-0491(98)10046-9. [DOI] [PubMed] [Google Scholar]
- 2.Baek IJ. Seo DS. Yon JM. Lee SR. Jin Y. Nahm SS. Jeong JH. Choo YK. Kang JK. Lee BJ. Yun YW. Nam SY. Tissue expression and cellular localization of phospholipid hydroperoxide glutathione peroxidase (PHGPx) mRNA in male mice. J Mol Histol. 2007;38:237–244. doi: 10.1007/s10735-007-9092-7. [DOI] [PubMed] [Google Scholar]
- 3.Baek YM. Hwang HJ. Kim SW. Hwang HS. Lee SH. Kim JA. Yun JW. A comparative proteomic analysis for capsaicin-induced apoptosis between human hepatocarcinoma (HepG2) and human neuroblastoma (SK-N-SH) cells. Proteomics. 2008;8:4748–4767. doi: 10.1002/pmic.200800094. [DOI] [PubMed] [Google Scholar]
- 4.Chae HZ. Robison K. Poole LB. Church G. Storz G. Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci U S A. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang XZ. Li DQ. Hou YF. Wu J. Lu JS. Di GH. Jin W. Ou ZL. Shen ZZ. Shao ZM. Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast Cancer Res. 2007;9:R76. doi: 10.1186/bcr1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JW. Dodia C. Feinstein SI. Jain MK. Fisher AB. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem. 2000;275:28421–28427. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- 7.Choi HJ. Kang SW. Yang CH. Rhee SG. Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat Struct Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury I. Mo Y. Gao L. Kazi A. Fisher AB. Feinstein SI. Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radic Biol Med. 2009;46:146–153. doi: 10.1016/reeradbiomed.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen G. Hochstein P. Glutathione peroxidase: the primary agent for the elimination of hydrogen peroxide in erythrocytes. Biochemistry. 1963;2:1420–1428. doi: 10.1021/bi00906a038. [DOI] [PubMed] [Google Scholar]
- 10.Derewenda ZS. Sharp AM. News from the interface: the molecular structures of triacylglyceride lipases. Trends Biochem Sci. 1993;18:20–25. doi: 10.1016/0968-0004(93)90082-x. [DOI] [PubMed] [Google Scholar]
- 11.Dierick JF. Kalume DE. Wenders F. Salmon M. Dieu M. Raes M. Roepstorff P. Toussaint O. Identification of 30 protein species involved in replicative senescence and stress-induced premature senescence. FEBS Lett. 2002;531:499–504. doi: 10.1016/s0014-5793(02)03604-9. [DOI] [PubMed] [Google Scholar]
- 12.Dierick JF. Wenders F. Chainiaux F. Remacle J. Fisher AB. Toussaint O. Retrovirally mediated overexpression of peroxiredoxin VI increases the survival of WI-38 human diploid fibroblasts exposed to cytotoxic doses of tert-butylhydroperoxide and UVB. Biogerontology. 2003;4:125–131. doi: 10.1023/a:1024154024602. [DOI] [PubMed] [Google Scholar]
- 13.Eismann T. Huber N. Shin T. Kuboki S. Galloway E. Wyder M. Edwards MJ. Greis KD. Shertzer HG. Fisher AB. Lentsch AB. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266–G274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fatma N. Kubo E. Sen M. Agarwal N. Thoreson WB. Camras CB. Singh DP. Peroxiredoxin 6 delivery attenuates TNF-alpha-and glutamate-induced retinal ganglion cell death by limiting ROS levels and maintaining Ca2+ homeostasis. Brain Res. 2008;1233:63–78. doi: 10.1016/j.brainres.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher AB. Dodia C. Lysosomal-type PLA2 and turnover of alveolar DPPC. Am J Physiol Lung Cell Mol Physiol. 2001;280:L748–L754. doi: 10.1152/ajplung.2001.280.4.L748. [DOI] [PubMed] [Google Scholar]
- 16.Fisher AB. Dodia C. Role of acidic Ca2+-independent phospholipase A2 in synthesis of lung dipalmitoyl phosphatidylcholine. Am J Physiol. 1997;272:L238–L243. doi: 10.1152/ajplung.1997.272.2.L238. [DOI] [PubMed] [Google Scholar]
- 17.Fisher AB. Dodia C. Role of phospholipase A2 enzymes in degradation of dipalmitoylphosphatidylcholine by granular pneumocytes. J Lipid Res. 1996;37:1057–1064. [PubMed] [Google Scholar]
- 18.Fisher AB. Dodia C. Chander A. Inhibition of lung calcium-independent phospholipase A2 by surfactant protein A. Am J Physiol. 1994;267:L335–L341. doi: 10.1152/ajplung.1994.267.3.L335. [DOI] [PubMed] [Google Scholar]
- 19.Fisher AB. Dodia C. Chander A. Jain M. A competitive inhibitor of phospholipase A2 decreases surfactant phosphatidylcholine degradation by the rat lung. Biochem J. 1992;288(Pt 2):407–411. doi: 10.1042/bj2880407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher AB. Dodia C. Feinstein SI. Ho YS. Altered lung phospholipid metabolism in mice with targeted deletion of lysosomal-type phospholipase A2. J Lipid Res. 2005;46:1248–1256. doi: 10.1194/jlr.M400499-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Fisher AB. Dodia C. Manevich Y. Chen JW. Feinstein SI. Phospholipid hydroperoxides are substrates for non-selenium glutathione peroxidase. J Biol Chem. 1999;274:21326–21334. doi: 10.1074/jbc.274.30.21326. [DOI] [PubMed] [Google Scholar]
- 22.Fisher AB. Dodia C. Yu K. Manevich Y. Feinstein SI. Lung phospholipid metabolism in transgenic mice overexpressing peroxiredoxin 6. Biochim Biophys Acta. 2006;1761:785–792. doi: 10.1016/j.bbalip.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Fisher AB. Jain M. Encyclopedia of Life Sciences (ELS) Chichester: John Wiley & Sons, Ltd.; 2009. Phospholipases: degradation of phospholipids in membranes and emulsions. [DOI] [Google Scholar]
- 24.Frank S. Munz B. Werner S. The human homologue of a bovine non-selenium glutathione peroxidase is a novel keratinocyte growth factor-regulated gene. Oncogene. 1997;14:915–921. doi: 10.1038/sj.onc.1200905. [DOI] [PubMed] [Google Scholar]
- 25.Fujii T. Fujii J. Taniguchi N. Augmented expression of peroxiredoxin VI in rat lung and kidney after birth implies an antioxidative role. Eur J Biochem. 2001;268:218–225. doi: 10.1046/j.1432-1033.2001.01843.x. [DOI] [PubMed] [Google Scholar]
- 26.Gao L. Tse SW. Conrad C. Andreadis A. Saitohin, which is nested in the tau locus and confers allele-specific susceptibility to several neurodegenerative diseases, interacts with peroxiredoxin 6. J Biol Chem. 2005;280:39268–39272. doi: 10.1074/jbc.M506116200. [DOI] [PubMed] [Google Scholar]
- 27.Hasanova N. Kubo E. Kumamoto Y. Takamura Y. Akagi Y. Age-related cataracts and Prdx6: correlation between severity of lens opacity, age and the level of Prdx 6 expression. Br J Ophthalmol. 2009;93:1081–1084. doi: 10.1136/bjo.2008.152272. [DOI] [PubMed] [Google Scholar]
- 28.Hiraoka M. Abe A. Shayman JA. Structure and function of lysosomal phospholipase A2: identification of the catalytic triad and the role of cysteine residues. J Lipid Res. 2005;46:2441–2447. doi: 10.1194/jlr.M500248-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Ho JN. Lee SB. Lee SS. Yoon SH. Kang GY. Hwang SG. Um HD. Phospholipase A2 activity of peroxiredoxin 6 promotes invasion and metastasis of lung cancer cells. Mol Cancer Ther. 2010;9:825–832. doi: 10.1158/1535-7163.MCT-09-0904. [DOI] [PubMed] [Google Scholar]
- 30.Iakoubova OA. Pacella LA. Her H. Beier DR. LTW4 protein on mouse chromosome 1 is a member of a family of antioxidant proteins. Genomics. 1997;42:474–478. doi: 10.1006/geno.1997.4762. [DOI] [PubMed] [Google Scholar]
- 31.Jain D. Dodia C. Bates SR. Hawgood S. Poulain FR. Fisher AB. SP-A is necessary for increased clearance of alveolar DPPC with hyperventilation or secretagogues. Am J Physiol Lung Cell Mol Physiol. 2003;284:L759–L765. doi: 10.1152/ajplung.00200.2002. [DOI] [PubMed] [Google Scholar]
- 32.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 33.Jia X. Veiseth-Kent E. Grove H. Kuziora P. Aass L. Hildrum KI. Hollung K. Peroxiredoxin-6—a potential protein marker for meat tenderness in bovine longissimus thoracis muscle. J Anim Sci. 2009;87:2391–2399. doi: 10.2527/jas.2009-1792. [DOI] [PubMed] [Google Scholar]
- 34.Kang SW. Baines IC. Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- 35.Kim HS. Manevich Y. Feinstein SI. Pak JH. Ho YS. Fisher AB. Induction of 1-cys peroxiredoxin expression by oxidative stress in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;285:L363–L369. doi: 10.1152/ajplung.00078.2003. [DOI] [PubMed] [Google Scholar]
- 36.Kim HS. Pak JH. Gonzales LW. Feinstein SI. Fisher AB. Regulation of 1-cys peroxiredoxin expression in lung epithelial cells. Am J Respir Cell Mol Biol. 2002;27:227–233. doi: 10.1165/ajrcmb.27.2.20010009oc. [DOI] [PubMed] [Google Scholar]
- 37.Kim TS. Dodia C. Chen X. Hennigan BB. Jain M. Feinstein SI. Fisher AB. Cloning and expression of rat lung acidic Ca(2+)-independent PLA2 and its organ distribution. Am J Physiol. 1998;274:L750–L761. doi: 10.1152/ajplung.1998.274.5.L750. [DOI] [PubMed] [Google Scholar]
- 38.Kim TS. Sundaresh CS. Feinstein SI. Dodia C. Skach WR. Jain MK. Nagase T. Seki N. Ishikawa K. Nomura N. Fisher AB. Identification of a human cDNA clone for lysosomal type Ca2+-independent phospholipase A2 and properties of the expressed protein. J Biol Chem. 1997;272:2542–2550. doi: 10.1074/jbc.272.4.2542. [DOI] [PubMed] [Google Scholar]
- 39.Kinnula VL. Lehtonen S. Kaarteenaho-Wiik R. Lakari E. Paakko P. Kang SW. Rhee SG. Soini Y. Cell specific expression of peroxiredoxins in human lung and pulmonary sarcoidosis. Thorax. 2002;57:157–164. doi: 10.1136/thorax.57.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinnula VL. Lehtonen S. Sormunen R. Kaarteenaho-Wiik R. Kang SW. Rhee SG. Soini Y. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196:316–323. doi: 10.1002/path.1042. [DOI] [PubMed] [Google Scholar]
- 41.Knoops B. Clippe A. Bogard C. Arsalane K. Wattiez R. Hermans C. Duconseille E. Falmagne P. Bernard A. Cloning and characterization of AOEB166, a novel mammalian antioxidant enzyme of the peroxiredoxin family. J Biol Chem. 1999;274:30451–30458. doi: 10.1074/jbc.274.43.30451. [DOI] [PubMed] [Google Scholar]
- 42.Krapfenbauer K. Engidawork E. Cairns N. Fountoulakis M. Lubec G. Aberrant expression of peroxiredoxin subtypes in neurodegenerative disorders. Brain Res. 2003;967:152–160. doi: 10.1016/s0006-8993(02)04243-9. [DOI] [PubMed] [Google Scholar]
- 43.Krapfenbauer K. Yoo BC. Fountoulakis M. Mitrova E. Lubec G. Expression patterns of antioxidant proteins in brains of patients with sporadic Creutzfeldt-Jacob disease. Electrophoresis. 2002;23:2541–2547. doi: 10.1002/1522-2683(200208)23:15<2541::AID-ELPS2541>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 44.Kubo E. Singh DP. Fatma N. Akagi Y. TAT-mediated peroxiredoxin 5 and 6 protein transduction protects against high-glucose-induced cytotoxicity in retinal pericytes. Life Sci. 2009;84:857–864. doi: 10.1016/j.lfs.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Kumin A. Huber C. Rulicke T. Wolf E. Werner S. Peroxiredoxin 6 is a potent cytoprotective enzyme in the epidermis. Am J Pathol. 2006;169:1194–1205. doi: 10.2353/ajpath.2006.060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumin A. Schafer M. Epp N. Bugnon P. Born-Berclaz C. Oxenius A. Klippel A. Bloch W. Werner S. Peroxiredoxin 6 is required for blood vessel integrity in wounded skin. J Cell Biol. 2007;179:747–760. doi: 10.1083/jcb.200706090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leavey PJ. Gonzalez-Aller C. Thurman G. Kleinberg M. Rinckel L. Ambruso DW. Freeman S. Kuypers FA. Ambruso DR. A 29-kDa protein associated with p67phox expresses both peroxiredoxin and phospholipase A2 activity and enhances superoxide anion production by a cell-free system of NADPH oxidase activity. J Biol Chem. 2002;277:45181–45187. doi: 10.1074/jbc.M202869200. [DOI] [PubMed] [Google Scholar]
- 48.Lee SP. Hwang YS. Kim YJ. Kwon KS. Kim HJ. Kim K. Chae HZ. Cyclophilin a binds to peroxiredoxins and activates its peroxidase activity. J Biol Chem. 2001;276:29826–29832. doi: 10.1074/jbc.M101822200. [DOI] [PubMed] [Google Scholar]
- 49.Lee TH. Yu SL. Kim SU. Kim YM. Choi I. Kang SW. Rhee SG. Yu DY. Characterization of the murine gene encoding 1-Cys peroxiredoxin and identification of highly homologous genes. Gene. 1999;234:337–344. doi: 10.1016/s0378-1119(99)00190-0. [DOI] [PubMed] [Google Scholar]
- 50.Lehtonen ST. Svensk AM. Soini Y. Paakko P. Hirvikoski P. Kang SW. Saily M. Kinnula VL. Peroxiredoxins, a novel protein family in lung cancer. Int J Cancer. 2004;111:514–521. doi: 10.1002/ijc.20294. [DOI] [PubMed] [Google Scholar]
- 51.Liu G. Feinstein SI. Wang Y. Dodia C. Fisher D. Yu K. Ho YS. Fisher AB. Comparison of glutathione peroxidase 1 and peroxiredoxin 6 in protection against oxidative stress in the mouse lung. Free Radic Biol Med. 2010;49:1172–1181. doi: 10.1016/j.freeradbiomed.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manevich Y. Feinstein SI. Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc Natl Acad Sci U S A. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manevich Y. Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 54.Manevich Y. Reddy KS. Shuvaeva T. Feinstein S. Fisher A. Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res. 2007;48:2306–2318. doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- 55.Manevich Y. Shuvaeva T. Dodia C. Kazi A. Feinstein SI. Fisher AB. Binding of peroxiredoxin 6 to substrate determines differential phospholipid hydroperoxide peroxidase and phospholipase A(2) activities. Arch Biochem Biophys. 2009;485:139–149. doi: 10.1016/j.abb.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manevich Y. Sweitzer T. Pak JH. Feinstein SI. Muzykantov V. Fisher AB. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proc Natl Acad Sci U S A. 2002;99:11599–11604. doi: 10.1073/pnas.182384499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martins-de-Souza D. Gattaz WF. Schmitt A. Novello JC. Marangoni S. Turck CW. Dias-Neto E. Proteome analysis of schizophrenia patients Wernicke's area reveals an energy metabolism dysregulation. BMC Psychiatry. 2009;9:17. doi: 10.1186/1471-244X-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McHugh P. Rogers G. Glubb D. Joyce R. Kenney M. Proteomic analysis of rat hippocampus exposed to the antidepressant paroxetine. J Psychopharmacol. 2010;24:1243–1251. doi: 10.1177/0269881109102786. [DOI] [PubMed] [Google Scholar]
- 59.Michiels C. Raes M. Toussaint O. Remacle J. Importance of Se-glutathione peroxidase, catalase, and Cu/Zn-SOD for cell survival against oxidative stress. Free Radic Biol Med. 1994;17:235–248. doi: 10.1016/0891-5849(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 60.Mo Y. Feinstein SI. Manevich Y. Zhang Q. Lu L. Ho YS. Fisher AB. 1-Cys peroxiredoxin knock-out mice express mRNA but not protein for a highly related intronless gene. FEBS Lett. 2003;555:192–198. doi: 10.1016/s0014-5793(03)01199-2. [DOI] [PubMed] [Google Scholar]
- 61.Monteiro G. Horta BB. Pimenta DC. Augusto O. Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci U S A. 2007;104:4886–4891. doi: 10.1073/pnas.0700481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munz B. Frank S. Hubner G. Olsen E. Werner S. A novel type of glutathione peroxidase: expression and regulation during wound repair. Biochem J. 1997;326(Pt 2):579–585. doi: 10.1042/bj3260579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murakami M. Kudo I. Phospholipase A2. J Biochem (Tokyo) 2002;131:285–292. doi: 10.1093/oxfordjournals.jbchem.a003101. [DOI] [PubMed] [Google Scholar]
- 64.Nagase T. Miyajima N. Tanaka A. Sazuka T. Seki N. Sato S. Tabata S. Ishikawa K. Kawarabayasi Y. Kotani H, et al. Prediction of the coding sequences of unidentified human genes. III. The coding sequences of 40 new genes (KIAA0081-KIAA0120) deduced by analysis of cDNA clones from human cell line KG-1 (supplement) DNA Res. 1995;2:51–59. doi: 10.1093/dnares/2.1.51. [DOI] [PubMed] [Google Scholar]
- 65.Nagy N. Malik G. Fisher AB. Das DK. Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2006;291:H2636–H2640. doi: 10.1152/ajpheart.00399.2006. [DOI] [PubMed] [Google Scholar]
- 66.Nevalainen TJ. 1-Cysteine peroxiredoxin: a dual-function ezyme with peroxidase and acidic Ca++-independent phospholipase A2 activities. Biochime. 2010 Feb 4; doi: 10.1016/j.biochi.2010.01.019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 67.Newton BW. Russell WK. Russell DH. Ramaiah SK. Jayaraman A. Liver proteome analysis in a rodent model of alcoholic steatosis. J Proteome Res. 2009;8:1663–1671. doi: 10.1021/pr800905w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Novoselov SV. Peshenko IV. Popov VI. Novoselov VI. Bystrova MF. Evdokimov VJ. Kamzalov SS. Merkulova MI. Shuvaeva TM. Lipkin VM. Fesenko EE. Localization of 28-kDa peroxiredoxin in rat epithelial tissues and its antioxidant properties. Cell Tissue Res. 1999;298:471–480. doi: 10.1007/s004419900115. [DOI] [PubMed] [Google Scholar]
- 69.Pak JH. Manevich Y. Kim HS. Feinstein SI. Fisher AB. An antisense oligonucleotide to 1-cys peroxiredoxin causes lipid peroxidation and apoptosis in lung epithelial cells. J Biol Chem. 2002;277:49927–49934. doi: 10.1074/jbc.M204222200. [DOI] [PubMed] [Google Scholar]
- 70.Peshenko IV. Novoselov VI. Evdokimov VA. Nikolaev Yu V. Shuvaeva TM. Lipkin VM. Fesenko EE. Novel 28-kDa secretory protein from rat olfactory epithelium. FEBS Lett. 1996;381:12–14. doi: 10.1016/0014-5793(96)00071-3. [DOI] [PubMed] [Google Scholar]
- 71.Peshenko IV. Novoselov VI. Evdokimov VA. Nikolaev YV. Kamzalov SS. Shuvaeva TM. Lipkin VM. Fesenko EE. Identification of a 28 kDa secretory protein from rat olfactory epithelium as a thiol-specific antioxidant. Free Radic Biol Med. 1998;25:654–659. doi: 10.1016/s0891-5849(98)00111-7. [DOI] [PubMed] [Google Scholar]
- 72.Peshenko IV. Shichi H. Oxidation of active center cysteine of bovine 1-Cys peroxiredoxin to the cysteine sulfenic acid form by peroxide and peroxynitrite. Free Radic Biol Med. 2001;31:292–303. doi: 10.1016/s0891-5849(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 73.Phelan SA. AOP2 (antioxidant protein 2): structure and function of a unique thiol-specific antioxidant. Antioxid Redox Signal. 1999;1:571–584. doi: 10.1089/ars.1999.1.4-571. [DOI] [PubMed] [Google Scholar]
- 74.Phelan SA. Beier DR. Higgins DC. Paigen B. Confirmation and high resolution mapping of an atherosclerosis susceptibility gene in mice on chromosome 1. Mamm Genome. 2002;13:548–553. doi: 10.1007/s00335-002-2196-1. [DOI] [PubMed] [Google Scholar]
- 75.Phelan SA. Johnson KA. Beier DR. Paigen B. Characterization of the murine gene encoding Aop2 (antioxidant protein 2) and identification of two highly related genes. Genomics. 1998;54:132–139. doi: 10.1006/geno.1998.5568. [DOI] [PubMed] [Google Scholar]
- 76.Phelan SA. Wang X. Wallbrandt P. Forsman-Semb K. Paigen B. Overexpression of Prdx6 reduces H2O2 but does not prevent diet-induced atherosclerosis in the aortic root. Free Radic Biol Med. 2003;35:1110–1120. doi: 10.1016/s0891-5849(03)00462-3. [DOI] [PubMed] [Google Scholar]
- 77.Power JH. Asad S. Chataway TK. Chegini F. Manavis J. Temlett JA. Jensen PH. Blumbergs PC. Gai WP. Peroxiredoxin 6 in human brain: molecular forms, cellular distribution and association with Alzheimer's disease pathology. Acta Neuropathol (Berl) 2008;115:611–622. doi: 10.1007/s00401-008-0373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Power JH. Nicholas TE. Immunohistochemical localization and characterization of a rat Clara cell 26-kDa protein (CC26) with similarities to glutathione peroxidase and phospholipase A2. Exp Lung Res. 1999;25:379–392. doi: 10.1080/019021499270141. [DOI] [PubMed] [Google Scholar]
- 79.Power JH. Shannon JM. Blumbergs PC. Gai WP. Nonselenium glutathione peroxidase in human brain: elevated levels in Parkinson's disease and dementia with lewy bodies. Am J Pathol. 2002;161:885–894. doi: 10.1016/S0002-9440(10)64249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quan C. Cha EJ. Lee HL. Han KH. Lee KM. Kim WJ. Enhanced expression of peroxiredoxin I and VI correlates with development, recurrence and progression of human bladder cancer. J Urol. 2006;175:1512–1516. doi: 10.1016/S0022-5347(05)00659-2. [DOI] [PubMed] [Google Scholar]
- 81.Ralat LA. Manevich Y. Fisher AB. Colman RF. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochemistry. 2006;45:360–372. doi: 10.1021/bi0520737. [DOI] [PubMed] [Google Scholar]
- 82.Ralat LA. Misquitta SA. Manevich Y. Fisher AB. Colman RF. Characterization of the complex of glutathione S-transferase pi and 1-cysteine peroxiredoxin. Arch Biochem Biophys. 2008;474:109–118. doi: 10.1016/j.abb.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roede JR. Stewart BJ. Petersen DR. Decreased expression of peroxiredoxin 6 in a mouse model of ethanol consumption. Free Radic Biol Med. 2008;45:1551–1558. doi: 10.1016/j.freeradbiomed.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 84.Schremmer B. Manevich Y. Feinstein SI. Fisher AB. Peroxiredoxins in the lung with emphasis on peroxiredoxin VI. Subcell Biochem. 2007;44:317–344. doi: 10.1007/978-1-4020-6051-9_15. [DOI] [PubMed] [Google Scholar]
- 85.Sevanian A. Muakkassah-Kelly SF. Montestruque S. The influence of phospholipase A2 and glutathione peroxidase on the elimination of membrane lipid peroxides. Arch Biochem Biophys. 1983;223:441–452. doi: 10.1016/0003-9861(83)90608-2. [DOI] [PubMed] [Google Scholar]
- 86.Shichi H. Demar JC. Non-selenium glutathione peroxidase without glutathione S-transferase activity from bovine ciliary body. Exp Eye Res. 1990;50:513–520. doi: 10.1016/0014-4835(90)90040-2. [DOI] [PubMed] [Google Scholar]
- 87.Shukla S. Pranay A. D'Cruz AK. Chaturvedi P. Kane SV. Zingde SM. Immunoproteomics reveals that cancer of the tongue and the gingivobuccal complex exhibit differential autoantibody response. Cancer Biomark. 2009;5:127–135. doi: 10.3233/CBM-2009-0604. [DOI] [PubMed] [Google Scholar]
- 88.Sorokina EM. Feinstein S. Fisher AB. Intracellular targeting of peroxiredoxin 6 to lysosomal organelles requires MAPK activity and binding to 14-3-3 epsilon. Am J Physiol: Cell Physiol. doi: 10.1152/ajpcell.00285.2010. (in review). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sorokina EM. Feinstein SI. Milovanova TN. Fisher AB. Identification of the amino acid sequence that targets peroxiredoxin 6 to lysosome-like structures of lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2009;297:L871–L880. doi: 10.1152/ajplung.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sparling NE. Phelan SA. Identification of multiple transcripts for antioxidant protein 2 (Aop2): differential regulation by oxidative stress and growth factors. Redox Rep. 2003;8:87–94. doi: 10.1179/135100003125001404. [DOI] [PubMed] [Google Scholar]
- 91.Strey CW. Spellman D. Stieber A. Gonatas JO. Wang X. Lambris JD. Gonatas NK. Dysregulation of stathmin, a microtubule-destabilizing protein, and up-regulation of Hsp25, Hsp27, and the antioxidant peroxiredoxin 6 in a mouse model of familial amyotrophic lateral sclerosis. Am J Pathol. 2004;165:1701–1718. doi: 10.1016/S0002-9440(10)63426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sung JH. Cho EH. Kim MO. Koh PO. Identification of proteins differentially expressed by melatonin treatment in cerebral ischemic injury—a proteomics approach. J Pineal Res. 2009;46:300–306. doi: 10.1111/j.1600-079X.2008.00661.x. [DOI] [PubMed] [Google Scholar]
- 93.Toppo S. Flohe L. Ursini F. Vanin S. Maiorino M. Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim Biophys Acta. 2009;1790:1486–1500. doi: 10.1016/j.bbagen.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 94.Wang X. Phelan SA. Forsman-Semb K. Taylor EF. Petros C. Brown A. Lerner CP. Paigen B. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem. 2003;278:25179–25190. doi: 10.1074/jbc.M302706200. [DOI] [PubMed] [Google Scholar]
- 95.Wang X. Phelan SA. Petros C. Taylor EF. Ledinski G. Jurgens G. Forsman-Semb K. Paigen B. Peroxiredoxin 6 deficiency and atherosclerosis susceptibility in mice: significance of genetic background for assessing atherosclerosis. Atherosclerosis. 2004;177:61–70. doi: 10.1016/j.atherosclerosis.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 96.Wang Y. Feinstein SI. Fisher AB. Peroxiredoxin 6 as an antioxidant enzyme: protection of lung alveolar epithelial type II cells from H2O2-induced oxidative stress. J Cell Biochem. 2008;104:1274–1285. doi: 10.1002/jcb.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y. Feinstein SI. Manevich Y. Ho YS. Fisher AB. Lung injury and mortality with hyperoxia are increased in peroxiredoxin 6 gene-targeted mice. Free Radic Biol Med. 2004;37:1736–1743. doi: 10.1016/j.freeradbiomed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y. Feinstein SI. Manevich Y. Ho YS. Fisher AB. Peroxiredoxin 6 gene-targeted mice show increased lung injury with paraquat-induced oxidative stress. Antioxid Redox Signal. 2006;8:229–237. doi: 10.1089/ars.2006.8.229. [DOI] [PubMed] [Google Scholar]
- 99.Wang Y. Manevich Y. Feinstein SI. Fisher AB. Adenovirus-mediated transfer of the 1-cys peroxiredoxin gene to mouse lung protects against hyperoxic injury. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1188–L1193. doi: 10.1152/ajplung.00288.2003. [DOI] [PubMed] [Google Scholar]
- 100.Wang Y. Phelan SA. Manevich Y. Feinstein SI. Fisher AB. Transgenic mice overexpressing peroxiredoxin 6 show increased resistance to lung injury in hyperoxia. Am J Respir Cell Mol Biol. 2006;34:481–486. doi: 10.1165/rcmb.2005-0333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Woo HA. Jeong W. Chang TS. Park KJ. Park SJ. Yang JS. Rhee SG. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J Biol Chem. 2005;280:3125–3128. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 102.Wu Y. Feinstein S. Manevich Y. Chowdhury I. Pak JH. Kazi A. Dodia C. Speicher DW. Fisher AB. Mitogen activated protein kinase-mediated phosphorylation of peroxiredoxin 6 regulates its phospholipid A2 activity. Biochem J. 2009;419:669–679. doi: 10.1042/BJ20082061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu YZ. Manevich Y. Baldwin JL. Dodia C. Yu K. Feinstein SI. Fisher AB. Interaction of surfactant protein A with peroxiredoxin 6 regulates phospholipase A2 activity. J Biol Chem. 2006;281:7515–7525. doi: 10.1074/jbc.M504525200. [DOI] [PubMed] [Google Scholar]
- 104.Yoo DR. Jang YH. Jeon YK. Kim JY. Jeon W. Choi YJ. Nam MJ. Proteomic identification of anti-cancer proteins in luteolin-treated human hepatoma Huh-7 cells. Cancer Lett. 2009;282:48–54. doi: 10.1016/j.canlet.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 105.Yoon S. Cong WT. Bang Y. Lee SN. Yoon CS. Kwack SJ. Kang TS. Lee KY. Choi JK. Choi HJ. Proteome response to ochratoxin A-induced apoptotic cell death in mouse hippocampal HT22 cells. Neurotoxicology. 2009;30:666–676. doi: 10.1016/j.neuro.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 106.Zhao L. Wang HP. Zhang HJ. Weydert CJ. Domann FE. Oberley LW. Buettner GR. L-PhGPx expression can be suppressed by antisense oligodeoxynucleotides. Arch Biochem Biophys. 2003;417:212–218. doi: 10.1016/s0003-9861(03)00342-4. [DOI] [PubMed] [Google Scholar]