Abstract
Aromatase inhibitors (AIs) are the major types of drugs to treat hormone-dependent breast cancer. Although these drugs work effectively, cancer still recurs in many patients after treatment as a result of acquired resistance to the AIs. To characterize the resistant mechanisms, a set of MCF-7aro cell lines that acquired resistance to the AIs was generated. Through an “Omics” approach, we found that the resistance mechanisms of the three AIs (anastrozole, letrozole, and exemestane) differ and activation of estrogen receptor alpha (ERα) is critical for acquired AI resistance. Our results reveal that growth factor/signal transduction pathways are upregulated after ERα-dependent pathways are suppressed by AIs, and ERα can then be activated through different crosstalk mechanisms.
Introduction
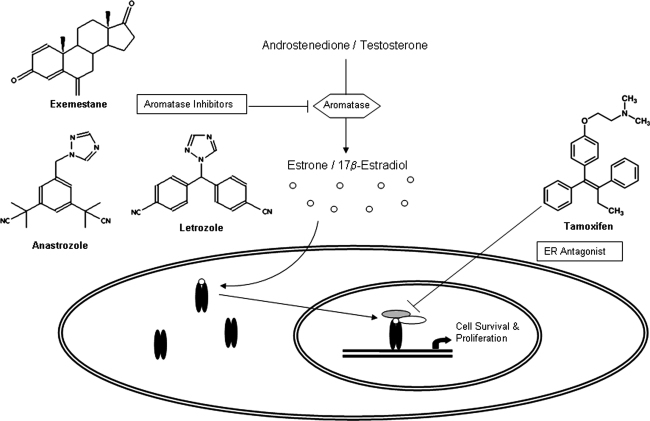
Approximately 60% of premenopausal and 75% of postmenopausal breast cancer patients have estrogen-dependent carcinomas. Antiestrogens and aromatase inhibitors (AIs) are the major types of drugs used to treat estrogen-dependent breast cancer. Antiestrogens [such as tamoxifen (TAM)] act as antagonists that block the binding of estrogen to ER (ER has two isoforms: ERα and ERβ; in this article, ER refers to ERα unless otherwise indicated). AIs [such as the third-generation AIs: anastrozole (ANA), letrozole (LET), and exemestane (EXE)] (Fig. 1) inhibit the aromatase enzyme that catalyzes estrogen biosynthesis. Based on results from several major Phase III clinical trials, these AIs are now considered important drugs for hormonal therapy of breast cancer in postmenopausal women (Baum et al., 2002; Coates et al., 2007; Coombes et al., 2004; Goss et al., 2003; Howell et al., 2005). AIs have been shown to be superior to tamoxifen with regard to disease progression, incidences of locoregional and distant relapses, and contralateral breast cancers.
FIG. 1.
Estrogen synthesis and targets for endocrine therapy. Aromatase is the key enzyme involved in the conversion of androgen to estrogen. Estrogen binds to ER in the cytoplasm and induces its activation and nuclear translocation. Aromatase inhibitors as well as the ER antagonist tamoxifen are shown.
Among three FDA-approved AIs, EXE is a steroidal inhibitor and an analogue of the androgen substrate. It is also a mechanism-based inhibitor in that aromatase converts it into an active derivative, leading to irreversible inactivation of the enzyme (Hong et al., 2007). Furthermore, irreversible binding of EXE triggers proteasome-mediated degradation of aromatase protein in cells (Wang and Chen, 2006). LET and ANA are not androgen analogues and are referred to as nonsteroidal inhibitors. These two AIs have a triazole functional group, which interacts with the heme prosthetic group of aromatase, and act as competitive inhibitors with respect to androgen substrates.
The three FDA-approved third-generation AIs are highly potent, specific, and effective drugs; however, cancer still recurs in many patients after treatment as a result of acquired resistance to the AIs. In acquired resistance, some patients respond to treatment well initially, but cancer recurs after a period of treatment. As part of our long-term goal of understanding the mechanisms of such acquired AI resistance and developing strategies to overcome it, we have generated cell line models, with the consideration of physiological relevance, in which AI resistance has been generated from long-term exposure of cells to AIs.
Because there is no ER+ and aromatase+ breast cancer cell line, MCF-7aro was generated by overexpressing aromatase in MCF-7 cells (Sun et al., 1997; Zhou et al., 1990), and was used to study responses to AIs. This enabled the generation of the first series of MCF-7aro cell lines that acquired resistance to each of the three AIs (Chen et al., 2006). These MCF-7aro-derived cell lines have been extensively characterized and verified as relevant models of acquired endocrine resistance (Masri et al., 2008, 2009a, 2009b, 2010). Furthermore, long-term estrogen deprivation MCF-7aro lines (LTEDaro) were generated and shown to represent a model of late stage acquired resistance that does not respond to treatment with any AI or tamoxifen (Masri et al., 2008, 2010). LTED cells have been used as a model of AI resistance by several laboratories (Lewis et al., 2005; Martin et al., 2003; Nicholson et al., 2004; Yue et al., 2002).
An Unbiased “Omics” Approach to Study the Mechanisms of Endocrine Resistance
AI-resistant breast cancers result from complex molecular changes and are challenging to cure. The most obvious mechanism of acquired resistance involves a selection process, that is, although estrogen production is suppressed by AIs, alternative regulatory pathways are upregulated to allow cancer to recur. When we initiated research into AI resistance (Chen et al., 2006), a decision was made to apply a nonbiased genome-wide approach to identify new genes or pathways that play roles in AI resistance. We were the first group to carry out gene expression profiling analysis on the entire series of AI-resistant cell lines (Masri et al., 2008) and ChIP-sequencing experiments for genome-wide analysis of ERα-binding sites in MCF-7aro (AI-responsive) and LTEDaro cells (Chen et al., 2009). One clinical feature associated with AI treatment is the lack of crossresistance among the three AIs (Lonning et al., 2009), suggesting that the different AIs use different resistance mechanisms or that AI-resistant cells developed supersensitive responses to ER (i.e., activation of ER with subphysiological concentrations of estrogen). Studies of our AI-resistant cell lines revealed for the first time that the resistance mechanisms of the three AIs differ and activation of ERα is critical for AI resistance, supporting our cell lines as valuable tools for studying the resistance mechanisms to these drugs. Briefly, growth factor/signal transduction pathways are upregulated after ER-dependent pathways are suppressed by AIs, and ER can then be activated through different crosstalk mechanisms.
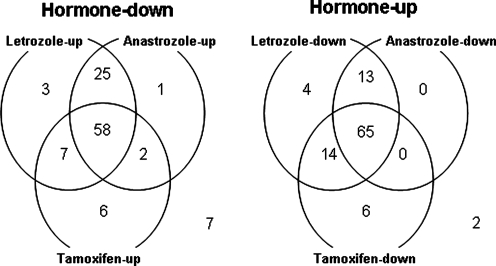
To first determine whether TAM and AIs work similar or not at the molecular level, we treated MCF-7aro cells with testosterone (T), 17β-estradiol (E2), T + AIs (LET and ANA), or T + TAM (Itoh et al., 2005). We found that T or E2 induced proliferation of MCF-7aro cells at a rate six times faster than the untreated cells. In addition, the T-induced proliferation of MCF-7aro cells was effectively suppressed by LET, ANA, or TAM. Microarray analyses, using Affymetrix Human Genome U133A GeneChips, were carried out using total RNA isolated from the control cells and five types of treated cells. At a false discovery rate of 0.05 and a minimum fold change criteria of 1.5, 104 genes upregulated and 109 genes downregulated by both T and E2 were identified (Fig. 2). More than 50% of these hormone-regulated genes were counterregulated by all three inhibitors, and more than 90% were counterregulated by at least one of the inhibitors. Comparison of the effect of each inhibitor on gene expression revealed that LET and ANA were more similar in terms of the genes they affected, compared to treatment with TAM. To validate the gene expression profiles identified from microarray analyses, the expression patterns of 13 representative genes were examined by Northern analysis. We found that the apoptosis pathway, MAPK cascade, and Wnt signaling pathway were affected by the treatments of T and inhibitors (Itoh et al., 2005). The results of this study provide a better understanding of the actions of AIs and TAM at the molecular level, which could serve as an important step in identifying unique expression patterns following drug treatment, and ultimately be useful in customizing patient treatment strategies for hormone-dependent breast cancer. Supporting the physiologic relevance of the findings from our preclinical studies, gene expression changes induced by AI treatment in normal postmenopausal breast tissue and malignant tissue (Kendall et al., 2008; MacKay et al., 2007) are consistent with gene expression profiles identified in our earlier studies on AI treated MCF-7aro cells (Itoh et al., 2005). EXE was not included in this set of microarray experiments because we did not have this inhibitor when the experiments were carried out. The results of microarray analysis of EXE-treated MCF-7aro will be discussed later.
FIG. 2.
Venn diagrams showing the numbers of genes responsive to individual inhibitors in hormone-regulated genes.
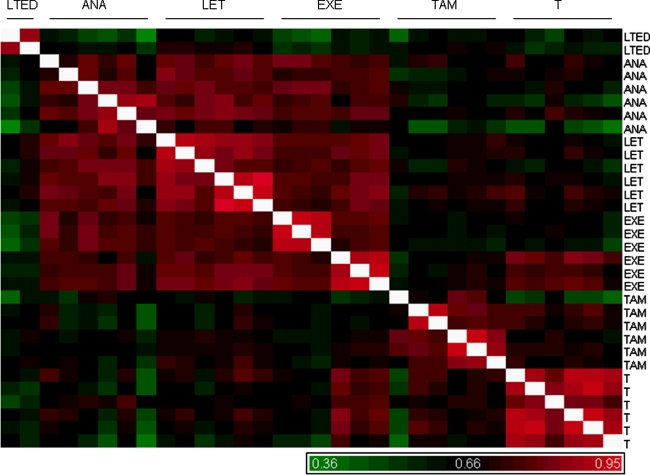
To elucidate mechanisms of acquired resistance to AIs, MCF-7aro cell lines resistant to LET (T+LET R), ANA (T+ANA R), and EXE (T+EXE R), as well as LTEDaro and TAM (T+TAM R) lines were generated. To generate these lines, cells were cultured in phenol red-free Eagle's MEM with nonessential amino acids, sodium pyruvate, and 10% charcoal/dextran-treated FBS with T (1 nM), T + LET (200 nM), T + ANA (1 μM), T + EXE (1 μM), or T + TAM (1 μM). The cells proliferated normally in the presence of T, but most died out in approximiately 2 weeks in the absence of T. In responding to AIs/TAM, the growth of cells cultured with T plus AIs/TAM was initially suppressed, then gradually resumed, and cells eventually grew at rates identical to cells cultured in the presence of T. As proper controls, MCF-7aro cells were also cultured in the presence of T only (i.e., AroT), in addition to medium alone, without T (i.e., LTEDaro). Six independent sets of each resistant line were generated. This is the first complete panel of endocrine therapy resistant cell lines, which were generated as multiple independent biological replicates for unbiased genome-wide analysis. To check the quality of our microarray analysis, a hierarchical clustering analysis of the data has been carried out (Chen et al., 2007). These lines generated high-quality Affymetrix microarray results. As a crucial quality control assessment, we are very pleased with our analysis in which replicates of each type of resistant lines do cluster together. As expected, our results indicate that data of T+LET R, T+ANA R, and T+EXE R lines are more similar than those of T+TAM R and T-treated lines. Although similarities are apparent (Fig. 3), the microarray results clearly demonstrated that gene signatures unique to AI-resistance were inherently different from LTEDaro and T+TAM R gene expression profiles (Masri et al., 2009a). We also prepared inhibitor-only resistant cell lines. The time to generate the different resistant cell lines varied among different inhibitors. Cells growing in the presence of testosterone and inhibitor as well as LTEDaro cells had similar generation times and were all established by 3 months. The inhibitor-only resistant cells had more variation among them. EXE R and ANA R were established in 2 months, whereas LET R and TAM R were established in 8 months and 5 months, respectively. It is important to point out again that acquired AI resistance occurs after a period of treatment. Therefore, gene expression analysis with cells treated with AIs for a few hours or a couple of days will not identify genes that play roles in acquired AI resistance (Chen et al., 2006).
FIG. 3.
Similarity matrix of resistant cell lines. All resistant cell lines were normalized with parental MCF-7aro and the correlation coefficients of 5007 significant genes were displayed as a similarity matrix, using Pearson's correlation. Significant genes were selected based on a fold change criteria of ± 1.2-fold and a p-value <0.05. Correlation coefficients ranged between 0.36 and 0.95, with red indicating good correlation and green representing less correlated lines.
A Global Way to View the Data—ER as the Major Player of Acquired AI Resistance
In addition to cell proliferation assays, aromatase and ER expression and activity levels in the resistant cell lines were examined in order to determine if these proteins play a role in resistance. The aromatase and ER mRNA levels in the resistant cell lines remained at similar levels as the original MCF-7aro cell line, except for the LTEDaro and AnaR cells in which ER transcript levels were elevated. Our results indicate that LTEDaro cell lines are similar to LTED/estrogen withdrawal cell lines generated in other laboratories, where ER expression is elevated. The aromatase protein level and activity in the T+LET R, T+ANA R, and T+TAM R cell lines are similar to the control T-treated cell lines. In addition, aromatase is still functional in these resistant lines and is responsive to the treatment of AIs, as measured by aromatase assay (Masri et al., 2008). These results indicate that AI resistance is not a result of change in aromatase expression or in its response to AIs. As expected, we detect a low level of aromatase activity and aromatase protein in the T+EXE R and EXE R cell lines because exemestane is a mechanism-based inhibitor and destabilizes the aromatase enzyme.
Based on hierarchical clustering, unique estrogen-responsive gene signatures varied depending on cell line, with some genes upregulated in all lines versus other genes upregulated only in the AI-resistant lines. Characterization of these resistant lines showed that LTEDaro, T+LET R, and T+ANA R cells contained a constitutively active ER that did not require estrogen for activation (Masri et al., 2008). Although EXE has previously been shown to act as an androgen (Ariazi et al., 2007), studies from our laboratory demonstrate the estrogen-like activity of EXE. Based on genome-wide microarray analysis, a high correlation was seen between EXE-Only (EXE O, hormone-free) and T-treated AI-resistant lines. In addition, the most upregulated genes in the EXE O lines were mostly estrogen-responsive genes. This estrogen-like activity of EXE was further validated using ER activity assays, where, in comparison to E2, EXE was able to induce ER activity, although at 1/1000 the potency of E2. Also, this EXE-mediated ER activity was blocked by the ER antagonist ICI182,780 (ICI) as well as the ERα-specific antagonist methyl-piperidino-pyrazole. Similarly, EXE induced proliferation of the breast cancer cell lines MCF-7, T47D and MCF-7aro, as well as activated transcription of known estrogen-responsive genes, that is, PGR, pS2, and amphiregulin (AREG). These results suggest that EXE does have weak estrogen-like activity (Lewis et al., 2005).
Because ER function was found to be upregulated, in the absence of estrogen, in a few resistant cell lines, we would expect that the expression of most estrogen-responsive genes remains high even in the presence of AI/tamoxifen. Interestingly, although genes like CCND1, CTSD, and TFF1 were found to be upregulated in all resistant cell lines, PGR was found to be upregulated in T-treated, T+ANA R, T+LET R, T+EXE R, EXE R, and T+TAM R, but not in LTEDaro and ANA R. These results would indicate that the expression of PGR in LTEDaro and ANA R is not regulated through ER. With the results generated so far, we think that we have four types of hormone-resistant cell lines. The first type includes LTEDaro and ANA R, which are ER overexpressing, with constitutively active ER, and with ER+/PR− phenotype. The second type includes T+ANA R and T+LET R, which are with constitutively active ER. The third type includes EXE R and T+EXE R, which contain ER that is estrogen-dependent or hormone responsive. The fourth type includes T+TAM R, which has the gene expression profile that is clearly different from those of LTEDaro and AI resistant cell lines. Using microarray analysis, we believe that our large panel of AI and tamoxifen-resistant lines will provide new insight into mechanistic differences between steroidal versus nonsteroidal aromatase inhibitors and how these pathways differ from tamoxifen resistance.
Experimental Confirmation of ER Function in AI-Resistant Cells
Further characterization of these resistant lines was performed using cell cycle analysis, immunofluorescence to visualize ER subcellular localization, as well as crossresistance studies to determine second-line inhibitor response (Masri et al., 2008). Using this well-defined model system, our studies provide important information regarding differences in resistance mechanisms to AIs, TAM, and LTEDaro, which are critical in overcoming resistance when treating hormone-responsive breast cancers. Our results indicate the resistance mechanisms of the four drugs are probably not identical, and our resistant-cell lines are valuable tools for studying the resistance mechanisms of these four drugs.
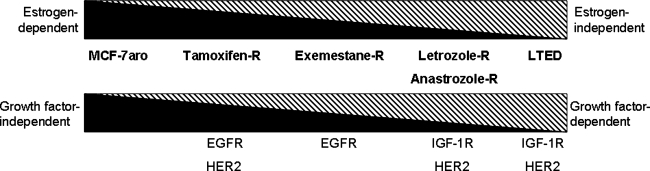
Based on results generated so far, we propose a model of acquired resistance that progresses from hormone-dependence (T+TAM R and T+EXE R) to hormone-independence (T+LET R and T+ANA R), eventually resulting in hormone-independence that does not rely on conventional ER signaling (LTEDaro) (Fig. 4) (Masri et al., 2008). Importantly, although the TAM-resistant cell lines responded to treatment with every AI and LTEDaro did not respond to any AI, all AI-resistant cell lines responded to treatment with another AI (Masri et al., 2008). Thus, our resistant cell lines behave similar to resistant tumors following AI treatment, that is, the lack of crossresistance among three AIs (Lonning, 2009).
FIG. 4.
Estrogen response and growth factor response of endocrine-resistance cell lines.
Immunofluorescence experiments were carried out to look at subcellular localization of ER in the resistant cell lines, in comparison to MCF-7aro (Masri et al., 2008). Upon treatment of MCF-7aro cells with E2, nuclear translocation of ER was apparent. This is different from LTEDaro and T+LET R lines, where ER staining, after E2 treatment, is seen in both the cytoplasm and the nucleus. Immunofluorescence analysis also revealed a unique morphology to the LTEDaro lines. The LTEDaro cells exhibit long axon-like extensions, or pseudopodia, as described by Santen et al. (2005). These pseudopodia were not seen in other resistant lines, or the parental MCF-7aro.
Using this well-defined model system, our studies provide important information regarding differences in resistance mechanisms to AIs, TAM, and LTEDaro, which are critical in overcoming resistance when treating hormone-responsive breast cancers. We hypothesize that EXE resistance results from the weak estrogen-like activity of EXE, LET/ANA resistance is mainly driven by ER hypersensitivity and growth factor (GF)-ER crosstalk, and the late stage of AI resistance (i.e., LTEDaro) results from GF–ER crosstalk.
We also carried out miRNA microarray analysis that identified 49 hormone-responsive miRNAs (Masri et al., 2009). Our study has revealed that miRNA-128a (associated with breast cancer aggressiveness) (Foekens et al., 2008) regulates TGF-β signaling in T+LET R cells.
Next Steps—Prevention and Treatment of AI Resistance
Affected by tumor microenvironment, it is not unexpected that multiple signaling pathways are upregulated in endocrine resistant tumors, that is, ER can be activated by different pathways. Therefore, inhibitors that target a single pathway will be only effective for some, but not all, endocrine resistant patients. Furthermore, secondary resistance may occur following the treatment with a highly selective drug. As discussed nicely in a recent review by Xu and Huang (2010), the complex and dynamic process of receptor tyrosine kinase coactivation during drug resistance needs treatment either by inhibitor combinations or by multitarget inhibitors. Recognizing such possible outcomes, we feel that effective treatment of endocrine resistance could result from the use of drugs with multiple targets. Heat-shock protein 90 (HSP90) is a chaperone protein that functions to assist other proteins, termed “client proteins,” in their proper folding. Many HSP90 client proteins include those involved in cell proliferation and survival. Therefore, HSP90 inhibitors were thought to be useful against AI resistance due to their capability of targeting multiple proteins simultaneously. As proof-of-principle studies, we examined the effect of 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin (17-DMAG), a HSP90 inhibitor, on our hormone-responsive and -resistant cell lines. Our experiments reveal that proliferation of hormone-responsive MCF7aro cells can be suppressed by 17-DMAG and letrozole in a synergistic manner. Furthermore, 17-DMAG was found to be very effective in suppressing the proliferation of both de novo and acquired endocrine resistant cells, but the treatment cannot resensitize the cells to letrozole. 17-DMAG induced apoptosis and G2 cell cycle arrest in both cell lines. Moreover, detailed mechanistic studies revealed decreased HER2, Akt, cyclin D1, and Bcl2 protein expression with 17-DMAG treatment (Wong and Chen, 2009). Results from our preclinical studies are being used as the basis of a clinical trial to evaluate the utility of HSP90 inhibitors to prevent/delay and treat/overcome AI resistance in patients with hormone-dependent breast cancer.
In summary, resistance to AIs is emerging as a complex phenomenon, based on new experimental information discussed in this article. Therefore, analysis of a large panel of resistant cell lines by microarray is an unbiased genome-wide examination of signaling pathways responsible for steroidal and nonsteroidal AI resistance. Acquired resistance to AIs is a hindrance in the clinic, and better understanding of the molecular mechanisms responsible for such occurrences would be beneficial for effectively treating hormone-dependent breast cancers. Our AI-resistant cell lines have been demonstrated to be valuable for both the determination of the resistance mechanisms and the evaluation of new drugs or approaches to help patients against recurring cancer following AI treatment.
Acknowledgments
The research findings discussed in this article were generated by Selma Masri, Ph.D., Sheryl Phung, M.S., Xin Wang, Ph.D., and Cynthie Wong, M.S. This work was supported by the NIH Grants CA44735 (to S.C.), ES08258 (to S.C.), and CA33572 (City of Hope Cancer Center grant).
Author Disclosure Statement
The author declares that no conflicting financial interests exist.
References
- Ariazi E.A. Leitao A. Oprea T.I. Chen B. Louis T. Bertucci A.M., et al. Exemestane's 17-hydroxylated metabolite exerts biological effects as an androgen. Mol Cancer Ther. 2007;6:2817–2827. doi: 10.1158/1535-7163.MCT-07-0312. [DOI] [PubMed] [Google Scholar]
- Baum M. Buzdar A.U. Cuzick J. Forbes J. Houghton J. Klijn J.G.M., et al. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trial. Lancet. 2002;359:2131–2139. doi: 10.1016/s0140-6736(02)09088-8. [DOI] [PubMed] [Google Scholar]
- Chen S. Masri S. Wang X. Phung S. Yuan Y.C. Wu X. What do we know about the mechanisms of aromatase inhibitor resistance? J Steroid Biochem Mol Biol. 2006;102:232–240. doi: 10.1016/j.jsbmb.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Masri S. Hong Y. Wang X. Phung S. Yuan Y., et al. New experimental models for aromatase inhibitor resistance. J Steroid Biochem Mol Biol. 2007;106:8–15. doi: 10.1016/j.jsbmb.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Masri S. Chan H. Petrossian K. Phung S. Yuan Y., et al. Molecular characterization of endocrine resistance. San Antonio Breast Cancer Symposium. 2009. Abs#407.
- Coates A.S. Keshaviah A. Thurlimann B. Mouridsen H. Mauriac L. Forbes J.F., et al. Five years of letrozole compared with tamoxifen as initial adjuvant therapy for postmenopausal women with endocrine-responsive early breast cancer: update of study BIG 1-98. J Clin Oncol. 2007;25:486–492. doi: 10.1200/JCO.2006.08.8617. [DOI] [PubMed] [Google Scholar]
- Coombes R.C. Hall E. Gibson L.J. Paridaens R. Jassem J. Delozier T., et al. A randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancer. N Engl J Med. 2004;350:1081–1092. doi: 10.1056/NEJMoa040331. [DOI] [PubMed] [Google Scholar]
- Foekens J.A. Sieuwerts A.M. Smid M. Look M.P. de Weerd V. Boersma A.W., et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss P.E. Ingle J.N. Martino S. Robert N.J. Muss H.B. Piccart M.J., et al. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N Engl J Med. 2003;349:1793–1802. doi: 10.1056/NEJMoa032312. [DOI] [PubMed] [Google Scholar]
- Hong Y. Yu B. Sherman M. Yuan Y.-C. Zhou D. Chen S. Molecular basis for the aromatization reaction and irreversible inhibition of human aromatase by exemestane. Mol Endocrinol. 2007;21:401–414. doi: 10.1210/me.2006-0281. [DOI] [PubMed] [Google Scholar]
- Howell A. Cuzick J. Baum M. Buzdar A. Dowsett M. Forbes J.F., et al. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years' adjuvant treatment for breast cancer. Lancet. 2005;365:60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- Itoh T. Karlsberg K. Kijima I. Yuan Y.C. Smith D. Ye J., et al. Letrozole-, anastrozole, and tamoxifen-responsive genes in MCF-7aro cells: a microarray approach. Mol Cancer Res. 2005;3:203–218. doi: 10.1158/1541-7786.MCR-04-0122. [DOI] [PubMed] [Google Scholar]
- Kendall A. Anderson H. Dunbier A.K. Mackay A. Dexter T. Urruticoechea A, et al. Impact of estrogen deprivation on gene expression profiles on normal postmenopausal breast tissue in vivo. Cancer Epidemiol. Biomarkers Prev. 2008;17:855–863. doi: 10.1158/1055-9965.EPI-07-2718. [DOI] [PubMed] [Google Scholar]
- Lewis J.S. Meeke K. Osipo C. Ross E.A. Kidawi N. Li T., et al. Intrinsic mechanism of estradiol-induced apoptosis in breast cancer cells resistant to estrogen deprivation. J Natl Cancer Inst. 2005;97:1746–1759. doi: 10.1093/jnci/dji400. [DOI] [PubMed] [Google Scholar]
- Lonning P.E. Lack of complete cross-resistance between different aromatase inhibitors; a real finding in search for an explanation? Eur J Cancer. 2009;45:527–535. doi: 10.1016/j.ejca.2008.10.019. [DOI] [PubMed] [Google Scholar]
- MacKay A. Urruticoechea A. Dixon J.M. Dexter T. Fenwick K. Ashworth A., et al. Molecular response to aromatase inhibitor treatment in primary breast cancer. Breast Cancer Res. 2007;9:R37. doi: 10.1186/bcr1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L.A. Farmer I. Johnston S.R. Ali S. Marshall C. Dowsett M. Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J Biol Chem. 2003;278:30458–30468. doi: 10.1074/jbc.M305226200. [DOI] [PubMed] [Google Scholar]
- Masri S. Phung S. Wang X. Wu X. Yuan Y.C. Wagman L., et al. Genome-wide analysis of aromatase inhibitor-resistant, tamoxifen-resistant, and long-term estrogen-deprived cells reveals a role for estrogen receptor. Cancer Res. 2008;68:4910–4918. doi: 10.1158/0008-5472.CAN-08-0303. [DOI] [PubMed] [Google Scholar]
- Masri S. Lui K. Phung S. Ye J. Zhou D. Wang X., et al. Characterization of the weak estrogen receptor alpha agonistic activity of exemestane. Breast Cancer Res Treat. 2009a;116:461–470. doi: 10.1007/s10549-008-0151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S. Liu Z. Phung S. Wang S.E. Yuan Y.-C. Chen S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res Treat. 2009b. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Masri S. Phung S. Wang X. Chen S. Molecular characterization of aromatase inhibitor-resistant, tamoxifen-resistant and LTEDaro cell lines. J Steroid Biochem Mol Biol. 2010;118:277–282. doi: 10.1016/j.jsbmb.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson R.I. Staka C. Boyns F. Hutcheson I.R. Gee J.M.W. Growth factor-driven mechanisms associated with resistance to estrogen deprivation in breast cancer: new opportunities for therapy. Endocrine-Related Cancer. 2004;11:623–641. doi: 10.1677/erc.1.00778. [DOI] [PubMed] [Google Scholar]
- Santen R.J. Song R.X. Zhang Z. Kumar R. Jeng M.H. Masamura S., et al. Adaptive hypersensitivity to estrogen: mechanisms and clinical relevance to aromatase inhibitor therapy in breast cancer treatment. J Steroid Biochem Mol Biol. 2005;95:155–165. doi: 10.1016/j.jsbmb.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Sun X.Z. Zhou D. Chen S. Autocrine and paracrine actions of breast tumor aromatase. A three-dimensional cell culture study involving aromatase transfected MCF-7 and T-47D cells. J Steroid Biochem Mol Biol. 1997;63:29–36. doi: 10.1016/s0960-0760(97)00068-x. [DOI] [PubMed] [Google Scholar]
- Wang X. Chen S. Aromatase destablizer: novel action of exemestane, an FDA approved aromatase inhibitor. Cancer Res. 2006;66:10281–10286. doi: 10.1158/0008-5472.CAN-06-2134. [DOI] [PubMed] [Google Scholar]
- Wong C. Chen S. HSP90 inhibitors: a new mode of therapy to overcome endocrine resistance. Cancer Res. 2009;69:8670–8677. doi: 10.1158/0008-5472.CAN-09-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A.M. Huang P.H. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70:3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W. Wang J.P. Conaway M. Masamura S. Li Y. Santen R.J. Activation of the MAPK pathway enhances sensitivity of MCF-7 breast cancer cells to the mitogenic effect of estradiol. Endocrinology. 2002;143:3221–3229. doi: 10.1210/en.2002-220186. [DOI] [PubMed] [Google Scholar]
- Zhou D. Pompon D. Chen S. Stable expression of human aromatase cDNA in mammalian cells—a useful system for aromatase inhibitor screening. Cancer Res. 1990;50:6949–6954. [PubMed] [Google Scholar]