Abstract
The development of reagents with high affinity and specificity to small molecules is crucial for the high-throughput detection of chemical compounds, such as toxicants or pollutants. Aptamers are short and single-stranded (ss) oligonucleotides able to recognize target molecules with high affinity. Here, we report the selection of ssDNA aptamers that bind to Bisphenol A (BPA), an environmental hormone. Using SELEX process, we isolated high affinity aptamers to BPA from a 1015 random library of 60 mer ssDNAs. The selected aptamers bound specifically to BPA, but not to structurally similar molecules, such as Bisphenol B with one methyl group difference, or 4,4′-Bisphenol with 2 methyl groups difference. Using these aptamers, we developed an aptamer-based sol–gel biochip and detected BPA dissolved in water. This novel BPA aptamer-based detection can be further applied to the universal and high-specificity detection of small molecules.
Introduction
Single-stranded (ss) DNA oligonucleotide aptamers can be used for molecular detection in many screening platforms. They can detect small molecules in solution, which is relevant for monitoring environmental pollutants, food toxicants, and disease-related metabolites (Fukata et al., 2006).
RNA or ssDNA aptamers can be obtained by SELEX process (Gold et al., 1997; Shi et al., 2007; Ahn et al., 2009). Aptamers are selected from an initial pool of ∼1015 molecules until they have high enough affinity, which typically ranges from micro-molar (μM) to nano-molar (nM) range, or even higher (Geiger et al., 1996; Guo et al., 2005; Shi et al., 2007; Pagano et al., 2008). Comparing to antibodies, aptamers are better capturing agents for small molecules, because (i) their shorter size more accurately discriminates functional groups between similar structures (Jenison et al., 1994), and (ii) aptamers targeting small molecules can be selected in vitro without the need of hapten, which is needed for selection of antibodies against molecules whose molecular weight is below 5,000 Da (Stevenson et al., 1970; Sheedy et al., 2007).
Bisphenol A (BPA) is a small carcinogenic molecule (MW = 228 Da), which is potentially dangerous to animals and humans (Schonfelder et al., 2002). They are defined as endocrine-disrupting compounds, which can mimic the action of hormone estrogen and disturb the estrogen-estrogen receptor binding process (hormonal pathways) (Diamanti-Kandarakis et al., 2009). Because of its threat to the environment and human health, there have been increasing needs for the detection and monitoring of BPA.
Until recently, BPA detection was done through chromatographic methods, such as gas and liquid chromatography (Stuart et al., 2005; Ballesteros-Gomez et al., 2009), or other conventional assay methods, such as immunoenzyme-based assays (Fukata et al., 2006). In particular, methods such as enzyme-linked immunosorbent assay (Freymuth et al., 1986; Zheng et al., 2008) showed insensitive assay, because BPA antibody has nonspecific binding, especially for similar molecules, such as Bisphenol B (BPB) (Ohkuma et al., 2002) or the analog, 4,4-Bis-(4-hydroxyphenyl) valeric acid (Marchesini et al., 2005).
Sol–gel material has a 3-dimensional (3D) structure and was originally developed for protein immobilization (Kim et al., 2006). Since aptamers have 3D structure similar to proteins, we realized sol–gel chip could be better format for aptamer immobilization than 2-dimensional (2D) surface-modified chips (Kim et al., 2006; Ahn et al., 2008, 2009).
In this study, we developed aptamers targeting BPA with nM affinity level. One of the selected aptamers had high affinity to BPA, but not to BPB (one methyl group difference), 4,4′-Bisphenol (2 methyl groups difference; BP), or 6F BPA (6 fluorine atoms difference; 6F). Using the high-affinity aptamers, we also developed a sol–gel biochip assay to detect BPA, and measured BPA level in water samples. This is the first successful demonstration of aptamer-based biochip assay for BPA detection. Thus, this aptamer-based detection strategy has a broad application range in small molecule detection. This innovative technology has potential relevance for a variety of applications, such as medical diagnostics, environmental control, and food safety.
Materials and Methods
Material preparation
For BPA aptamer selection, BPA (4,4′-dihydroxy-2,2-diphenylpropane; Sigma-Aldrich) was dissolved in 50% dimethylformamide at a final concentration of 20 mM. Epoxy-activated Sepharose 6B resin (GE Healthcare Bio-Sciences Corp.) was used to immobilize BPA via ether linkages to hydroxyl groups. Then, acridine yellow affinity column (Bio-Rad) was used for housing BPA coupled resin.
To prepare a random ssDNA library, a collection of the sequences 5′-GGGCCGTTCGAACACGAGCATG-N60-GGACAGTACTCAGGTCATCCTAGG-3′ was chemically synthesized (Genotech Inc.).
BPA similar structures—BPB, 6F, and BP—were purchased from TCI.
For the aptamer chip preparation, we used the SolB™ (www.pclchip.com PCL Inc.) for immobilizing materials and cyanine 3 (Cy3)-labeled rabbit secondary antibodies (Abcam) for positive controls.
BPA aptamers in vitro selection
First, to immobilize BPA, the epoxy-activated resin with coupling buffer (50% dimethylformamide, pH 13.0) was mixed with 20 mM BPA. BPA-resin coupling occurred overnight. Then, the coupled resin was washed and hydroxyl groups that remained unoccupied were blocked. In parallel, a separate aliquot of naked resin was coated with ethanolamine, the aptamer selection negative control. By measuring the amounts of unbound BPA using UV spectrometry (absorption at 280 nm), we can estimate the amount of BPA bound to resin.
To prepare aptamer library, synthesized ssDNAs were amplified by asymmetric polymerase chain reaction (PCR). The initial pool had 1015 ssDNA oligonucleotides (Shi et al., 2007; Sevilimedu et al., 2008; Ahn et al., 2009). Twelve cycles of aptamer selection and amplification were performed as described before (Niazi et al., 2008), but with some modifications. In detail, the BPA-coupled resin was washed with binding buffer (25 mM Tris-HCl, 100 mM NaCl, 25 mM KCl, 10 mM MgCl2, and 5% DMSO, pH 8.0) before each SELEX round. The random ssDNA library pool was introduced at round 1 (R1). Then, in every selection round, 19 μM of BPA was mixed with random ssDNA pool and incubated at room temperature for 1 hour. Washing the resin with binding buffer, unbound ssDNA was removed. After each round, aptamers were eluted by 50 mM BPA solution in binding buffer. The eluted ssDNAs were precipitated, washed, and dissolved in distilled water. After R3 (Round 3), a negative selection step removed nonspecific ssDNA. R11 and R12 eluted aptamer pools were cloned, with pGEM-T easy vector system (Promega), and sequenced (Solgent Inc.).
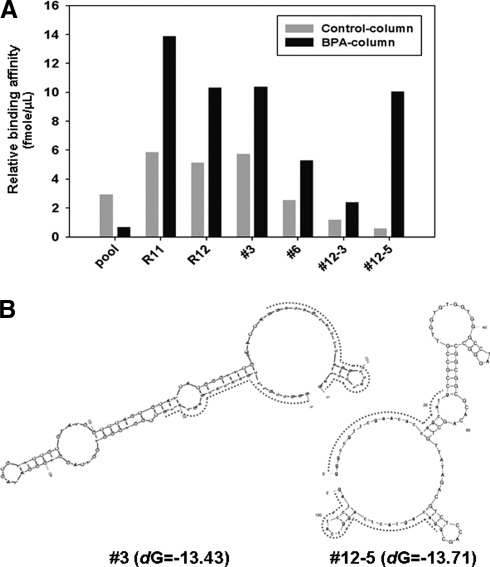
For measurement of binding affinities in Fig. 2A, real-time PCR (ABI) was performed. First, 280 pmol of pool, R11, R12, and selected aptamers were incubated both with BPA column and no-BPA column. Then, eluted aptamers were quantified by real-time PCR according to manufacturer's recommendation. Finally, isolated aptamer secondary structure analysis was done with a free energy minimization algorithm using Mfold software (ZUKER, 2003).
FIG. 2.
BPA-specific aptamers selection. (A) SELEX selection of ssDNAs bound to BPA. By real-time polymerase chain reaction, binding affinity was measured for initial ssDNA pool, R11 (SELEX Round 11) and R12 (SELEX Round 12) elutes, and individual aptamers (#3 and #6 from R11; #12–3 and #12–5 from R12). Control columns were used for each aptamer. (B) Secondary structures prediction (Mfold software) for the highest affinity aptamers, #3 and #12–5. [Capital letter: N60 random sequence region, Lowercase letter and dotted line: constant sequence region.]
Dissociation constants (Kd) determination
To calculate Kd, we performed aptamer-based binding assays using the equilibrium filtration method (Niazi et al., 2008). Kds were calculated by plotting the percent of bound BPA versus ssDNA aptamer concentration. Then, data points were fit into nonlinear regression analysis, according to the following equation and Sigmaplot 10.0 software
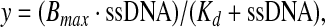 |
where y is the degree of saturation and Bmax is the number of maximum BPA binding sites. Kds of BPB, 6F, and BP were measured the same manner.
Aptamer binding to BPA in sol–gel chip
To assess the affinity of selected aptamers, we used sol–gel biochip system (Kim et al., 2006). In detail, the selected aptamers were mixed with SolB reagents individually and micro-spotted in 96-well plate (polymethyl methacrylate) using OmniGrid Accent Microarrayer (Digilab). Then, the aptamers were immobilized within sol–gel spots by gelation. For the sandwich-based binding assay, each aptamer was labeled with Cy3, using terminal deoxynucleotidyl transferase (Fermentas) and Cy3-dUTP (GeneChem Inc.) according to manufacturer's recommendation. To each well of the 96-well plate spotted, equimolar amount of BPA (1 μM) and free Cy3-labeled aptamers (1 μM) were added and incubated along with the control well (no BPA). Aptamers bound to sol–gel spots were subjected to sandwich assay with BPA and free Cy3-labeled aptamers. Then, each well was heavily washed with washing solution (1xPBS containing 0.1% Tween; Sigma). To qualify the BPA binding to aptamer, the assayed spots were analyzed with a Multi-Image Analyzer (Fujifilm). Each plate had a negative (without aptamers) and positive control (with labeled antibodies described in Materials and Methods above).
Results and Discussion
BPA aptamer selection and detection
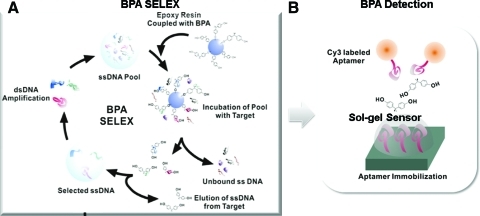
Figure 1 shows the experimental procedure of BPA aptamer selection and detection. In brief, we used SELEX process to select BPA-specific aptamers from a pool of 1015 ssDNAs (Fig. 1A) (Niazi et al., 2008). This step enabled us to isolate several aptamers with high affinity to BPA molecules. For the sandwich assay, we needed a pair of capturing and labeling aptamers, which would form sandwich structures only with BPA. The capturing aptamer was fixed onto the sol–gel chip and the labeling aptamer, with Cy3-fluorescence, was added with or without BPA molecules to check binding specificity (Fig. 1B).
FIG. 1.
BPA aptamer selection and detection. (A) BPA aptamer selection: From the aptamer library (1015 ssDNAs), using SELEX, we selected aptamers specifically bound to BPA. (B) BPA detection with selected aptamers: Using 3-dimensional aptamer sol–gel chips, water samples contaminated with BPA were analyzed by fluorescence microscopy. ss, single-stranded; BPA, bisphenol A. Color images available online at www.liebertonline.com/oli
We should note that, according to the method of BPA immobilization to resin described at Materials and Methods, we could immobilize 16 to 23 μmol of BPA to the resin for this entire BPA SELEX. After 12 rounds of SELEX, as shown in Table 1, we were able to fish out aptamer candidates.
Table 1.
Sequences of the Selected Aptamer Groups
| Groupa | Freqeuncyb | ID | Sequencec |
|---|---|---|---|
| 1 | 11 | #31 | CGGCCCTAGGATGACCTGAGTACTGTCCCTCACCCCTACTTCCGCCACTGGCCCAACAGC |
| #23 | TGCCTAGGATGACCTGAGTACTGTCCAGGCTCCGACCTTGTCCCTGCCGCCACTCTCCCA | ||
| #47 | GCGGACGGGCTCGGCTCACCTAGGATGACCTGAGTACTGTCCCCGTGGCGCTAATTCGGG | ||
| #50 | CGGCCCGCCCCTAGGATGACCTGAGTACTGTCCGCGGGACGGTATCGCTGAGACAGGTGC | ||
| #41 | CGGCAGCCCTAGGATGACCTGAGTACTGTCCGCGAAAGACTCCATGGTACCCGGTGCTTA | ||
| #27 | GGGGGCGTCGNCCTAGGATGACCTGAGTACTGTCCGCACNCAGGGAGGATGCATTGAC | ||
| #45 | GTGTCCCCACGTCCTAGGATGACCTGAGTACTGTCCAATGCCGCTCCTCCCGATGCAGAC | ||
| #11 | CTCTTCNCTCCAATTCGTAAGATGACCTGAGGTCTGCCCAACGGTGTTTAGAACCCCTTG | ||
| #12–3 | CGCAGCGCGCCCCTGAGTACTGTCCGCCCAACGGTGTGACGGCCCTGCGATCAACGATTG | ||
| #12–4 | GGGCCGTCCTAGGATGACCTGAGTACTGTCCGCCCAACGGTGTGACGGCCCTGCGATCAA | ||
| #22 | CCTCGCCCTGAGTACTGTCCCCCGTCCGTCCGGTGAGGGCCACTATCGCTAACTGATCA | ||
| 2 | 4 | #4 | AGGCCGTTGGTGTGGTGGGCCTAGGGCCGGCGGCGCACAGCTGTTATAGACGTCTCCAGC |
| #12–5 | CCGCCGTTGGTGTGGTGGGCCTAGGGCCGGCGGCGCACAGCTGTTATAGACGTCTCCAGC | ||
| #6 | CCGCCGTTGGTGTGGTGGGCCTAGGGCCGGCGGCGCACAGCTGTTATAGACGCCTCCAGC | ||
| #12–7 | CCGCCGTTGGTGTGGTGGGCCCAGGGCCGGCGGCGCACAGCTGTTATAGACGCCTCCAGC | ||
| 3 | 3 | #12–2 | TGACGGTGGCGTGGAGGGCGCGTATCAATCGTTGATCGCAGGGCCGTCATACCGTTGGAG |
| #12–9 | TGACGGTGGCGTGGAGGGCGCGTATCAATCGTTGATCGCAGGGCCGTCATACCGTTGGGGG | ||
| #12–6 | TGACGGTGGCGTGGAGGGCGCGTATCAATCGTTGATCGCAGGGCCGTCATACCGGTCGGG | ||
| 4 | 3 | #2 | GCCGACAGGGCATGGGACGCTATACAGCGGTGTCAATCGAATTCCCGCGGCCGCCATGCGG |
| #14 | GGTCCCCGCAGCTCATACGGCGCTCCAGCGTAATCGAATTCCCGCGGCCGCCATGCGGCC | ||
| #46 | GCGAGTGGCCCATCAGCAGAGCGTAATCCCCACGCACATCGAGTGCCCCCGGCCGGTGCT | ||
| 5 | 2 | #12 | GTATTGTCATTCATATCCTCGTGCTTGCTGTCCTCACCCCACCCACCAGAATGGAAA |
| #13 | CCTGGTATTGTCTTGCCAATCCTCGCCCTGGCTGTCTTACCCCTCCCCACCCGCCTGAAG | ||
| 6 | 2 | #48 | GTCGACTCGCGGGTACCGTGCTCAATGTCCCAATCCGGGGAAGCGTTTAGACCCGCAGCCCAC |
| #40 | GTCGCCACTGCGGGTACCGTGCTTGGGCNACCGATGNACCNTGNNACCGTGTTTNGCC | ||
| 7 | 2 | #3 | CCGGTGGGTGGTCAGGTGGGATAGCGTTCCGCGTATGGCCCAGCGCATCACGGGTTCGCACCA |
| #32 | GGGCGGTGGGTGGCGAGTTGTGAGACGCTGGAGGAGGTTGCTGCCCCCGGCACATTGGGA | ||
| Constant sequences | 5′-GGGCCGTTCGAACACGAGCATG-(N)60-GGACAGTACTCAGGTCATCCTAG-3′ | ||
Groups: Similar homologous aptamers classified as the same group.
Frequency: Number of individual sequences hitting the same group.
Sequences: Only variable region of selected aptamer groups. Shaded sequences indicate the conserved region within each group. The constant sequences (primer region) are shown at the bottom.
BPA aptamer selection
Figure 2A shows the binding affinity of aptamers isolated by SELEX using BPA-columns (marked as “BPA column”). As a control experiment, for each aptamer, we performed the same affinity test using resins not coupled with BPA (marked as “Control column”). In the initial pool, aptamers' binding affinity to BPA molecules was very low, but increased dramatically after repeated SELEX rounds, especially in round 11 and 12 (Fig. 2A—pool, R11 and R12). Then, we measured the affinities of individual selected aptamers and found that aptamers #3 (R11) and #12–5 (R12) displayed the highest affinity toward BPA (Fig. 2A—#3, #6, #12–3 and #12–5). The secondary structure models of aptamers #3 and #12–5 were predicted with Mfold software (Fig. 2B). No apparent sequence or secondary structure similarity between the 2 was noticed.
BPA aptamer and BPA molecule-specific interaction
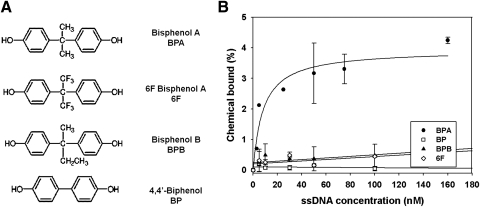
Table 1 lists all aptamer sequences selected from SELEX process. To further evaluate the specificity of the BPA aptamers, we measured the affinities (Kd) of aptamer #3 to BPA and structurally similar molecules such as 6F BPA (6F), BPB, and 4,4′-Biphenol (BP) (Fig. 3A), using the equilibrium filtration method (Niazi et al., 2008). Although they have similar structures, they do not exhibit environmental toxicity like BPA. The results show that aptamer #3 has highly selective binding only to BPA, but not to other BP family molecules (Fig. 3B). The measured Kd of aptamer #3 for BPA, 6F, BPB, and BP are 8.3 nM, 208 mM, 139 mM, and 139 mM, respectively. The affinity of aptamer #3 targeting BPA is comparable to that of a high-affinity antibody (3–10 nM) targeting large biomolecules (Niazi et al., 2008). Considering that antibodies to small molecules typically display much lower affinities than to macromolecules such as proteins (Kim et al., 2008), aptamer #3, with its nM-level Kd for small molecules, can be considered to be a very strong BPA binder. Further, aptamer #3 exhibited much stronger affinity to BPA than to BPB, which has very similar structure. In contrast, at least one well-characterized BPA antibody has been shown to have cross affinity with BPB (Ohkuma et al., 2002; Marchesini et al., 2005). Thus, the aptamers isolated by our study are superior over corresponding antibodies, and can be used for highly-selective detection of BPA.
FIG. 3.
Selected aptamers affinity to BPA. (A) BP molecules family. (B) Aptamer #3 specific binding to BPA [Kds: BPA = 8.3 nM; 6F = 208 mM; Bisphenol B (BPB) = 139 mM; and BP = 139 mM].
BPA detection with aptamer sol–gel biochip
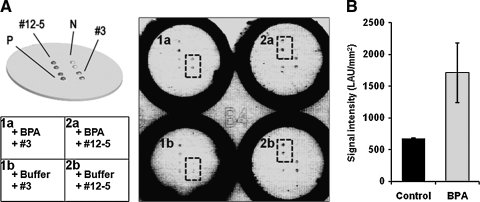
As explained above (Fig. 1B), the direct measurement assay requires a pair of BPA aptamers. For that, we used sol–gel chips to isolate the BPA aptamer pair suitable for sandwich assay (Fig. 4). We chose BPA aptamers #3 and #12–5, and performed specific and nonspecific binding experiments for all possible pairs. Figure 4A shows the inverse contrast fluorescence images of Cy3-labeled aptamer adsorbed on sol–gel droplets. With BPA molecules, the pair #3-#3 formed #3-BPA-#3 sandwich structures (dotted rectangle in Fig. 4A-1a), and without BPA there was no binding (dotted rectangle in Fig. 4A-1b). The pair #12–5-#12–5 had nonspecific interaction with or without BPA (dotted rectangles in Fig. 4A-2a, 2b). These results show that our 2-step screening process can be used to identify aptamer pairs suitable for sandwich assays. Because we used the same aptamer set (#3-#3) for the sandwich assay, this assay system can only be applied in a limited concentration range. An equal molar ratio of free aptamers and BPA (1 μM) was optimal for the analytical detection of BPA.
FIG. 4.
BPA sandwich assay with aptamers #3 and #12–5 in 3-dimensional sol–gel biochip. (A) On each well, duplicate sol–gel spots, including aptamers #3 and #12–5, were printed along with positive (P: cyanine 3-labeled rabbit secondary antibodies) and negative (N: without proteins) controls (upper left sketch): 1a and 2a wells were incubated with BPA and labeled aptamers #3 and #12–5, respectively, and 1b and 2b wells were incubated only with labeled aptamers #3 and #12–5, respectively. Sandwich assay results (right panel) showed that specific interaction only occurs with aptamer #3 (1a). Aptamer #12–5 had nonspecific signals (2a and 2b). (B) This assay was repeated 8 times. The average fluorescent signal intensities of control (A-1b) and BPA (A-1a) were shown (LAU/min2 is the unit number from fluorescent scanner described in the Materials and Methods section).
Considering that aptamers and antibodies have similar 3D structures, sol–gel biochips are relevant high-sensitivity detection platforms for aptamer-based assays (Kim et al., 2006; Ahn et al., 2008). Previously, we used sol–gel biochips for antibody-based protein detection with sensitivity at femto molar (fM) range (Lee et al., 2007). Now, our new aptamer-based detection method has nM range sensitivity (Fig. 3B). We also performed sandwich assay using 2D biochips with optical detection, but they exhibited low sensitivity compared with sol–gel chips (data not shown).
We believe that, like antigen–antibody interactions, aptamers can interact with BPA only when they keep proper spatial orientation. Sol–gel chip 3D format (i) helps to maintain the aptamer in its active conformation and (ii) holds more aptamers within the same surface area than 2D chips. These features enhance stability and sensitivity of aptamer sensing. Therefore, we believe that sol–gel chips are suitable platforms for aptamer screening and detection (Park et al., 2009).
Conclusion
Our work is the first report of successful BPA molecule detection using aptamers. We report highly specific ssDNA oligonucleotide aptamers and an aptamer-based sol–gel chip assay for sensitive and selective detection of small molecule pollutants.
Chemical SELEX strategy is very attractive to select aptamers that target small molecules, because counter SELEX can be used to increase specificity against molecules with similar structure. Thus, aptamers can capture small molecules better than antibodies, due to their higher ability to discriminate small structural differences, such as single functional group. In our case, the selected aptamer #3 can bind specifically to BPA, and not to BPB or other structurally similar molecules.
Further, we applied #3 aptamers for different sensor platform, such as swCNT-FET or capacity-based sensor. Using these sensors, we demonstrated the detection limit of 1 pM and single-carbon-atomic resolution (Lee et al., 2011). These results demonstrate that specific aptamers of small molecules can be used in many sensor applications, such as environmental monitorization and food safety.
Acknowledgments
The authors acknowledge funding from the Ministry of Knowledge Economy (10032113), Korea Ministry of Environment as “The Eco-technopia 21 project” (2010-10002-0065-0/2010-09001-0076-0) the National Research Laboratory program of the NRF grant (No. 20100008018) of the MEST. This work also partially supported by Agricultural Research Center (ARC, 710003-03-1-SB120). SH also thanks the NRF grant (No. 2011-0000390) of the MEST and the System 2010 program of the MKE. DKL acknowledges support from Global Research Laboratory grant from MEST.
Author Disclosure Statement
No competing financial interests exist.
References
- AHN J.Y. CHO M. LEE S. PARK J. HONG S. SHIN S. JEONG M. LEE D.K. KIM S. Sol-gel material optimization for aptamer biosensors. Mol. Cell. Toxicol. 2008;4:100–105. [Google Scholar]
- AHN J.Y. KIM E. RYU J.C. KIM S. Selection of Aptamers in SELEX process. Toxicol. Environ. Health. Sci. 2009;1:1–7. [Google Scholar]
- BALLESTEROS-GOMEZ A. RUBIO S. PEREZ-BENDITO D. Analytical methods for the determination of bisphenol A in food. J Chromatogr. A. 2009;1216:449–469. doi: 10.1016/j.chroma.2008.06.037. [DOI] [PubMed] [Google Scholar]
- DIAMANTI-KANDARAKIS E. BOURGUIGNON J.P. GIUDICE L.C. HAUSER R. PRINS G.S. SOTO A.M. ZOELLER R.T. GORE A.C. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocr. Rev. 2009;30:293–342. doi: 10.1210/er.2009-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREYMUTH F. QUIBRIAC M. PETITJEAN J. AMIEL M.L. POTHIER P. DENIS A. DUHAMEL J.F. Comparison of two new tests for rapid diagnosis of respiratory syncytial virus infections by enzyme-linked immunosorbent assay and immunofluorescence techniques. J. Clin. Microbiol. 1986;24:1013–1016. doi: 10.1128/jcm.24.6.1013-1016.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUKATA H. MIYAGAWA H. YAMAZAKI N. MORI C. Comparison of Elisa- and LC-MS-based methodologies for the exposure assessment of bisphenol A. Toxicol. Mech. Methods. 2006;16:427–430. doi: 10.1080/15376520600697404. [DOI] [PubMed] [Google Scholar]
- GEIGER A. BURGSTALLER P. VON DER ELTZ H. ROEDER A. FAMULOK M. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLD L. BROWN D. HE Y. SHTATLAND T. SINGER B.S. WU Y. From oligonucleotide shapes to genomic SELEX: novel biological regulatory loops. Proc. Natl. Acad. Sci. U.S.A. 1997;94:59–64. doi: 10.1073/pnas.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO K. WENDEL H.P. SCHEIDELER L. ZIEMER G. SCHEULE A.M. Aptamer-based capture molecules as a novel coating strategy to promote cell adhesion. J. Cell. Mol. Med. 2005;9:731–736. doi: 10.1111/j.1582-4934.2005.tb00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JENISON R.D. GILL S.C. PARDI A. POLISKY B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- KIM S. KIM Y. KIM P. HA J.M. KIM K. SOHN M. YOO J.S. LEE J. KWON J.A. LEE K.N. Improved sensitivity and physical properties of sol-gel protein chips using large-scale material screening and selection. Anal. Chem. 2006;78:7392–7396. doi: 10.1021/ac0520487. [DOI] [PubMed] [Google Scholar]
- KIM Y.S. LEE S.J. GU M.B. Electrochemical aptamer-based biosensors. Biochip J. 2008;2:175–182. [Google Scholar]
- LEE J. JO M. KIM T. AHN J.Y. LEE D.K. KIM S. HONG S. Aptamer sandwich-based carbon nanotube sensors for single-carbon-atomic-resolution detection of non-polar small molecular species. Lab. Chip. 2011;11:52–56. doi: 10.1039/c0lc00259c. [DOI] [PubMed] [Google Scholar]
- LEE S. KIM Y.S. JO M. JIN M. LEE D.K. KIM S. Chip-based detection of hepatitis C virus using RNA aptamers that specifically bind to HCV core antigen. Biochem. Biophys. Res. Commun. 2007;358:47–52. doi: 10.1016/j.bbrc.2007.04.057. [DOI] [PubMed] [Google Scholar]
- MARCHESINI G.R. MEULENBERG E. HAASNOOT W. IRTH H. Biosensor immunoassays for the detection of bisphenol A. Anal. Chim. Acta. 2005;528:37–45. [Google Scholar]
- NIAZI J.H. LEE S.J. KIM Y.S. GU M.B. ssDNA aptamers that selectively bind oxytetracycline. Bioorg. Med. Chem. 2008;16:1254–1261. doi: 10.1016/j.bmc.2007.10.073. [DOI] [PubMed] [Google Scholar]
- OHKUMA H. ABE K. ITO M. KOKADO A. KAMBEGAWA A. MAEDA M. Development of a highly sensitive enzyme-linked immunosorbent assay for bisphenol A in serum. Analyst. 2002;127:93–97. doi: 10.1039/b103515k. [DOI] [PubMed] [Google Scholar]
- PAGANO B. MARTINO L. RANDAZZO A. GIANCOLA C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008;94:562–569. doi: 10.1529/biophysj.107.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK S.M. AHN J.Y. JO M. LEE D.K. LIS J.T. CRAIGHEAD H.G. KIM S. Selection and elution of aptamers using nanoporous sol-gel arrays with integrated microheaters. Lab. Chip. 2009;9:1206–1212. doi: 10.1039/b814993c. [DOI] [PubMed] [Google Scholar]
- SCHONFELDER G. WITTFOHT W. HOPP H. TALSNESS C.E. PAUL M. CHAHOUD I. Parent bisphenol A accumulation in the human maternal-fetal-placental unit. Environ. Health Perspect. 2002;110:A703–A707. doi: 10.1289/ehp.110-1241091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEVILIMEDU A. SHI H. LIS J.T. TFIIB aptamers inhibit transcription by perturbing PIC formation at distinct stages. Nucleic Acids Res. 2008;36:3118–3127. doi: 10.1093/nar/gkn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEEDY C. MACKENZIE C.R. HALL J.C. Isolation and affinity maturation of hapten-specific antibodies. Biotechnol. Adv. 2007;25:333–352. doi: 10.1016/j.biotechadv.2007.02.003. [DOI] [PubMed] [Google Scholar]
- SHI H. FAN X. SEVILIMEDU A. LIS J.T. RNA aptamers directed to discrete functional sites on a single protein structural domain. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3742–3746. doi: 10.1073/pnas.0607805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEVENSON G.T. EISEN H.N. JONES R.H. The problem of hapten persistently bound to antibody. Biochem. J. 1970;116:151–153. doi: 10.1042/bj1160151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUART J.D. CAPULONG C.P. LAUNER K.D. PAN X. Analyses of phenolic endocrine disrupting chemicals in marine samples by both gas and liquid chromatography-mass spectrometry. J. Chromatogr. A. 2005;1079:136–145. doi: 10.1016/j.chroma.2005.03.075. [DOI] [PubMed] [Google Scholar]
- ZHENG J. ZHANG K. ZHAO S.Q. [Study on spectral and immune identification of artificial antigen of bisphenol A] Guang. Pu. Xue. Yu. Guang. Pu. Fen. Xi. 2008;28:1583–1586. [PubMed] [Google Scholar]
- ZUKER M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]