Abstract
Peroxiredoxins (Prxs), some of nature's dominant peroxidases, use a conserved Cys residue to reduce peroxides. They are highly expressed in organisms from all kingdoms, and in eukaryotes they participate in hydrogen peroxide signaling. Seventy-two Prx structures have been determined that cover much of the diversity of the family. We review here the current knowledge and show that Prxs can be effectively classified by a structural/evolutionary organization into six subfamilies followed by specification of a 1-Cys or 2-Cys mechanism, and for 2-Cys Prxs, the structural location of the resolving Cys. We visualize the varied catalytic structural transitions and highlight how they differ depending on the location of the resolving Cys. We also review new insights into the question of how Prxs are such effective catalysts: the enzyme activates not only the conserved Cys thiolate but also the peroxide substrate. Moreover, the hydrogen-bonding network created by the four residues conserved in all Prx active sites stabilizes the transition state of the peroxidatic SN2 displacement reaction. Strict conservation of the peroxidatic active site along with the variation in structural transitions provides a fascinating picture of how the diverse Prxs function to break down peroxide substrates rapidly. Antioxid. Redox Signal. 15, 795–815.
Scope and Purpose
Peroxiredoxins (Prxs) are now recognized as the family of peroxidases that is broadly important in both antioxidant protection and cellular signaling pathways (84). Much ongoing work is elucidating the role of Prxs throughout biology, and many excellent reviews (25) have been published summarizing our current understanding of various aspects of Prxs, including covalent modifications (2) and signaling (28), and their importance in systems such as mitochondria (16), plants (22), yeast (18), and Caenorhabditis elegans (63). As for all enzymes, function flows directly from structure, and in this case, structural knowledge makes critical contributions to illuminating Prx function in both its antioxidant and cellular signaling roles. The goal of this review is to survey and organize the current structural information known about the Prxs; since our previous review published in 2007 (42), the number of known Prx structures has doubled. This review does not simply provide an up-to-date reference guide, pointing readers to the original publications for additional insights, but also presents essential principles of Prx function that can be derived from these structures.
Introduction
Peroxiredoxins (Prxs) are ubiquitous peroxidases that use a conserved Cys residue to reduce peroxide substrates. Although they are not as well known as catalase and glutathione peroxidase, many Prxs have high expression levels [up to 1% or more of cellular proteins (84)] and fast catalytic rates on the order of ∼107 M−1s−1 (70). Based on these qualities, competitive kinetic analyses have predicted that under normal cellular conditions, eukaryotic Prxs will be responsible for the reduction of ∼90% of mitochondrial H2O2 (16), and, in terms of initial reactivity, almost 100% of cytoplasmic H2O2 (84). These striking numbers make clear that Prxs are the dominant player in the protection of cells from oxidative stress. However, despite these numbers, Prxs were not recognized as a broadly important peroxidase family until the 1990s (9). A major reason for this is that early assays used high H2O2 concentrations that inactivated the abundant eukaryotic Prxs; in the case of human PrxII, the half-life for inactivation in the presence of 1 mM H2O2 and reductant is just 20 s (90). It was the recognition of the structural explanation for this sensitivity that led to the proposal that, in addition to their protective role, some Prxs are uniquely involved in regulating non–stress-related redox signaling pathways (87). Since that report, it has become well accepted that H2O2 is a second messenger produced by cellular NADPH oxidases and is involved in the signaling pathways for a wide variety of growth factors and cytokines [reviewed in (28)].
Prxs appear to have a common ancestor with a variety of other redox proteins that are all described as having a thioredoxin (Trx) fold (14, 45). Thus, the cousins of Prxs include Trxs, glutaredoxins (Grxs), cytochrome maturation proteins, glutathione-S-transferases (GSTs), protein disulfide bond isomerases, and glutathione peroxidases (Gpxs). Interestingly, single mutations to Escherichia coli AhpC confer on it the ability to act as a Grx-like deglutathionylating disulfide reductase (89). Among these Trx-related superfamily members, all known Prxs include four very highly conserved residues, one of which is the active site Cys, or peroxidatic Cys (CP). The CP is equivalent to the second Cys in the CXXC motif of Trx and is the residue that reacts directly with the peroxide substrate in Prxs (26). Based on sequence, the Prx family separates into six distinct subfamilies. Five are large and easily recognized (see Fig. 2 of ref. 45), and we refer to them here as Prx1, Prx6, Prx5, Tpx (thiol peroxidase), and BCP (bacterioferritin comigratory protein). The sixth is a small group represented by the protein Mycobacterium tuberculosis AhpE, which is not so easily classified, given the few representatives (25, 42).
FIG. 2.
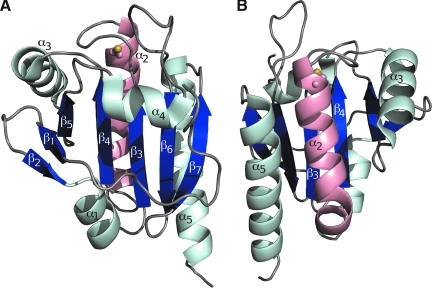
The common-core secondary structural elements of Prxs. (A) A representative FF Prx showing the α-helices (pale cyan and pink) and β-strands (dark blue) common to all known Prxs. The CP (ball-and-stick with sulfur atom colored yellow) is located in the first turn of α2 (pink). The structure shown is a monomeric Prx from the BCP subfamily (entry 63 in Table 1). (B) Helix α2 lies in a cradle with the base formed by β-strands β3 and β4 and the sides formed by the flanking helices α3 and α5. Compared with (A), the view shown is from the backside (i.e., ∼180 degrees rotated around the y-axis). Coloring as in (A); figure prepared using Pymol (20). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
As is often the case for fields that develop with time, one aspect of the Prx field that is still a cause for confusion is the nomenclature. Because many names and naming conventions were developed before much was known about structures and sequence features, the naming schemes are sometimes quite misleading. For instance, within what is now called the Prx1 subfamily, protein names include Prx1, Prx2, Prx3, Prx4, TXNPx, TryP, AhpC, and 2Cys. As another example, the term “thiol peroxidase” not only is used for Prxs in what is now called the Tpx subfamily, but also is sometimes used to describe the entire Prx family, is the given name for specific enzymes in the Prx1, Prx5, and BCP subfamilies, and is also used for nonselenium Gpxs (18). Additionally, the mechanistic division of Prxs into “1-Cys,” “typical 2-Cys,” and “atypical 2-Cys” (88) contributes to confusion because all three types of Prxs are found in more than one subfamily, suggesting many independent evolutionary origins of these features. The “typical” and “atypical” 2-Cys nomenclature is a historical remnant, with “typical” referring to the Prxs with the resolving Cys residue (CR) in the C-terminal helix (which were discovered first), and “atypical” referring to all other 2-Cys Prxs that have any other position for the CR. As this nomenclature is not based on sequence similarity, but rather is a general mechanistic scheme that is shared across different subfamilies, we will not use it here and will instead differentiate 2-Cys Prxs simply by the location of their CR. In the following sections, we provide an overview of the Prx catalytic cycle, present a survey of the breadth of structures determined for Prxs, outline the structural features common to Prxs, and then discuss the structure–function features unique to the individual Prx subfamilies.
Universal Features of the Prx Catalytic Cycle
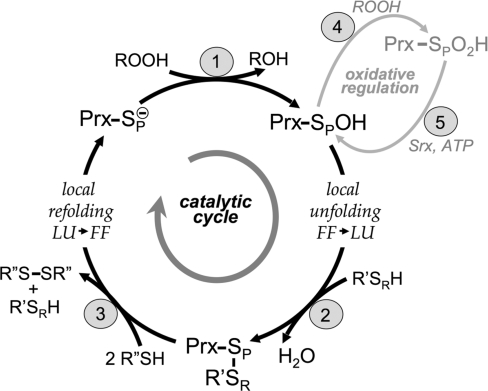
All Prxs have in common a catalytic cycle that uses a conserved active-site Cys residue, called the peroxidatic Cys (CP), to reduce peroxide substrates directly. Catalysis involves the three main chemical steps of (1) peroxidation, (2) resolution, and (3) recycling, with steps 2 and 3 requiring local conformational changes (Fig. 1). Throughout this review, the peroxidatic Cys will be designated as CP, with SP referring to the sulfur atom of the side chain. Similarly, the resolving thiol, which forms a disulfide with the CP (described later), is indicated as SR when referring to the sulfur atom and as CR when referring to the residue in the case that the thiol is provided by a Cys residue (as is often true).
FIG. 1.
The Prx catalytic cycle. Peroxide reduction by Prxs involves three main chemical steps of (1) peroxidation, (2) resolution, and (3) recycling. Two distinct protein conformations are involved in the cycle: FF (fully folded, active-site intact) and LU (locally unfolded, disulfide between the CP and the CR). The local unfolding event is required for disulfide bond formation in step 2, as is the local refolding event to reform the peroxide-binding active site after the disulfide is reduced in step 3. Oxidative regulation (gray, steps 4 and 5) is seen in sensitive, eukaryotic floodgate-type 2-Cys Prxs. Inactivation of the Prx by overoxidation of the CP (step 4) is peroxide dependent. The inactivated form can be rescued through an ATP-dependent reaction catalyzed by sulfiredoxin (Srx) (step 5). The generic Prx is represented as a monomer, with SP designating the sulfur atom of the CP. The CR (from R′ with SR designating the sulfur atom) can be supplied by a different protein (1-Cys mechanism) or by a second Cys within the same Prx, either on the same chain or on the other subunit of a B-type dimer (2-Cys mechanism). Different proteins, including Trx and AhpF, have been identified as R″ in step 3.
The catalytic cycle begins with the peroxide substrate binding in the fully folded (FF) active site; in this conformation, the enzyme has a fully formed, peroxide-binding active site, and the CP thiolate is activated and ready to react with substrate (see later). Peroxidation (step 1) involves a nucleophilic attack of the CP thiolate on the peroxide substrate to release the corresponding alcohol (or water), whereas the CP itself becomes oxidized to sulfenic acid (SPOH). Although substrate preferences vary in different Prxs, they have been found to react with H2O2, alkyl hydroperoxides and peroxynitrite (32, 45, 79, 88). Resolution (step 2) occurs when the resolving thiol (SRH), present either on the Prx itself (2-Cys mechanism) or on another protein or small molecule (1-Cys mechanism; see later), attacks the SPOH to release a water molecule and form a disulfide (Prx-SP –SR-R′). For this attack to occur, the SPOH moiety must move out of the protected, FF active-site pocket through a conformational change involving, at a minimum, the local unfolding of the active site to give a locally unfolded (LU) conformation (local unfolding FF → LU in Fig. 1). It is expected that the FF and LU conformations are in dynamic equilibrium until the formation of the disulfide in step 2 locks the protein in an LU conformation and prevents the FF conformation from reforming. Recycling (step 3) occurs when the disulfide is reduced by another protein or small-molecule thiol, regenerating the free thiols SPH and SRH. For many Prxs, this step is known to involve a thioredoxin (Trx) or a specialized Trx-like protein or domain such as the N-terminal domain of the bacterial enzyme AhpF (69, 88). Once the disulfide is reduced, the FF active site refolds, and in doing so, the Prx is prepared for another round of catalysis (local refolding LU → FF in Fig. 1).
For all Prxs, disulfide bond formation in step 2 of the “normal” (productive peroxide breakdown) catalytic cycle is in competition with additional reactions with peroxide that result in further oxidation of the CP. Because the FF conformation has an intact peroxide binding site, the SPOH group can rotate so that a lone electron pair of the SP atom is in position to attack peroxide (72). Thus, in this side reaction (gray in Fig. 1), additional substrate molecules can react with the SPOH in the FF conformation (step 4) to form first sulfinic (SPO2H) and, in some cases, sulfonic (SPO3H) acid. The terminal oxidation state varies for different Prxs and is thought to be dependent on the geometry of the active site (72). These overoxidized forms are unable to react with the SR to form a disulfide and be readily returned to the reduced state, and thus represent inactive forms of the enzyme. Interestingly, for some eukaryotic Prxs, evolution appears to have selected for structural features that favor SPO2H acid formation (87). For these “floodgate”-type Prxs, the singly overoxidized (SPO2H) Prxs can be reduced and reactivated in an ATP-dependent reaction with sulfiredoxin (Srx, step 5) and perhaps also by sestrin (39). This oxidative regulation pathway is thought to be physiologically relevant in peroxide signaling events (18, 28), functioning as a way to turn off temporarily the peroxidase activity of the Prxs and allow the peroxide to build up locally for signaling (see later).
Summary of Structural Investigations
Since our 2007 review (42), the field has seen an exciting doubling of the number of known Prx structures, bringing the total to 71 deposited Prx structures as of the February 2, 2010, release of the Protein Data Bank (Table 1). A recent, high-resolution (1.45 Å) structure of human PrxV determined by our group [HsPrxV, entry 39, (29)] is also included in this analysis. The 72 available structures represent 35 distinct Prxs and include examples from each of the six subfamilies: 22 Prx1s, 15 Prx6s, 12 Prx5s, 12 Tpxs, eight BCPs, and two AhpEs. All possible redox states for the CP have been observed (SPH, SPOH, SPO2H, SPO3H, SP–SR), and FF and LU structures of the same Prx have been determined for at least one member of the Prx1, Prx5, Tpx, and BCP subfamilies. No LU conformation has been determined for the Prx6 or AhpE subfamilies. In addition to providing many views of the FF and LU conformations, two structures of human Prx1 (HsPrxI, entries 2 and 3 in Table 1) have been solved as complexes with Srx, and 18 structures have either a substrate or substrate analogue bound in the active-site pocket (entries 24–26, 29, 35–43, 46, 56, 66, 68, and 70 in Table 1). All of the structures have been determined with x-ray crystallography except for the FF and LU forms of Bacillus subtilis Tpx (BsTpx, entries 59 and 60 in Table 1), which were solved with NMR (50). Thirteen of the structures have been determined by structural genomics groups, and although three of these have been mentioned in a publication, none of them has been the primary focus of an original publication. In several structures, chains forming a dimer are in different redox states (entries 41, 43, 44, 64, and 70 in Table 1). It is unknown whether the asymmetry in the crystal reflects asymmetry in the solution chemistry, although recent computational studies have identified potential asymmetry in one Prx (91). One structure of particular note is of Chromobacterium violaceum Tpx (CvTpx, entry 62 in Table 1). The highest-scoring BLAST hits are members of the Tpx subfamily (compared with E. coli Tpx, CvTpx has 23% sequence identity and a Cα RMSD of 1.7 Å over 166 residues); however, the published sequence of this protein does not contain any Cys residues, and without a CP, the protein cannot be active as a peroxidase that uses the Prx mechanism. Other notable sequence differences suggest that this protein may be a unique homologue that has lost its peroxidase function and does something else, but more work must be done before definite conclusions can be made.
Table 1.
Deposited Structures of Prxs
| ID | Structurea | Oligomericbstate | Redoxcstate | Formd | Mutation | Bounde | Res.f(Å) | PDB code | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Prx1 | |||||||||
| 1 | HsPrxII | (α2)5 | SO2H(51;172’) | FF | - | - | 1.7 | 1QMV | (74) |
| 2 | HsPrxI | α2/Bg | SS(52-Srx;173’) | LUaltd | C71S/83E/173S | - | 2.6 | 2RII | (38) |
| 3 | HsPrxI | α2/Bg | SO2Hh(52;173’) | LUaltd | Multiplei | - | 2.1 | 3HY2 | (37) |
| 4 | RnPrxI | (α2)5 | SHj(52;173’) | FF | C52S | - | 2.9 | 2Z9S | (52) |
| 5 | RnPrxI | α2/Bg | SS(52-173’) | LUaltd | C83S | - | 2.6 | 1QQ2 | (31) |
| 6 | HsPrxIV | (α2)5 | SH(124;245’) | FF | - | - | 1.8 | 2PN8 | - |
| 7 | BtPrxIII | (α2)6k | SH(47;168’) | FF | C168S | - | 3.3 | 1ZYE | (7) |
| 8 | TcTXNPx | (α2)5 | SH(52;173’) | FF | - | - | 2.8 | 1UUL | (68) |
| 9 | CfTryP | (α2)5 | SH(52;173’) | LUaltd | - | - | 3.2 | 1E2Y | (1) |
| 10 | HpAhpC | (α2)5 | SS(49-169’) | LUC-term′ | - | - | 3.0 | 1ZOF | (64) |
| 11 | PyPrxI | (α2)4 | SH(44;164’) | LUC-term′ | - | - | 2.3 | 2H01 | (80) |
| 12 | Pv2Cys | (α2)5 | SS(50-170’) | LUC-term′ | - | - | 2.5 | 2H66 | (80) |
| 13 | Pv2Cys | (α2)5 | SH(50;170’) | FF | - | - | 2.5 | 2I81 | - |
| 14 | PfTrx-Px2 | α2/Bg | SS(67-187’) | LUaltd | - | - | 1.8 | 2C0D | (6) |
| 15 | MtAhpC | (α2)6 | SS(61-174’) | LUaltd | C176S | - | 2.4 | 2BMX | (27) |
| 16 | AxAhpC | (α2)5 | SS(47-166’) | LUC-term′ | - | - | 2.9 | 1WE0 | (44) |
| 17 | StAhpC | (α2)5 | SHj(46;165’) | FF | C46S | - | 2.2 | 1N8J | (87) |
| 18 | StAhpC | (α2)5 | SS(46-165’) | LUC-term′ | - | - | 2.5 | 1YEP | (86) |
| 19 | StAhpC | (α2)5 | SS(46-165’) | LUC-term′ | T77D | - | 2.3 | 1YEX | (65) |
| 20 | StAhpC | (α2)5 | SS(46-165’) | LUC-term′ | T77I | - | 2.5 | 1YF0 | (65) |
| 21 | StAhpC | (α2)5 | SS(46-165’) | LUC-term′ | T77V | - | 2.6 | 1YF1 | (65) |
| 22 | StAhpC | (α2)5 | SRl(46-AAn;165’) | LUC-term′ | C165S | - | 4.0 | 3EMP | (30) |
| Prx6 | |||||||||
| 23 | HsPrxVI | α2/B | SOH(47) | FF | C91S | - | 2.0 | 1PRX | (10) |
| 24 | AmPRDX6 | α2/B | SHj(45;183’) | FF | C45S | BEZ | 1.6 | 2V2G | (75) |
| 25 | AmPRDX6 | α2/B | SHj(45;183’) | FF | C45S | BEZ | 2.0 | 2V32 | (75) |
| 26 | AmPRDX6 | α2/B | SHj(45;183’) | FF | C45S | BEZ | 2.4 | 2V41 | (75) |
| 27 | Py1Cys | α2/B | SH(47) | FF | - | - | 2.3 | 1XCC | (80) |
| 28 | ApTpx | (α2)5 | SO3H(50;213’) | FF | - | - | 2.3 | 2CV4 | (53) |
| 29 | ApTpx | (α2)5 | SH(50;213’) | FF | C207S | EDOm | 2.0 | 1X0R | (57) |
| 30 | ApTpx | (α2)5 | SH(50;213’) | FFn | C207S | - | 2.4 | 2E2G | (56) |
| 31 | ApTpx | (α2)5 | SO2H(50;213’) | FF | C207S | - | 2.6 | 2E2M | (56) |
| 32 | ApTpx | (α2)5 | SO3H(50;213’) | FF | C207S | - | 2.4 | 2NVL | (56) |
| 33 | ApTpx | (α2)5 | SOH-No(50;213’) | FF | C207S | - | 1.8 | 2ZCT | (56) |
| 34 | ApTpx | (α2)5 | SH(50;213’) | FF | - | - | 2.2 | 3A5W | (55) |
| 35 | ApTpx | (α2)5 | SHj(50;213’) | FF | C50S | ACT | 1.9 | 3A2X | (55) |
| 36 | ApTpx | (α2)5 | SHj(50;213’) | FF | C50S | PERp | 2.3 | 3A2W | (55) |
| 37 | ApTpx | (α2)5 | SH(50;213’) | FF | C207S | PER | 1.7 | 3A2V | (55) |
| Prx5 | |||||||||
| 38 | HsPrxV | α2/Aq | SH(47;151) | FF | - | BEZ | 1.5 | 1HD2 | (19) |
| 39 | HsPrxV | α2/A | SH(47;151) | FF | - | D1D | 1.5 | 3MNG | (29) |
| 40 | HsPrxV | α2/A | SH(47;151) | FF | - | BEZ | 2.0 | 1H4O | (19) |
| 41 | HsPrxV | α2/A | SH/SS(47;47-151’) | FF/LUaltd | - | BEZ | 2.0 | 1OC3 | (24) |
| 42 | HsPrxV | α2/A | SHj(47;151) | FF | C47S | BEZ | 1.7 | 1URM | (24) |
| 43 | HsPrxV | α2/A | SH/SS(47;47-151) | FF/LUα5 | - | BEZ | 1.9 | 2VL2 | (76) |
| 44 | HsPrxV | α2/A | SH/SS(47;47-151) | FF/LUα5 | - | - | 1.8 | 2VL3 | (76) |
| 45 | HsPrxV | α2/A | SS(47-151) | LUα5 | C72S | - | 2.7 | 2VL9 | (76) |
| 46 | PtPrxD | α2/A | SH(51) | FF | - | SO4 | 1.6 | 1TP9 | (23) |
| 47 | HiHyPrxV | α2r/A | SH(49) | FF/LUaltd | - | - | 2.8 | 1NM3 | (43) |
| 48 | PsPrxII | α2/A | SH(59) | FF | - | - | 2.8 | 2PWJs | - |
| 49 | PfAOP | α2/A | SO3H(59) | FF | - | - | 1.8 | 1XIY | (72) |
| Tpx | |||||||||
| 50 | EcTpx | α2/A | SHj(61;95) | FF | C61S | - | 1.8 | 3HVV | (30) |
| 51 | EcTpx | α2/A | SS(61-95) | LUα3 | - | - | 2.2 | 1QXH | (11) |
| 52 | EcTpx | α2/A | SS(61-95) | LUα3 | - | - | 1.8 | 3HVS | (30) |
| 53 | EcTpx | α2/A | SS(61-95) | LUα3 | - | - | 2.8 | 3I43 | (30) |
| 54 | EcTpx | α2/A | SS(61-61’;95) | LUaltd | C82/95S | - | 2.1 | 3HVX | (30) |
| 55 | HiTpx | α2/A | SS(59-93) | LUα3 | - | - | 1.9 | 1Q98 | - |
| 56 | MtTpx | α2/A | SHj(60;93) | FF | C60S | ACT | 2.1 | 1Y25 | (77) |
| 57 | MtTpx | α2/A | SSt(60-93) | LUα3 | - | - | 1.8 | 1XVQ | (71) |
| 58 | SpTpx | α2/A | SH(58;92) | FF | - | - | 2.3 | 1PSQ | - |
| 59 | BsTpx | α2u/A | SH(60;94) | FF | - | - | NMR | 2JSZ | (50) |
| 60 | BsTpx | α2u/A | SS(60-94) | LUα3 | - | - | NMR | 2JSY | (50) |
| 61 | AaTpx | α2/A | SH(61;95) | FF | - | - | 1.9 | 2YZH | - |
| 62 | CvTpxv | - | - | FF | - | - | 1.8 | 3KEB | - |
| BCP | |||||||||
| 63 | ScnTPx | α/- | SHj(107;112) | FF | C107/112S/K123E | - | 1.8 | 2A4V | (12) |
| 64 | ApBCP | α2/Aw | SH/SS(49;49-54) | FF/LUα2 | - | - | 2.3 | 2CX4 | - |
| 65 | ApBCP | α2/A | SS(49-54) | LUα2 | - | - | 2.6 | 2CX3 | - |
| 66 | SsBcp1 | α/- | SHj(45;50) | FF | C45/50S | CIT | 2.2 | 3DRN | (17) |
| 67 | StoBcp | α2/A | SS(44;49) | LUaltd | - | - | 1.6 | 2YWN | - |
| 68 | XcBcp | α/- | SHj(48;84) | FF | C48/84S | FMT | 1.5 | 3GKM | (49) |
| 69 | XcBcp | α/- | SS(48-84) | LUα3 | - | - | 1.8 | 3GKK | (49) |
| 70 | XcBcp | α/- | SHj/SS(48;84-84’) | FF/LUaltd | C48A | BIH | 1.5 | 3GKN | (49) |
| AhpE | |||||||||
| 71 | MtAhpE | α2/Ax | SH(45) | FF | - | - | 1.9 | 1XXU | (48) |
| 72 | MtAhpE | α2/Ax | SOH(45) | FF | - | - | 1.9 | 1XVW | (48) |
Structures included in Table 1 are from the February 2, 2010, release of the Protein Data Bank plus an additional DTT-bound HsPrxV determined by our own group (entry 39). Within a subfamily, Prxs are in order of decreasing sequence identity relative to the one that is listed first. Organism abbreviations are as follows: Aa, Aquifex aeolicus; Am, Arenicola marina; Ap, Aeropyrum pernix; Ax, Amphibacillus xylanus; Bs, Bacillus subtilis; Bt, Bos taurus; Cf, Crithidia fasciculata; Cv, Chromobacterium violaceum; Ec, Escherichia coli; Hi, Haemophilus influenzae; Hp, Helicobacter pylori; Hs, Homo sapiens; Mt, Mycobacterium tuberculosis; Pf, Plasmodium falciparum; Ps, Pisum sativum; Pt, Populus trichocarpa; Pv, Plasmodium vivax; Py, Plasmodium yoelii; Rn, Rattus norvegicus; Sc, Saccharomyces cerevisiae; Sp, Streptococcus pneumoniae; Ss, Sulfolobus solfataricus; St, Salmonella typhimurium; Sto, Solfolobus tokodaii; Tc, Trypanosoma cruzi; Xc, Xanthomonas campestris.
All octamers, decamers, and dodecamers are made up of both A- and B-type dimer interfaces. For dimeric structures, the type of dimer interface is indicated (A or B).
The redox state of the CP is given, as well as the residue numbers for the CP and, in the case of 2-Cys Prxs, the CR. CR residues contributed by the second chain of the dimer are indicated with a prime.
The conformation of the active site is indicated as FF for fully folded and LU for locally unfolded, with subscripts indicating where the CR is located (see Fig. 5). Noncanonic LU conformations are labeled with the subscript “alt” for alternate for the following reasons: 2, this is a Prx-Srx complex, with a disulfide formed between the CP and residue 99 of Srx; 3, this is a Prx-Srx complex; 5, the CP-loop has shifted, presumably related to decamer dissociation (86); 9, the 10 chains display different LU conformations, as none is locked in place by a disulfide; 14, the CP-loop has shifted, presumably related to decamer dissociation; 15, α2 has shifted ∼8 degrees; 41, normally an intramolecular disulfide, in this structure, the disulfide is formed between the CP and the CR of separate chains; 47, α2 is perturbed in one chain of the structure, possibly as an intermediate resembling the LU conformation of this 1-Cys Prx5; 54, the disulfide is formed between the CP residues of two chains, linking two A-type dimers together; 67, residues 44–50 are not modeled because of weak electron density; however, the conformation of α2 is most similar to the LU state; and 70, the disulfide is formed between the CR residues of two chains.
Compounds bound in the active site are listed by their three-letter atom code: ACT, acetate; BEZ, benzoate; BIH, naphthalene-2,6-disulfonic acid (DNS); CIT, citrate; D1D, dithiothreitol; EDO, 1,2-ethanediol; FMT, formic acid; GOL, glycerol; PER, hydrogen peroxide; SO4, sulfate.
The resolution (Å) of the crystal structures is rounded to the nearest tenth.
The structure is a B-type dimer in the crystal structure, but the protein is thought to function as a decamer.
A Cys → Asp mutant of the CP mimics the SPO2H form.
Multiple mutations were necessary to capture the complex and include CP52D, C71S, C83E, A86E, C173S, and K185C.
A Cys → Ser or a Cys → Ala mutant of the CP mimics the reduced state.
The concatameric interaction of the dodecamers is thought to be an artifact of crystallization.
The CP is modified with S-acetanilide (AAn).
Ethanediol is bound in one of two general conformations in only five of the 10 chains.
The authors refer to this as a “preoxidation” conformation, defined by the movement of the conserved Arg away from the CP and the movement toward the CP of the His involved in the hypervalent intermediate.
The hypervalent SP forms a covalent bond to a nearby His residue.
Three of the 10 chains have a bound H2O2 molecule, whereas the remaining have a bound glycerol that adopts one of two general conformations.
Originally described as a monomer when published by the authors but later acknowledged as an A-type dimer (24).
The glutaredoxin domains interact to make the protein a dimer of dimers.
Residues 10 through 32 are modeled poorly.
Although the sulfur atom of the CP is not visible in the electron density, the conformations of α2 and α3 match the LU conformation seen in other Tpx subfamily members.
The NMR structure was determined as a monomer; however, we expect that it exists as a dimer.
This protein may not be an active Prx.
The A-type dimer formed by chains A and B is linked by a disulfide, but this does not appear to change the A-type dimer interface.
The authors described the structure as an (α2)4 octamer, but we suspect the octamer is an artifact of high protein concentration.
Structural Features Common to All Prxs
Overall structure
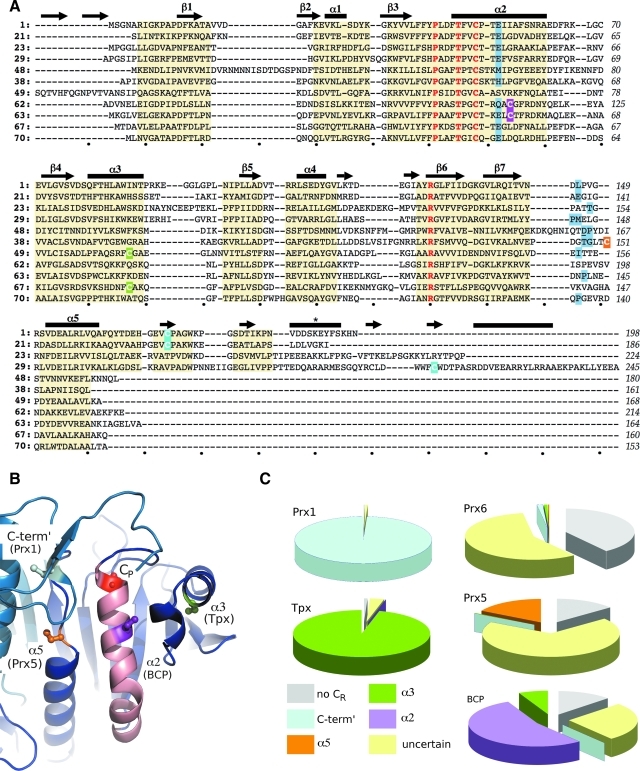
Prxs have a compact, globular protein structure based on a Trx fold (14). The highly spatially conserved, common-core tertiary structure of Prxs contains seven β-strands (β1 through β7) and five α-helices (α1 through α5); a central twisted β-sheet formed by five β-strands (β5-β4-β3-β6-β7) is covered by β1-β2-α1 and α4 on one face, and by α2, α3, and α5 on the opposite face (Fig. 2). Interactions between the β1-β2 hairpin and β5 cause the central sheet to sometimes be referred to as seven-stranded. In approximately half of the known Prx structures, α1 is a 310-helix. In the FF conformation, the conserved CP residue is located in the first turn of α2, and the CP-loop is formed by residues in the loop immediately preceding α2 (Figs. 2 and 3A and B). In all but the Tpx subfamily, a kink in α2 is followed by an additional one or two turns of the helix. Also, although α5 begins in the approximate same position in all structures, it varies in length from two to five turns. As can be seen in a sequence alignment of representative Prxs (Fig. 4A), sequence insertions are generally found at the N- and C-termini and in loops between the conserved secondary structure elements. In some publications, the secondary structure elements have been numbered differently because of the presence of additional elements not conserved across the entire family. For example, in some structures in which α1 is a 310-helix, the helix containing the CP is referred to as α1, and in the Tpx subfamily, an N-terminal insertion of two β-strands shifts the numbering of the remaining strands. The naming scheme in Fig. 2 represents a universal numbering scheme for the entire family that is based on the conserved core elements and can be used consistently to describe features across different Prxs.
FIG. 3.
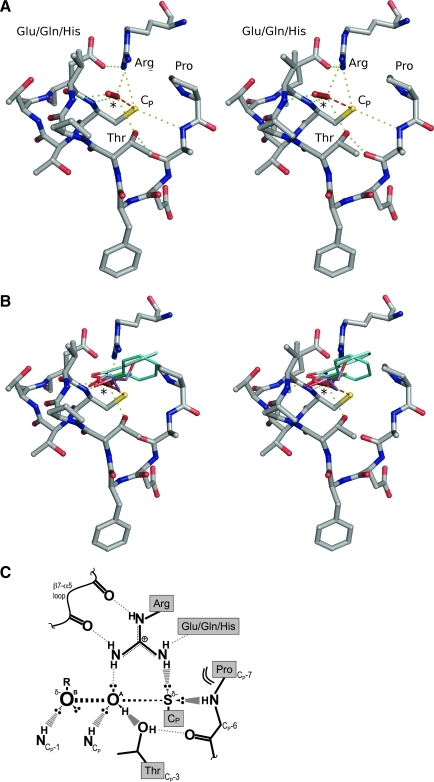
The peroxidatic active site. (A) Stereoview of the FF Prx active site with a bound H2O2 molecule. Shown are the highly conserved contiguous CP-loop and the first turn of α2 plus the active-site Arg and an associated Glu/Gln/His supporting residue. The proximity to the CP and the hydrogen-bonding interactions (green dotted lines) highlight the importance of the conserved Pro, Thr, and Arg in binding and activating the peroxide substrate (*) and in activating the CP sulfhydryl for attack of the substrate oxygen atom (orange dashed line). The Glu/Gln/His residue, although not 100% conserved across all Prxs, is important as a hydrogen-bond acceptor positioning the conserved Arg. This figure was created by using ApTpx (entry 37 in Table 1), colored by atom (C, gray; N, blue; O, red; S, yellow). (B) Stereoview of an overlay of the H2O2-bound Prx from (A) with benzoate (cyan tones, entries 38 and 24 in Table 1), acetate (green tones, entries 35 and 56 in Table 1), ethanediol (light blue, entry 29 in Table 1) and glycerol (violet, entry 36 in Table 1), as seen bound in other Prx structures. Protein atoms are shown only for the Prx bound to H2O2, and protein coloring and hydrogen bonds to H2O2 (*) are as in (A). (C) Cartoon representation of the active-site transition-state conformation. The stabilizing interactions between key atoms from the backbone and the four conserved residues, and with the H2O2 substrate, are indicated. In the transition state, a bond is forming between the S atom of the CP and the OA of H2O2, and a bond is breaking between the OA and OB atoms of H2O2. The geometry of the active site is ideal for stabilizing the larger distance between the OA and OB atoms as the bond is broken. (A, B) were prepared by using Pymol (20). (C) is based on a figure from Hall et al. (29). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
FIG. 4.
Variations in Prx sequences. (A) Structure-based sequence alignment of representative Prxs. Residues that have a common main-chain path among all Prxs are highlighted by a yellow background. Secondary structure elements are indicated above the alignment with the common-core Prx elements labeled as in Fig. 2, and other elements present in only some Prxs are shown but not labeled. The four residues conserved in all Prxs are colored red, and the CR position of each 2-Cys Prx is highlighted by a purple, green, orange, or cyan background for a CR placed in α2, α3, α5, or the C-terminus, respectively (B). *The YF-motif helix associated with some Prxs sensitive to overoxidation. Residues involved in backbone-mediated passing chain stabilization of the conserved Arg are given a blue background; in one case, Asp 163 of PfAOP (underlined residue in line 5 sequence) stabilizes the Arg via its side chain. Structures are referenced by index number from Table 1 and include a sensitive Prx1 (1), a robust Prx1 (21), a 1-Cys Prx6 (23), a 2-Cys Prx6 (29), a 1-Cys Prx5 (49), a 2-Cys Prx5 (38), a Tpx (50), a 1-Cys BCP (63, monomeric), a 2-Cys BCP (64, CR in α2, dimeric), a 2-Cys BCP (68, CR in α3, monomeric), and an AhpE (71). The last residue of each line is numbered and dots below the alignment mark every 10 spaces. (B) The four prototypical locations for the CR [colored as in (A) and labeled by location and the subfamily it is commonly associated with] are mapped onto a composite structure based on StAhpC (entry 21 in Table 1). The conserved CP (red) is also shown. The two chains of the B-type dimer are colored in dark and light blue, and helix α2 is colored pink. (C) Pie charts based on ∼3,500 Prx sequences showing the frequency at which the CR is in a given location for each subfamily. Wedges are colored by CR position consistent with (A) and (B), by using the notation in (B): no CR (gray), C-term′ (cyan), α5 (orange), α3 (green), α2 (purple), and uncertain (pale yellow). The exact positions are defined as follows: C-term′ aligns with residue 172 in HsPrxII (entry 1 in Table 1); α5 aligns with residue 151 (or −2 residues) in HsPrxV (entry 38 in Table 1); α3 aligns with residue 95 in EcTpx (entry 50 in Table 1); and α2 aligns with residue 112 in ScnTPx (entry 63 in Table 1). Sequences marked “uncertain” have additional Cys residues present, but none aligns exactly with one of the known locations. (B) was prepared by using Pymol (20). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
A conserved cradle for α2
Helix α2 contains the universally conserved CP and is thus necessarily involved in the local unfolding event required for catalysis. Looking at how α2 packs in the structure, it can be thought of as a baby in a cradle: the bed of the cradle is formed by β-strands β3 and β4, and the walls by helices α3 and α5 (Fig. 2B). Although different changes occur to α2 during unfolding for each subfamily (see later), in every subfamily, the cradle around α2 is highly important in stabilizing both the FF and LU conformations, as well as in facilitating the switch between the two conformations. As was seen in the Tpx subfamily from which the cradle concept was first derived (30), a subfamily-specific pattern of residue conservation lines the cradle and stabilizes discrete conformations of α2. It is expected that each subfamily will have a distinct conservation pattern around the cradle that, when identified, will assist in understanding the local unfolding transitions of α2.
Peroxide activation by the fully folded peroxidatic active site
The FF conformation is required for productive binding of peroxide substrates. In this highly spatially conserved active-site conformation, the CP is at the bottom of a pocket, surrounded by the three additional conserved residues, Pro, Thr, and Arg (Figs. 3A, 3C, and 4A). The Pro, Thr, and CP are found in a contiguous segment with a universally conserved PXXXTXXCP sequence motif. The conservation of this segment across all six subfamilies with no gaps implies that the catalytic efficiency is exquisitely sensitive to the constellation of these residues. Here we refer to this eight-residue segment as the CP-loop, a term that was coined to denote the region that undergoes conformational change during catalysis in the Prx1 subfamily (86). It is of interest that, from an evolutionary perspective, the Thr in the CP-loop (which is substituted as a Ser in ∼3% of sequences) replaces the first Cys in the CXXC motif of a Trx-like ancestral protein (26), implying that this position is important in both the Prx and Trx chemistry, but with a changed role (14).
Among all FF structures, little variation is found in the CP-loop conformation itself, but the conserved Arg side chain that is contributed to the active site from strand β6 has greater variation. In the large majority of structures containing an SPH or a CP → Ser mutation, the Arg adopts what we represent here as the canonic, catalytically productive conformation (29). In FF structures with SPOH, SPO2H, or SPO3H in the active site, the Arg may be present in different conformations (e.g., Table 1, entries 32 and 72). The roles of these conserved residues (in addition to the CP) had not been well defined until recently (29); the previous consensus has been that the Arg lowers the pKa of the CP and stabilizes the CP thiolate, that the Thr may also contribute to a lowered CP-pKa value and play a role as a proton shuttle, and that the Pro shields the CP from water and positions the subsequent peptide nitrogen to donate a hydrogen bond to the CP (42, 88).
At the time of our 2007 review (42), no substrate-bound complexes were known. We proposed, however, that the benzoate bound in the active site of human Prx5 (HsPrxV, entry 38 in Table 1) mimics peroxide binding, and we used it to model how H2O2 would bind in the active-site pocket. Now, the first peroxide-bound Prx structure has been determined (Fig. 3A, entry 37 in Table 1), and it confirms that the benzoate carboxylate does indeed accurately mimic peroxide binding (Fig. 3B). Furthermore, in the updated list of Prx structures, additional structures (entries 24–26, 29, 35, 36, 38–43, and 56 in Table 1) have bound acetate, benzoate, dithiothreitol, ethanediol, or glycerol molecules with oxygen atoms mimicking those of H2O2. Some other Prx structures in Table 1 have sulfate, citrate, or formate, or an alternate conformation of ethanediol or glycerol bound in the active site, with one of the ligand oxygen atoms placed in the position occupied by one of the oxygen atoms of H2O2 (entries 46, 66, 68, 29, and 36, respectively, in Table 1).
Through exploration of the details of hydrogen-bonding interactions in the active site of the Michaelis complex and the various ligand-bound Prx structures, insight into the catalytic power of the Prxs has recently been obtained (29). In terms of the protein atoms, hydrogen bonding, involving both the protein backbone and side-chain atoms, positions the key residues (i.e., the conserved Pro, Thr, Arg, and CP) and activates and stabilizes the CP-thiolate for peroxidation (Fig. 3C). For discussion of the H2O2 molecule, we designated the two peroxide oxygen atoms as “OA” and “OB,” with OA being the atom attacked by the CP, and OB being the oxygen of the hydroxide (or alkoxide) leaving group (Fig. 3C). In terms of the chemistry involved, this redox reaction is actually a simple in-line SN2 nucleophilic displacement reaction with the CP thiolate as the nucleophile, OA as the electrophilic center, and OB as the group to be displaced. In the H2O2-bound structure, the H2O2 molecule is well positioned in the active site for the peroxidatic in-line SN2 reaction. The OA atom is positioned 3.4 Å away from the SP atom, with an SP▪▪▪OA–OB angle of 172 degrees, and is stabilized by four hydrogen bonds.
This analysis provided the first clearly presented proposal for the roles of conserved residues in catalysis and a compelling structure-based explanation for the catalytic power of Prxs (29). For catalysis, of most interest is not the interactions in the Michaelis complex itself, but the transition state, because in classic enzyme catalysis, the active site should be optimally complementary to the transition state of the reaction. As is illustrated in Fig. 3, we see how indeed the Prx active site is exquisitely organized to stabilize the transition state, which will have a partial bond formed between the SP and OA, and a partial bond broken between OA and OB. From the geometry of the hydrogen bonds to the H2O2 molecule seen in the ground state (Fig. 3A), it is clear that each of these hydrogen bonds will align more favorably as atoms move to the positions they will adopt in the transition state (Fig. 3C).
A major take-home point from this analysis is how the active site is not simply activating the CP residue to be a potent nucleophile; but an equally (if not more) important contribution to catalysis is the activation of the peroxide itself to be attacked. Indeed, the conserved Arg and Thr residues and two backbone amide hydrogens are specifically interacting with the peroxide. With this in mind, the roles of the conserved residues can be identified: the Pro shields the activated CP-thiolate from unwanted reactions and positions the following two residues for hydrogen bonding. The Thr positions and activates the OA atom of the peroxide substrate; the Arg positions and activates both the CP-thiolate and the OA atom of the peroxide substrate. The strong role of peroxide activation in catalysis helps explain how, in many cases, Prxs can undergo facile overoxidation reactions, even though the SPOH and SPO2H sulfur atoms must be much weaker nucleophiles than the original thiolate.
Aside from the catalytic chemistry, another requirement of the active site of some Prxs is the recognition of the alkyl moiety of organic peroxide substrates. From crystal structures, some insight comes from the binding of benzoate, acetate, ethanediol, glycerol, and citrate molecules. All of these structures show the alkyl moiety pointing up and away from the conserved Thr, suggesting that the carbon atom directly bound to OB would be directed toward the opening of the active-site pocket, whereas the remaining lone electron pair is aligned into the pocket toward the conserved Thr (Fig. 3C). Additionally, from the binding of a DNS (naphthalene-2,6-disulfonic acid) molecule in XcBcp (entry 70 in Table 1) close to the active-site pocket, a longer alkyl chain can be predicted to bind in a conformation that extends from the OB atom toward a cleft (49) that is partially formed by variable subfamily-specific features in the loop between α4 and β6.
pKa analyses
Prxs have catalytic rates with peroxide substrates on the order of 107 M−1s−1 at neutral pH (59, 65, 67). Stabilization of the CP as a thiolate through lowering of its pKa from a typical value of 8.4 or greater is an important element of its reactivity. Hydrogen-bonding interactions seen in the active site are consistent with the stabilization of the negatively charged thiolate of the CP (Fig. 3A and C; see later).
The pKa values have been measured for only a few Prx proteins, but all exhibit or suggest values below 7. Because Prxs undergo local unfolding to yield a more accessible CP, and some approaches rely on the variation of Cys alkylation rates with pH, standard pKa measurements are frequently complicated by the need to ensure that the pKa value measured is for the FF, active conformation, and not the LU form of the protein (59). pKa values have been determined by using functional assays across a range of pH values to measure competition with horseradish peroxidase (HRP) for Salmonella typhimurium AhpC (pKa = 5.9) (59) and Saccharomyces cerevisiae Tsa1 (pKa = 5.4) and Tsa2 (pKa = 6.0) (62). The pKa of human Prx2 has also been estimated to be between 5 and 6, based on its tendency to be oxidized by H2O2 across a range of pH values (67). The pKa for human Prx3 was suggested to be lower than 5, given the lack of a decrease in activity for this protein at low pH by using HRP/catalase competition and gel-shift assay (15). pKa values lower than 6 were also estimated for M. tuberculosis AhpE (a 1-Cys Prx) (34) and human PrxV, based on H2O2-dependent fluorescent changes (79).
Although some proteins containing redox-reactive Cys residues exhibit even lower pKa values [e.g., E. coli glutaredoxin-3 (pKa < 5.5) (61), protein tyrosine phosphatases (pKa < 5) (21), and DsbA (pKa = 3.5) (58)], a pKa of 6 is sufficiently low for 91% of the CP to be deprotonated at pH 7. Once the thiolate is formed, its nucleophilicity actually decreases as its pKa is lowered (82, 83), indicating that a very low pKa would be expected to decrease Prx activity. It should also be noted that the lowered pKa of the CP to yield predominantly thiolate at the active site can account for rates of only roughly 20 M−1s−1, based on studies with small-model thiol-containing compounds, leaving another ∼105 to 106 rate enhancement imparted by other features of the Prx active site (84). This underscores the importance of what we described earlier as the exquisitely oriented binding and activation of peroxide in the Prx active site.
Local unfolding of the peroxidatic active site
Structures of the LU conformation have been determined for four of the six Prx subfamilies; no examples exist for members of the Prx6 and AhpE subfamilies. Distinct from the FF conformation (see earlier) that is highly consistent across all Prxs, the LU conformation has a disulfide formed between the CP and the CR (Figs. 5 and 6), and so its details depend on the position of the CR (see next section). Nevertheless, all LU structures have in common a structural change in α2 and in the CP-loop to move the CP out of the protected, peroxide-binding active-site pocket and into an exposed position that is close enough to the CR to form a bond. For Prxs that function with a 2-Cys mechanism, structural changes are also observed in the region where the CR is located. Whereas changes occur in regions around the CP and the CR, the rest of the protein structure remains remarkably unchanged. As the details of local unfolding are unique, depending on the position of the CR, they are individually discussed later (see next section).
FIG. 5.
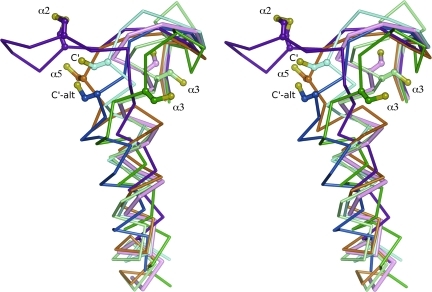
Local unfolding changes the conformation of α2 and the CP-loop. Comparison of the canonic FF structure (pink, entry 1 in Table 1) with LU representatives from each subfamily shows that the LU conformations of the CP-loop and helix α2 vary by subfamily. Shown in stereo and viewed as in Fig. 2B are the LU conformations of a Prx1 (light blue, entry 17 and dark blue, entry 15 in Table 1), a Prx5 (orange, entry 44 in Table 1), a Tpx (dark green, entry 53 in Table 1), an α2-BCP (purple, entry 64 in Table 1), and an α3-BCP (pale green, entry 69 in Table 1). Labels indicate the location of the CR (α2, α3, and α5, as in Fig. 4; C′ and C′-alt for the CR near the C-terminus as in Fig. 4 with “alt” for the ∼8-degree shift from the canonic conformation, as described). No LU example is given from the Prx6 or AhpE subfamilies. The CP residue in each structure is shown as ball-and-stick, with the sulfur atom colored yellow. The LU structures are all disulfide-bonded forms, even though the CR is shown only for the α2-BCP example. Figure was prepared by using Pymol (20). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
FIG. 6.
Conformational changes for disulfide formation are localized to the positions surrounding the CP and the CR. Stereoview of the interpolated structural changes colored by rainbow between the FF (blue) and LU (red) conformations for a representative from each major subfamily: (A) Prx1 (StAhpC, entries 17 and 18 in Table 1); (B) Prx5 (HsPrx5, entries 38 and 44 in Table 1); (C) Tpx (EcTpx, entries 50 and 52 in Table 1); (D) α2-BCP (ApBCP, entry 64 in Table 1); and (E) α3-BCP (entries 68 and 69 in Table 1). The interpolations show how most of the protein structure does not change during the local unfolding transition. In (A), the C-terminus is truncated at residue 165 because of disorder in the rest of the chain, although in the FF conformation, residues through 186 are ordered. In (E), residues 78–80 are omitted, as they are disordered in the LU conformation. The CP and the CR are shown as sticks with sulfur atoms colored yellow, and the calculated intermediate structures are partially transparent. Interpolations are calculated by using the Yale morph server (47) and visualized by using Pymol (20). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Features Varying Between Prx Subfamilies
Quaternary structure
The four key Prx catalytic residues come from a single chain, so in theory, Prxs could be monomeric. However, Prxs that are naturally present and active as monomers have been observed only in the BCP subfamily. Although Tpxs were reported to be functional monomers (11), it is now clear that they function as dimers (3, 30). All other Prxs are known to form dimers, and in some cases, higher-order octameric, decameric, and dodecameric oligomer structures involving only two types of dimer interfaces.
The two distinct dimer interfaces that account for all of the oligomeric associations seen in Prxs are referred to as the A- and B-type dimer interfaces (72) (Fig. 7). B-type dimers (“B” for β-strand) are formed by interactions at the β-sheets in a head-to-tail fashion to form an extended 10-stranded β-sheet (Fig. 7B). All Prxs with B-type interfaces have in common a C-terminal extension that reaches across the twofold axis to make extensive interactions that help stabilize the B-type dimer; in the Prx1 subfamily, the CR is located across the B-type interface from its partner CP, and B-type dimers have not been observed to dissociate. A-type dimers (“A” for alternate) are formed by a tip-to-tip association of equivalent parts of the two chains involving β1, β2, and the loops preceding α2, α3, and α4 (Fig. 7A). Because this is seen in nearly all Prxs and is thought to be linked with catalytic activity (see later), it is thought to be the more ancestral dimerization surface; thus “A” could stand for either “alternate” or “ancestral” (72).
FIG. 7.
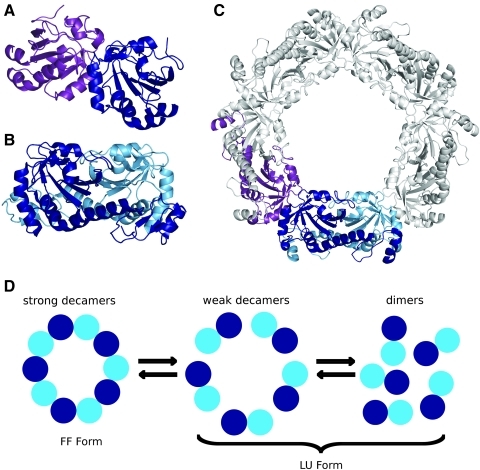
Quaternary structures of Prxs. For some Prxs, the basic monomeric structure shown in Fig. 2 can form (A) A-type dimers, interacting near α3, or (B) B-type dimers, interacting at the β-sheet to form an extended 10-stranded β-sheet. (C) Some members of the Prx1 and Prx6 subfamilies form decameric structures through the interaction of five B-type dimers via the A-type dimer interface. Subunit coloring for the A-type dimer (purple and dark blue) and the B-type dimer (dark blue and light blue) are used in the decamer to show how it is composed of these two types of interactions. (D) The oligomerization of the decamers is redox dependent. In the Prx1 and Prx6 subfamilies, reduced and overoxidized Prxs form decamers, with the A-type dimer interface stabilizing the FF active site. The structural change with disulfide formation destabilizes the A-type dimer interface, and the decamer falls apart to B-type dimers. Octamers and dodecamers have also been observed (see Table 1) and are thought to be functionally equivalent to the decamer. (A–C) were prepared by using Pymol (20). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
For most Prxs that form B-type dimers (Prx1 and Prx6 subfamily members), higher-order oligomers are formed with four, five, or six B-type dimers associating through the A-type dimer interface to form a toroid-like doughnut structure (Fig. 7C). For these enzymes, evidence suggests that during the catalytic cycle, dissociation occurs at the A-type dimer interface, causing a redox state–linked transition between doughnuts and dimers (86) (see later). The tighter ring structure of the octamer and the expanded ring structure of the dodecamer are due to small shifts at the B-type dimer interface. We expect that octameric, decameric, and dodecameric Prxs will function equivalently, and so for simplicity, any properties of decameric Prxs discussed in the remainder of this review are expected to refer to octamers and dodecamers as well. The distribution of dimerization types and characteristics of oligomerization specific to the Prx subfamilies are discussed in each subfamily section. In brief, known members of the Prx1 and Prx6 subfamilies form B-type dimers, with most oligomerizing to decamers through the A-type interface, known members of the Prx5, Tpx, and AhpE subfamilies only form A-type dimers, and BCP subfamily members exist as either monomers or as A-type dimers.
To complete this categorization of higher-order structures, four Prxs are listed in Table 1 that require additional explanation. First is the structure of Bos taurus PrxIII (entry 7 in Table 1), which in the crystal was seen to be a concatenated pair of dodecamers (7). The authors state that in solution, single dodecamers also exist (as were seen for M. tuberculosis AhpC; entry 15 in Table 1), and that they are “unsure if [the concatenated] assembly has any physiological relevance.” The second structure is the hybrid Grx-Prx from Haemophilus influenzae (entry 47 in Table 1), which is a tetramer in the crystal; it is made up of two dimeric Prxs that form a tetramer through dimerization interactions of the Grx domains of the protein (43); thus as far as Prx is concerned, it is a dimer. Third is B. subtilus Tpx (entries 59 and 60 in Table 1), whose structure solved by NMR was determined as a monomer, and no comment or measurement of the true oligomeric state is made (50). Because residues at the core of the Tpx dimer interface are conserved in this protein sequence, we expect that, like all other members of the Tpx subfamily, the protein is dimeric, and that the dimer structure went unnoticed in the NMR experiments. Last is M. tuberculosis AhpE (entries 71 and 72 in Table 1), which was reported to be an octamer (48), but two reasons lead us to suspect that the octamer is an artifact of crystallization rather than a physiologically relevant state. Most important, gel filtration at high concentration showed the majority of the protein was a dimer, with only a little octamer present; less conclusive but still of interest, the interface building the octamers was not very extensive and did not involve the other known (B-type) interface.
Location and Conservation of the CR
As shown in Fig. 1, the catalytic mechanism of all Prxs requires a second thiol (i.e., SR) for resolution of the SPOH. For some Prxs, referred to as “1-Cys” Prxs, a small molecule or a second protein contributes the SR. For all other Prxs, referred to as “2-Cys” Prxs, the SR is contributed by a second Cys residue (i.e., CR), that comes from within the Prx. Based on the prototypical Prx studied for each subfamily, a strong association of certain positions for the CR with each subfamily has arisen. In 1998, the first Prx structure was published (10), a 1-Cys Prx6 subfamily member (HsPrxVI) that defined the prototype for the subfamily. A year later, the first Prx1 structure published (entry 5 in Table 1) had the CR located near the C-terminus, with an intersubunit disulfide between the CP and the CR of the two chains of a B-type dimer. Then, in 2003, the first Tpx structure (entry 51 in Table 1) showed the formation of an intrasubunit disulfide, with the CR in α3 of the same chain. The prototypical Prx5 structure was HsPrxV (entry 41 in Table 1), and it revealed a CR associated with α5. Finally, the prototypical BCP structure (entry 63 in Table 1) had the CR in α2 itself, just five residues beyond the CP. These four positions for the CR (C-term′, α2, α3 and α5), and their prototypic association with a given subfamily is shown in Fig. 4B.
Interestingly, as is often the case, the division of the Prx family by sequence similarity into the Prx1, Prx6, Prx5, Tpx, BCP, and AhpE subfamilies (45) does not coincide with the divisions based on the existence or the positions of the CR. Specifically, from structurally known Prxs, it has already been seen that Prx6, Prx5, BCP, and AhpE subfamilies include both 1-Cys and 2-Cys members, and even for those that are 2-Cys Prxs, the structure of XcBcp (entry 69 in Table 1) that has its CR in α3, shows that the position of the CR can vary even within a subfamily. To shed further light on this, we draw here on results from a survey of more than 3,500 Prx sequences (58a). That survey confirms the appropriateness of splitting the Prx family into six subfamilies, with the AhpE subfamily containing only ∼25 members, and the other Prx subfamilies each containing between 300 and 1,100 members. As shown in Fig. 4C, this work (58a) provides a broader perspective on the variation of the CR within each subfamily. In the analysis, Prx sequences with no Cys residues besides the CP were identified as 1-Cys (“no CR” in Fig. 4C). Other Prxs were labeled as “uncertain” with respect to their CR if they contained additional Cys residues, but none matched one of the known prototypic positions. It is still possible that additional CR locations will be determined as more Prxs are characterized, but we expect that most of the enzymes identified as “uncertain” are actually 1-Cys Prxs, because many characterized Prxs have sporadically placed Cys residues that are not involved in catalysis [e.g., HsPrxV (19), PfAOP (72), EcTpx (30)].
Assuming that all of the “uncertain” instances are indeed 1-Cys Prxs, the variation can be summarized quite simply: in the Prx1 and Tpx subfamilies, 96% or more of the members resemble the prototypes with the CR in the C-term and α3 locations, respectively; in the Prx6 subfamily, ∼98% are 1-Cys; in the Prx5 subfamily, only ∼17% have the CR in the prototypical α5 location, whereas the rest are 1-Cys; in the BCP subfamily, ∼54% resemble the prototype with the CR in α2, ∼39% are 1-Cys, and ∼7% have the CR in α3 at the location associated with the Tpx prototype (Fig. 4C). This analysis of the CR conservation patterns indicates that the structural diversity seen in the 72 known Prx structures is a good sampling of the diversity existing across the entire family. In the remaining sections, we discuss the specific structural features of each subfamily, including comments on the structural transition each goes through during catalysis.
Before moving on, a couple of cases involving the CR deserve special comment. First is Saccharomyces cerevisiae Ahp1 (ScAhp1), which is a member of the Prx5 subfamily. Published evidence indicates that ScAhp1 is a 2-Cys Prx with its CR residue located immediately preceding α4 (36). From a structural point of view, it is difficult to envision how this CR could form a disulfide within a single-chain or A-type dimer. An alternate explanation is that ScAhp1 is a 1-Cys Prx (as is seen for other Prx5 subfamily members) and that the disulfide observed is an artifact resulting from a reaction involving a surface-exposed Cys of one chain and the SPOH of another; such nonphysiologic disulfides have been seen in a few of other Prx structures (entries 41, 54, and 70 in Table 1). Of second note is the Prx1 from Mycobacterium tuberculosis (MtAhpC, entry 15 in Table 1), which has two Cys residues near the C-terminus, Cys174 and Cys176. Although Cys174 has been identified as the primary CR (27, 46), Cys176 is able to substitute as a CR when Cys174 has been mutated, retaining ∼30% of the activity of the wild-type AhpC (46). Similar mechanistic flexibility with lowered catalytic rates has been observed for other CR mutants, although the alternate CR has not been identified [e.g., (17, 49)].
Subfamily Prx1
Overview
The Prx1 subfamily members appear to be the most widespread and highly expressed of the Prxs, distributed among archaea, bacteria, and eukaryotes. This subfamily includes the yeast TSA proteins, several plant Prxs, tryparedoxin peroxidases, and the bacterial AhpC proteins, as well as the human Prxs I to IV. The subfamily has been referred to as the “A” group (32, 78) or the “typical 2-Cys” group. Almost all known members contain the CR near the C-terminus in the prototypical position equivalent to the CR in HsPrxII (line 1 in Fig. 4A). S. typhimurium AhpC has been shown to prefer hydrogen peroxide to the bulkier peroxides (65). Some Prx1 subfamily members appear to have physiological roles that extend beyond that of a simple peroxidase, having been linked with important roles in cellular signaling events and, in some cases, acting as a molecular chaperone. In these roles, they have been seen to undergo regulation by both overoxidation and phosphorylation (28).
The 22 known structures of Prx1 subfamily members represent 13 different proteins, including both FF and LU conformations, as well as distinct LU conformations seen in two structures of a Prx–Srx complex. Single Prx1 subfamily members with both FF and LU conformations known are those from rat (RnPrx1, entries 4 and 5 in Table 1), Plasmodium vivax (Pv2Cys, entries 12 and 13 in Table 1), and S. typhimurium (StAhpC, entries 17 and 18 in Table 1).
Compared with the common core structure of the Prxs, Prx1 subfamily members contain an approximate 40- to 50-residue extension at the C-terminus. All known subfamily members adopt a B-type dimer interface with the C-terminal extension reaching across the dimer, forming contacts with the other chain (Fig. 7B). In most cases, the B-type dimers of the Prx1 subfamily members associate to form doughnut-like assemblies that are most often decameric, with five B-type dimers associating through the A-type dimer interface (Fig. 7C). In the FF form, the CR residue is buried within the folded C-terminal extension ∼14 Å away from the CP. For the disulfide to form, the CP-loop and first turn of α2 unfold to expose the CP, whereas the C-terminal extension unfolds to expose the CR (Fig. 6A). The result is that the C-terminal extension becomes largely disordered and is not visible in crystal structures. Three structures have slightly different LU conformations compared with the canonic form for the subfamily: in RnPrxI (entry 5 in Table 1), the CP-loop has collapsed to a more-condensed structure that Wood et al. (86) have speculated is related to its dissociation from a decamer to a dimer and that may serve to enhance the recycling of the CP and CR thiols. In PfTrx-Px2 (entry 14 in Table 1), changes in the CP-loop are presumably also related to decamer dissociation, and in MtAhpC (entry 15 in Table 1), an ∼8-degree shift in α2 is found in addition to the unwinding of the first turn of α2.
A link between decamer assembly and the catalytic cycle
The decameric assembly of Prx1 proteins is dynamic, with dimers and decamers existing in an equilibrium affected by redox state, phosphorylation, protein concentration, pH, or ionic strength [reviewed in (4)]. As concentration, pH, and ionic strength are not expected to vary much in vivo, it is thought that redox state and phosphorylation are the dominant factors that will influence the oligomeric state of the Prxs within the cell.
The sensitivity of oligomerization to redox state was first shown by Wood et al. (86) and confirmed by Guimaraes et al. (27). Disulfide formation weakens the decamer-building interactions so that the decamer dissociates to B-type dimers; all other forms of the enzyme (SPH, SPOH, SPO2H, SPO3H) appear to exist as stable decamers (Fig. 7D). The proposed physical explanation for the link between disulfide formation and decamer destabilization is that the FF active site (especially the CP-loop) buttresses the decamer building surface (A-type interface), so that when the LU active site is locked in place by disulfide formation, the decamer is destabilized. Thus, during the catalytic cycle (Fig. 1), these Prxs undergo a change from decamers to dimers and back to decamers. This explanation implies that for these Prxs, the stability of the FF active site (and hence catalytic activity) is linked with decamerization (65). This link was confirmed by a study showing that mutants of StAhpC designed to weaken decamer formation were 100-fold less active than wild-type enzymes, solely due to a Km effect (65).
Sensitive and robust Prx1 subfamily members
Although the features discussed thus far are shared by all Prx1 subfamily members, the Prx1 subfamily can be divided into two distinct groups based on their sensitivity to inactivation by a second substrate molecule reacting with the SPOH before disulfide formation (i.e., resolution) can occur (Fig. 1). Many eukaryotic subfamily members, including HsPrxI and HsPrxII, are very sensitive to inactivation (90), whereas others, such as StAhpC, are robust (87). The structural origin of this difference was shown to be the presence of a C-terminal helix containing a conserved “YF” motif (asterisk in Fig. 4A), which packs against the first turn of helix α2 on the side opposite the active-site pocket and hinders the local unfolding of the CP-loop (Fig. 8) (87). This structural explanation has been confirmed by protein engineering of two Prxs from Schistosoma mansoni (73), which showed that adding the C-terminal helix to a robust enzyme made it sensitive, and that deleting the C-terminal helix from a sensitive enzyme made it robust.
FIG. 8.
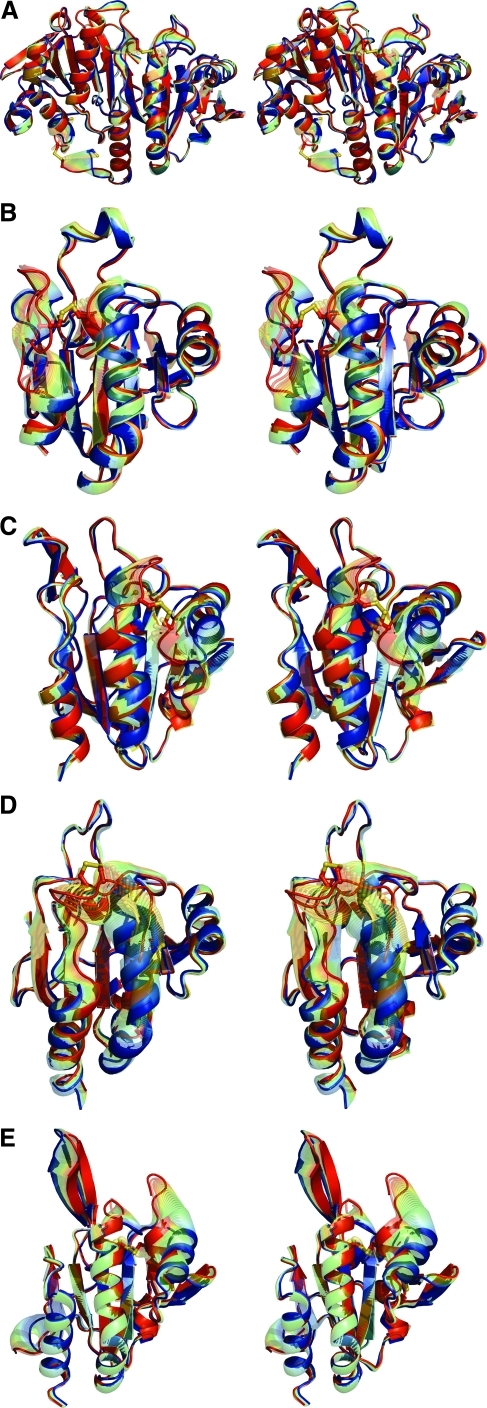
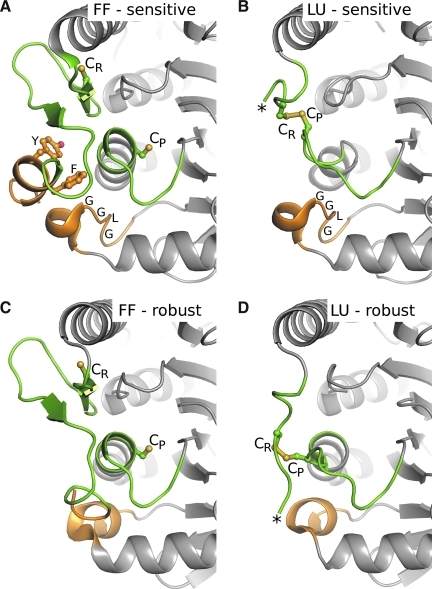
The structural difference between robust and sensitive Prx1s. Comparison of a sensitive Prx1 (A and B, RnPrx1, entries 5 and 4 in Table 1) and a robust Prx1 (C and D, StAhpC, entries 21 and 17 in Table 1) in the FF (left panels) and LU (right panels) conformations reveals the structural feature causing sensitivity. The two regions with differences in sequence that correlate with sensitive versus robust Prxs are a loop with an inserted GGLG motif and a C-terminal extension that forms a helix with a “YF” motif; they are colored orange. The regions that undergo conformational change during local unfolding are colored green (except for the C-terminus, which is orange). In sensitive Prx1s, the conserved C-terminal helix containing the YF motif and the adjacent GGLG motif bury the N-terminal end of α2, stabilizing the FF conformation. This hinders local unfolding, slowing disulfide-bond formation and thus enhancing the competing overoxidation pathway. Comparison of (A) with (C) shows the structural differences that result from the absence of GGLG and YF motifs in robust Prx1s. The CP and CR are shown as ball-and-stick with sulfur atoms colored yellow. *The end of the ordered C-terminus in the LU conformations (with additional residues being disordered). Figure prepared by using Pymol (20). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
To provide a rationale for the reason that sensitivity to substrate-based inactivation has been selected for during evolution, Wood et al. (87) proposed that it allows these Prxs to act as peroxide floodgates that allow the hydrogen peroxide concentration to build up in the vicinity of NADPH oxidase enzymes that are turned on by the binding of various hormones to cellular receptors. This proposal was one of many developments that stimulated increasing acceptance over the last decade that peroxide does serve as a classic second messenger in many hormonal signaling pathways. Although the mechanisms by which sensitive Prx1 subfamily members are involved in cellular signaling pathways are still unfolding, it has become abundantly clear that these Prxs are more than just simple antioxidant enzymes (28, 60). In addition to inactivation caused directly by overoxidation, posttranslational modifications can also regulate the ability of Prxs to reduce peroxide substrates [reviewed in (2)]. In recent work, phosphorylation of Prx1 on the Tyr of the YF motif in the C-terminal helix was linked with its loss of peroxidase activity and was speculated to be a necessary step allowing H2O2 accumulation during signal propagation (85). In addition to the proposal that the inhibition of peroxidase activity is a key mode of regulation, some evidence suggests that Prx overoxidation also allows them to function as molecular chaperones (13, 35).
Regeneration of overoxidized Prx1
As mentioned earlier, under conditions in which high concentrations of hydroperoxide substrates are present, as well as sufficient reducing capacity to support multiple turnovers, some eukaryotic Prxs are susceptible to overoxidation of the CP to form SPO2H and SPO3H. These species are inactive in peroxide reduction and can no longer be regenerated by the “normal” catalytic cycle. Instead, a repair enzyme known as sulfiredoxin (Srx), characterized best from yeast and human systems, catalyzes the “retroreduction” of SPO2H within selected members of the Prx1 subfamily (step 5 in Fig. 1) (5, 39). Extensive x-ray crystallographic analyses coupled with isotopic exchange and mass spectrometry experiments have revealed many details regarding the Srx–Prx interaction (entries 2 and 3 in Table 1) and the chemical steps involved in this unusual chemistry (38, 37, 40). In brief, the SPO2H, which is buried within the FF active-site pocket of decameric or even higher-molecular-weight aggregate forms of the Prx, must be made accessible for repair. Srx accomplishes this through extensive reorganization of the C-terminal tail of the substrate Prx, which then wraps around the backside of an Srx monomer, stabilizing the Prx-Srx “embrace.” Within this conformation, the CP side chain comes into close proximity of the γ-phosphate of ATP, and a residue within the CP-loop of Prx1, Phe 50, docks into a surface pocket of Srx, further stabilizing the repair complex. This reaction is dependent on the presence Mg2+, ATP, and a thiol-containing reductant to regenerate the active Prx. Experiments to date support the requirement for formation of a phosphoryl ester intermediate at the CP of the Prx that is subsequently dephosphorylated and reduced by the combined action of an active site Cys in Srx and a thiol reductant like Trx or glutathione. No evidence exists for Srx-mediated reduction of SPO2H-containing substrates other than Prxs.
Subfamily Prx6
The first Prx structure determined was human Prx6 (HsPrxVI) (10), a 1-Cys Prx from which this subfamily takes its name. Originally identified as the “B” group (32), Prx6 subfamily members are found in archea, bacteria, and eukaryotes and are almost exclusively 1-Cys Prxs (Fig. 4C). Similar to the Prx1 subfamily, Prx6s contain a C-terminal extension of ∼50 to 80 residues compared with members of the other four subfamilies (Fig. 4A). Although the Prx1 and Prx6 subfamilies may be combined because of their somewhat similar sequences (∼21% to 34% identity) and the presence in both of a C-terminal extension and B-type dimers (14), the two subfamilies have distinct sequence patterns, with the most visible distinguishing trait being a 15-to 40-residue longer C-terminal extension in the Prx6 subfamily. The direct reductant of the Prx6s is still unclear; whereas S. cerevisiae Prx1p is reduced by Trx (66), HsPrxVI is not (41). GSTπ may catalyze the glutathione-dependent reduction of HsPrxVI (51), and ascorbate has also been reported to reduce some Prx6 proteins (54).
Fifteen known Prx6 structures represent four different Prxs, all in the FF conformation and with the CP in a variety of oxidation states. Both 1-Cys and 2-Cys subfamily members are represented. The unique structural trait seen across the Prx6 subfamily compared with the common core Prx structure is the long C-terminal extension mentioned earlier. The basic structure is a B-type dimer, with the C-terminal extension making extensive contacts across the dimer interface. The spatial organization of the C-terminal extension is different from that for the Prx1 subfamily members, as the first helix of the extension is in a different position, and the following β-hairpin of Prx6s fills approximately the same space as the C-terminal helix in the Prx1 subfamily. Whereas all known subfamily members form B-type dimers, not all form the higher-order decameric structures seen for Prx1 subfamily members. This feature does not seem to be linked to the catalytic mechanism, as both 1-Cys and 2-Cys subfamily members have been characterized as only B-type dimers, and compared with Prx1 enzymes, they presumably have adaptive features that stabilize the FF active site, even in the absence of the A-type dimer (see preceding sections). The inability of some Prx6s to form A-type dimers may be caused by a longer insertion between α4 and β6 that blocks the potential A-type dimer interface (75). No known structure exists for an LU member of the Prx6 subfamily, but the structural similarity with the Prx1 subfamily has led to the suggestion that for 2-Cys Prx6 subfamily members with a CR in the C-terminal extension, an intersubunit disulfide will be formed between the CP and the CR across the B-type dimer interface and that the structural rearrangements will involve unfolding of the C-terminal domain to allow CP and CR residues, which are ∼15 Å apart in the FF conformation, to be close enough for the disulfide bond to form. For 1-Cys Prx6s, in principle, the C-terminal extension need not unfold, but its fate is unknown.
The most structurally well characterized Prx6 subfamily member is the Aeropyrum pernix (ApTpx), for which 10 structures have been solved. From the structural analyses, the catalytic cycle of this Prx has been proposed to include a hypervalent sulfur intermediate, which involves a covalent bond between the SP and a Nδ1 atom of a nearby His (residue 42 in ApTpx) (55, 56). It is not known whether this is a normal part of catalysis for Prx6 enzymes or a nonphysiologic side reaction that occurs in the crystal when the normally rapid progression to the disulfide form is slowed. In any case, this mechanism cannot be relevant to other subfamilies, as the His involved is not conserved outside of the Prx6 subfamily.
Subfamily Prx5
This subfamily is named after human Prx5 (HsPrxV), a mitochondrial, peroxisomal, and cytoplasmic Prx that was the first member of this subfamily to be structurally characterized (19). Also referred to as the “D” group, Prx5 subfamily members are found in mammals, fungi, bacteria, and higher plants (32). Unique to this subfamily are fused Prx-Grx proteins, for which the linking of the Prx and its reductant may facilitate catalytic turnover. For subfamily members that are not fused to a Grx protein, Trx is the typical reductant. HsPrxV is somewhat less reactive with H2O2 than is PrxI or PrxII at 3 × 105 M−1s−1, but exhibits considerable reactivity (106 to 107 M−1s−1) toward organic hydroperoxides and another signaling-relevant oxidant, peroxynitrite (79). Surprisingly, only ∼17% of the known subfamily members are 2-Cys, resembling the prototype HsPrxV with the CR placed in the loop just before α5 (Fig. 4). All members appear to form A-type dimers independent of redox state.
From the Prx5 subfamily, 12 known structures represent five different Prxs. The best-characterized subfamily member is HsPrxV, a 2-Cys Prx for which both FF and LU conformations have been determined. Four FF structures of 1-Cys Prx5s have also been solved. Although no LU 1-Cys structure has been determined, one chain of HiHyPrxV (entry 47 in Table 1) displays a conformation for α2 that is perturbed from the FF conformation (43) and may be an interesting intermediate structure providing insight into the structural transitions of the 1-Cys Prx5 subfamily members.
Compared with the common core Prx structure, the FF conformation for Prx5 subfamily members contains a bulge in helix α2, an insertion between α4 and β6 that forms a short helix and, like the Tpx subfamily, has a shorter α5 helix of about two turns. The bulge in α2 or “α-aneurysm” (72), is the most distinctive feature of this subfamily, caused by an insertion of one residue into the helix two residues after the CP and associated also with a conserved Pro four residues later (Fig. 4A). The insertion between α4 and β6 both forms part of the substrate-binding pocket and is involved in the A-type dimer interface. Similar to the Tpx subfamily (see preceding section), the substrate-binding pocket involves residues from both chains of the dimer, and this makes Prx5 subfamily members obligate dimers.
HsPrxV is used as the model system for the structural transitions that occur with local unfolding for prototypical 2-Cys Prx5 subfamily members. In the FF conformation, the CP and the CR are located ∼14 Å apart. During local unfolding, the main movements are an opening of the first two turns of α2 (including the bulge) to move the CP toward α5, and the unwinding of the first turn of α5 to change the conformation of the loop between β7 and α5, bringing the CR toward α2 (Fig. 6B). The α-aneurysm bulge is no longer present in the LU conformation, but it is plausible that the presence of the inserted residue and bulge are important for stabilizing the extended loop structure that now includes these residues. It is unknown whether similar structural changes occur in α2 for the 1-Cys subfamily members; interestingly, the α-aneurysm is conserved in 1-Cys subfamily members, suggesting an importance of the bulge beyond aiding the formation of the disulfide with the CR in α5.
In addition to the x-ray crystallography characterizations, NMR backbone dynamics studies have been performed on two members of the PrxV subfamily, Populus trichocarpa PrxD (PtPrxD) (23) and S. cerevisiae Ahp1 (ScAhp1) (78). In agreement with the crystal structures, these dynamics data are consistent with an overall ordered protein that forms dimers. Interestingly, although neither NMR analysis shows significant changes in dynamic motion in the region of α2, both have missing assignments principally for residues that are part of the CP-loop, suggesting that the CP-loop residues are involved in intermediate exchange.
Subfamily Tpx
The Tpx subfamily, originally referred to as the “E” group (32), comprises bacterial peroxidases and contains the original protein designated by the name “thiol peroxidase” or p20 (8). Tpx subfamily members are typically reduced by Trx. E. coli Tpx has been shown to exhibit a much lower Km for cumene hydroperoxide (a bulky, hydrophobic substrate) than for hydrogen peroxide (9 μM compared with 1.7 mM, respectively) (3). Almost all identified Tpx subfamily members function with a 2-Cys mechanism with the CR in the prototypical location in the C-terminal turn of helix α3; about 1% of the subfamily appear to function as 1-Cys Prxs (Fig. 4).
For the Tpx subfamily, 13 known structures represent seven different Prxs. As described earlier, the structure solved by a structural genomics group for CvTpx (entry 62 in Table 1) does not contain a CP and is therefore not an active Prx, but simply a Prx homologue. The most well-studied subfamily member is E. coli Tpx (EcTpx), for which the structure of the LU form was first described by Choi et al. (11). Recently, structures of EcTpx in the FF and LU conformations, as well as a transitional conformation, were combined with a sequence-conservation analysis, leading both to evidence that Tpxs are obligate dimers and to a description of the conformational changes that the Tpxs undergo during catalysis (30). Compared with the core common to all Prxs, the unique feature of Tpx subfamily members is an N-terminal β-hairpin that is involved in forming a hydrophobic collar around the active-site pocket that likely tailors substrate specificity to alkyl peroxides. Residues involved in the collar come from this β-hairpin and residues 58, 126, 127, 130, and 153 in one chain and residues 34, 35, and 89 in the other chain of the dimer (30).
In terms of conformational changes during catalysis, local unfolding mostly alters the CP-loop, α2, and α3, which allows the CP and CR thiols to move from ∼13 Å apart to form a disulfide (Fig. 6C). During the changes, α2 twists and tilts to adopt a new conformation in the cradle, and the CP-loop shifts to expose the CP; for α3, the main change is an unraveling of the C-terminal turn of the helix and shift of the turn after α3. The majority of the conserved cradle residues form a featureless, hydrophobic surface on which α2 can rotate. Among many side-chain shifts, a notable one is a well-conserved aromatic residue just preceding the CR that slides into the protein core vacated by the unfolding of α2. A fascinating feature is what could be called an arginine-trigger that links the unfolding of α2 to the destabilization and consequent local unfolding of α3. This involves a well-conserved Arg located in α2 (residue CP + 5) that makes a salt bridge with a glutamate in helix α3 that is disrupted by the unfolding of α2 (30).
All characterized Tpx subfamily members exist as A-type dimers that do not dissociate with changes in redox state (3). The in-depth analysis of the Tpx subfamily sequences revealed that the most highly conserved residues all cluster at the A-type dimer interface. Interestingly, the turns leading to helices α2 and α3, the two regions that locally unfold during catalysis, are buried at the conserved dimer interface. It was proposed that the Tpxs are obligate dimers, primarily because the dimer interface acts as an essential anchor, tethering the bases of α2 and α3 and maintaining protein stability while allowing the dynamics required for the local unfolding and refolding transitions (30).
Subfamily BCP
The BCP subfamily, originally designated the “C” group (32), appears to be the most diverse. Known members vary in oligomeric state and the presence or location of the CR, existing as monomers and A-type dimers with either 1-Cys or 2-Cys mechanisms, and with two distinct locations seen so far for the CR. Although the majority of the BCP subfamily members are bacterial, BCPs have also been found in archaea and eukaryotes; the plant homologues are referred to as PrxQ. With regard to nomenclature, the BCP subfamily has been broken into an α-group, containing the prototypical 2-Cys Prxs with the CPXXXXCR motif, and a β-group with no CR that function as 1-Cys Prxs (∼10% of the members) (81), but now that more sequences are known, this nomenclature is not adequate for describing the entire subfamily. The broad diversity among members of this subfamily and the existence of monomeric structures lead us to propose that the BCP subfamily is the modern grouping that most resembles the ancestral protein from which the current Prxs diverged.
For the BCP subfamily, eight known structures represent five different Prxs. For members that function with a 2-Cys mechanism, the most common location for the CR is in the prototypical location in α2, five residues C-terminal to CP. This CPXXXXCR motif is seen in ∼55% of the members. For another significant group (∼7%), the CR is in α3 at the same position as the prototypical CR location in the Tpx subfamily (Fig. 4). Fortuitously, both a FF and a LU conformation have been solved for two members, Aeropyrum pernix BCP (ApBCP) with the CR in the more common α2 position and Xanthomonas campestris BCP (XcBCP) with the CR in the α3 position. These two pairs of structures give insight into the structural transitions of more than 60% of the subfamily.
Compared with the core common to all Prxs, the FF BCP structure contains a longer loop between α4 and β6 that forms a β-hairpin and a longer α5 with four or five turns. Whereas in most Prxs, the conserved Arg is stabilized by one or two backbone carbonyls from the loop between β7 and α5, this loop is shorter in most BCP structures, and only the ApBCP structure shows a stabilizing hydrogen bond formed between the Pro142 carbonyl and the conserved Arg side chain. The subfamily members that do not form a stabilizing hydrogen bond to the Arg are monomeric; the stabilization may be an evolved trait that is absent in the more-ancient BCP structures. Comparison of the known monomeric and dimeric BCPs with the CR in α2 (entries 63 and 64 in Table 1) reveals an insertion between α4 and β6 (Fig. 4A) that may disrupt the ability to form A-type dimers; however, this insertion is even longer in the characterized dimeric BCP subfamily member with the CR in α3 (entry 68 in Table 1), suggesting that the relation between sequence, structure, and oligomerization in the BCP subfamily must be further explored. Presumably, as is expected for Prx6 subfamily members that do not form A-type dimers, the CP-loop and FF active site of monomeric BCPs will be stabilized by alternate means.
The LU conformations are, of course, quite different between BCP subfamily members with CR locations in either α2 and α3. For BCP subfamily members with the CPXXXXCR motif, the CP and the CR are 12 Å apart in the FF conformation, one and a half turns away from each other in α2. Local unfolding causes the entire helix to be pulled upward as four residues are looped out to form a β-hairpin structure that accommodates the disulfide between the CP and the CR. Only two turns remain of what used to be α2, and these adopt a 310-helical structure. The newly formed hairpin points toward the α5-side of the cradle, and, to accommodate the structural rearrangement, the first loop of α5 unravels, and the entire α5 helix tilts ∼5 degrees away from the top of α2 (Fig. 6D). An Arg and adjacent Asp residue, conserved in α2 among the BCPs with the CPXXXXCR motif, form hydrogen bonds that are important in stabilizing the two conformations of α2 (17).
For 2-Cys BCP subfamily members with the CR in α3, the LU conformation is surprisingly distinct from that observed for members of the Tpx subfamily with the CR in the same location. Based on XcBCP, local unfolding does not involve a change in the position of α2 in the cradle, but simply a shift in the rotamer of the CP side chain and an unraveling of the N-terminal two turns of α3. The local unfolding of α3 moves the CR from its original position 14 Å away from the CP to disulfide bonding distance (Fig. 6E). The β-hairpin between α4 and β6 is longer in the structures from XcBCP and is thought to play a part in regulating substrate channel accessibility (49). The fact that a different local unfolding structural transition can occur for Prxs with the same CR position supports the hypothesis that the CR positions independently evolved multiple times during the divergence of the Prx family.
Subfamily AhpE
AhpE proteins do not clearly classify with any other Prx subfamilies. Bioinformatic analysis of the sequence database identified only 25 putative members of this group, all 1-Cys Prxs found in mycobacterial and closely related bacterial species (K. Nelson et al., unpublished observations). Although AhpE separates as its own unique subfamily, it shares ∼30% sequence identity with members of the Prx1 subfamily and 25% identity with members of the BCP subfamily and has high structural similarity based on Dali scores (33) (Z scores near 24) to both. Kinetic characterization of AhpE form Mycobacterium tuberculosis indicates that it reacts faster with peroxynitrite than with H2O2 (∼107 vs. ∼105 M−1s−1), and its physiologic reductant is still unknown (34).
Only two structures have been determined for members of the AhpE subfamily, both of the same Prx (MtAhpE) as an A-type dimer in the FF conformation. Compared with the common Prx core, unique features of the AhpE subfamily structure are an extended loop at the N-terminus, a β-hairpin loop between α4 and β6, and a longer loop between β7 and α5. The active-site pocket is small and positively charged. Interestingly, the extended N-terminus packs against β7, blocking the formation of the B-type dimers. Although the authors state that MtAhpE forms octamers, we suspect the octamers are a result of crystal packing rather than being physiologic. The only significant difference among the representative structures is in the active site. Oxidation of the CP to SPOH causes the conserved Arg and Thr side chains to swing away from their positions that stabilize the thiolate form of the CP. To accommodate the changed Arg position, the loop between β7 and α5 shifts about 4 Å, opening up the active site (48). These rearrangements may be an early part of the local unfolding associated with resolution of this 1-Cys Prx, although additional conformational changes are expected in α2 that will move the CP to a more accessible position to an incoming protein or small-molecule thiol.
Outlook
The 12 years of work since the first Prx structure was solved have produced a strong foundation of structural knowledge that spans much of the broad diversity of this family. This foundation will support ongoing biochemical and biomedical research on all types of Prxs and will advance our understanding of the roles of Prxs in both oxidative-stress protection and H2O2 signaling. Additionally, with the insights gained from the clear organization of the Prx family into six subfamilies and descriptions of structural changes with catalysis, consistent nomenclature can now be used to describe the field, making it more accessible and understandable to a broader audience. This review highlights a major advance that can now be made regarding the understanding of substrate binding and catalysis, with the proposal of specific roles for each of the conserved active-site groups in transition-state stabilization. The primary need for further structural information is now the determination of complexes of 1-Cys and 2-Cys Prxs with the partner protein responsible for their reduction. These structures may show features that allow specific recognition of Prxs and will continue to expand our understanding of the bigger picture of Prx catalysis and regulation.
Abbreviations Used
- AhpE
a Prx subfamily named alkyl hydroperoxidase component E
- AhpF
alkyl hydroperoxide reductase component F
- BCP
a Prx subfamily named bacterioferritin comigratory protein
- CP
peroxidatic Cys
- CR
resolving Cys
- DNS
naphthalene-2,6-disulfonic acid
- FF
fully folded
- Gpx
glutathione peroxidase
- Grx
glutaredoxin
- GST
glutathione-S-transferase
- HRP
horseradish peroxidase
- LU
locally unfolded
- Prx
peroxiredoxin
- Prx1
a Prx subfamily
- Prx5
a Prx subfamily
- Prx6
a Prx subfamily
- SP
sulfur atom of the peroxidatic Cys
- SPOH
sulfenic acid
- SPO2H
sulfinic acid
- SPO3H
sulfonic acid
- SR
sulfur atom of the resolving thiol
- Srx
sulfiredoxin
- Tpx
a Prx subfamily named thiol peroxidase
- Trx
thioredoxin
Acknowledgments
The authors' research program on peroxiredoxins is supported by National Institutes of Health grant RO1 GM050389. Bioinformatics work was also supported by NSF grant MCB 0517343 to Jacquelyn S. Fetrow. We thank Todd Lowther for descriptions of the Prx-Srx complexes.
References
- 1.Alphey MS. Bond CS. Tetaud E. Fairlamb AH. Hunter WN. The structure of reduced tryparedoxin peroxidase reveals a decamer and insight into reactivity of 2-Cys-peroxiredoxins. J Mol Biol. 2000;300:903–916. doi: 10.1006/jmbi.2000.3881. [DOI] [PubMed] [Google Scholar]
- 2.Aran M. Ferrero DS. Pagano E. Wolosiuk RA. Typical 2-Cys peroxiredoxins: modulation by covalent transformations and noncovalent interactions. FEBS J. 2009;276:2478–2493. doi: 10.1111/j.1742-4658.2009.06984.x. [DOI] [PubMed] [Google Scholar]
- 3.Baker LM. Poole LB. Catalytic mechanism of thiol peroxidase from Escherichia coli: sulfenic acid formation and overoxidation of essential CYS61. J Biol Chem. 2003;278:9203–9211. doi: 10.1074/jbc.M209888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barranco-Medina S. Lazaro JJ. Dietz KJ. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009;583:1809–1816. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 5.Biteau B. Labarre J. Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 6.Boucher IW. McMillan PJ. Gabrielsen M. Akerman SE. Brannigan JA. Schnick C. Brzozowski AM. Wilkinson AJ. Muller S. Structural and biochemical characterization of a mitochondrial peroxiredoxin from Plasmodium falciparum. Mol Microbiol. 2006;61:948–959. doi: 10.1111/j.1365-2958.2006.05303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Z. Roszak AW. Gourlay LJ. Lindsay JG. Isaacs NW. Bovine mitochondrial peroxiredoxin III forms a two-ring catenane. Structure (Camb) 2005;13:1661–1664. doi: 10.1016/j.str.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Cha MK. Kim HK. Kim IH. Thioredoxin-linked "thiol peroxidase" from periplasmic space of Escherichia coli. J Biol Chem. 1995;270:28635–28641. doi: 10.1074/jbc.270.48.28635. [DOI] [PubMed] [Google Scholar]
- 9.Chae HZ. Robison K. Poole LB. Church G. Storz G. Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci U S A. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi HJ. Kang SW. Yang CH. Rhee SG. Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat Struct Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 11.Choi J. Choi S. Cha MK. Kim IH. Shin W. Crystal structure of Escherichia coli thiol peroxidase in the oxidized state: insights into intramolecular disulfide formation and substrate binding in atypical 2-Cys peroxiredoxins. J Biol Chem. 2003;278:49478–49486. doi: 10.1074/jbc.M309015200. [DOI] [PubMed] [Google Scholar]
- 12.Choi J. Choi S. Chon JK. Cha MK. Kim IH. Shin W. Crystal structure of the C107S/C112S mutant of yeast nuclear 2-Cys peroxiredoxin. Proteins. 2005;61:1146–1149. doi: 10.1002/prot.20704. [DOI] [PubMed] [Google Scholar]
- 13.Chuang MH. Wu MS. Lo WL. Lin JT. Wong CH. Chiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci U S A. 2006;103:2552–2557. doi: 10.1073/pnas.0510770103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Copley SD. Novak WR. Babbitt PC. Divergence of function in the thioredoxin fold suprafamily: evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry. 2004;43:13981–13995. doi: 10.1021/bi048947r. [DOI] [PubMed] [Google Scholar]
- 15.Cox AG. Peskin AV. Paton LN. Winterbourn CC. Hampton MB. Redox potential and peroxide reactivity of human peroxiredoxin 3. Biochemistry. 2009;48:6495–6501. doi: 10.1021/bi900558g. [DOI] [PubMed] [Google Scholar]
- 16.Cox AG. Winterbourn CC. Hampton MB. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem J. 2010;425:313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- 17.D'Ambrosio K. Limauro D. Pedone E. Galdi I. Pedone C. Bartolucci S. De Simone G. Insights into the catalytic mechanism of the Bcp family: functional and structural analysis of Bcp1 from Sulfolobus solfataricus. Proteins. 2009;76:995–1006. doi: 10.1002/prot.22408. [DOI] [PubMed] [Google Scholar]
- 18.D'Autreaux B. Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 19.Declercq JP. Evrard C. Clippe A. Stricht DV. Bernard A. Knoops B. Crystal structure of human peroxiredoxin 5, a novel type of mammalian peroxiredoxin at 1.5 A resolution. J Mol Biol. 2001;311:751–759. doi: 10.1006/jmbi.2001.4853. [DOI] [PubMed] [Google Scholar]
- 20.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 21.Denu JM. Dixon JE. Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr Opin Chem Biol. 1998;2:633–641. doi: 10.1016/s1367-5931(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 22.Dietz KJ. Jacob S. Oelze ML. Laxa M. Tognetti V. de Miranda SM. Baier M. Finkemeier I. The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot. 2006;57:1697–1709. doi: 10.1093/jxb/erj160. [DOI] [PubMed] [Google Scholar]
- 23.Echalier A. Trivelli X. Corbier C. Rouhier N. Walker O. Tsan P. Jacquot JP. Aubry A. Krimm I. Lancelin JM. Crystal structure and solution NMR dynamics of a D (type II) peroxiredoxin glutaredoxin and thioredoxin dependent: a new insight into the peroxiredoxin oligomerism. Biochemistry. 2005;44:1755–1767. doi: 10.1021/bi048226s. [DOI] [PubMed] [Google Scholar]
- 24.Evrard C. Capron A. Marchand C. Clippe A. Wattiez R. Soumillion P. Knoops B. Declercq JP. Crystal structure of a dimeric oxidized form of human peroxiredoxin 5. J Mol Biol. 2004;337:1079–1090. doi: 10.1016/j.jmb.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Flohé L. In: Peroxiredoxin systems. Harris JR, editor. New York: Springer; 2007. [Google Scholar]
- 26.Fomenko DE. Gladyshev VN. Identity and functions of CxxC-derived motifs. Biochemistry. 2003;42:11214–11225. doi: 10.1021/bi034459s. [DOI] [PubMed] [Google Scholar]
- 27.Guimaraes BG. Souchon H. Honore N. Saint-Joanis B. Brosch R. Shepard W. Cole ST. Alzari PM. Structure and mechanism of the alkyl hydroperoxidase AhpC, a key element of the Mycobacterium tuberculosis defense system against oxidative stress. J Biol Chem. 2005;280:25735–25742. doi: 10.1074/jbc.M503076200. [DOI] [PubMed] [Google Scholar]
- 28.Hall A. Karplus PA. Poole LB. Typical 2-Cys peroxiredoxins: structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall A. Parsonage D. Poole LB. Karplus PA. Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization. J Mol Biol. 2010;402:194–209. doi: 10.1016/j.jmb.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall A. Sankaran B. Poole LB. Karplus PA. Structural changes common to catalysis in the Tpx peroxiredoxin subfamily. J Mol Biol. 2009;393:867–881. doi: 10.1016/j.jmb.2009.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirotsu S. Abe Y. Okada K. Nagahara N. Hori H. Nishino T. Hakoshima T. Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product. Proc Natl Acad Sci U S A. 1999;96:12333–12338. doi: 10.1073/pnas.96.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann B. Hecht HJ. Flohe L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 33.Holm L. Kaariainen S. Rosenstrom P. Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hugo M. Turell L. Manta B. Botti H. Monteiro G. Netto LE. Alvarez B. Radi R. Trujillo M. Thiol and sulfenic acid oxidation of AhpE, the one-cysteine peroxiredoxin from Mycobacterium tuberculosis: kinetics, acidity constants, and conformational dynamics. Biochemistry. 2009;48:9416–9426. doi: 10.1021/bi901221s. [DOI] [PubMed] [Google Scholar]
- 35.Jang HH. Lee KO. Chi YH. Jung BG. Park SK. Park JH. Lee JR. Lee SS. Moon JC. Yun JW. Choi YO. Kim WY. Kang JS. Cheong GW. Yun DJ. Rhee SG. Cho MJ. Lee SY. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Jeong JS. Kwon SJ. Kang SW. Rhee SG. Kim K. Purification and characterization of a second type thioredoxin peroxidase (type II TPx) from Saccharomyces cerevisiae. Biochemistry. 1999;38:776–783. doi: 10.1021/bi9817818. [DOI] [PubMed] [Google Scholar]
- 37.Jonsson TJ. Johnson LC. Lowther WT. Protein engineering of the quaternary sulfiredoxin.peroxiredoxin enzyme.substrate complex reveals the molecular basis for cysteine sulfinic acid phosphorylation. J Biol Chem. 2009;284:33305–33310. doi: 10.1074/jbc.M109.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonsson TJ. Johnson LC. Lowther WT. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 2008;451:98–101. doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jonsson TJ. Lowther WT. The peroxiredoxin repair proteins. In: Flohé L, editor; Harris JR, editor. Peroxiredoxin Systems. New York: Springer; 2007. pp. 115–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonsson TJ. Tsang AW. Lowther WT. Furdui CM. Identification of intact protein thiosulfinate intermediate in the reduction of cysteine sulfinic acid in peroxiredoxin by human sulfiredoxin. J Biol Chem. 2008;283:22890–22894. doi: 10.1074/jbc.C800124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang SW. Baines IC. Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- 42.Karplus PA. Hall A. Structural survey of the peroxiredoxins. In: Flohé L, editor; Harris JR, editor. Peroxiredoxin Systems. New York: Springer; 2007. pp. 41–60. [DOI] [PubMed] [Google Scholar]
- 43.Kim SJ. Woo JR. Hwang YS. Jeong DG. Shin DHk. Kim K. Ryu SE. The tetrameric structure of Haemophilus influenza hybrid Prx5 reveals interactions between electron donor and acceptor proteins. J Biol Chem. 2003;278:10790–10798. doi: 10.1074/jbc.M209553200. [DOI] [PubMed] [Google Scholar]
- 44.Kitano K. Kita A. Hakoshima T. Niimura Y. Miki K. Crystal structure of decameric peroxiredoxin (AhpC) from Amphibacillus xylanus. Proteins. 2005;59:644–647. doi: 10.1002/prot.20412. [DOI] [PubMed] [Google Scholar]
- 45.Knoops B. Loumaye E. Van Der Eecken V. Evolution of the peroxiredoxins. In: Flohé L, editor; Harris JR, editor. Peroxiredoxin Systems. New York: Springer; 2007. pp. 27–40. [DOI] [PubMed] [Google Scholar]
- 46.Koshkin A. Knudsen GM. Ortiz De Montellano PR. Intermolecular interactions in the AhpC/AhpD antioxidant defense system of Mycobacterium tuberculosis. Arch Biochem Biophys. 2004;427:41–47. doi: 10.1016/j.abb.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 47.Krebs WG. Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S. Peterson NA. Kim MY. Kim CY. Hung LW. Yu M. Lekin T. Segelke BW. Lott JS. Baker EN. Crystal structure of AhpE from Mycobacterium tuberculosis, a 1-Cys peroxiredoxin. J Mol Biol. 2005;346:1035–1046. doi: 10.1016/j.jmb.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 49.Liao SJ. Yang CY. Chin KH. Wang AH. Chou SH. Insights into the alkyl peroxide reduction pathway of Xanthomonas campestris bacterioferritin comigratory protein from the trapped intermediate-ligand complex structures. J Mol Biol. 2009;390:951–966. doi: 10.1016/j.jmb.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 50.Lu J. Yang F. Li Y. Zhang X. Xia B. Jin C. Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase. Biochem Biophys Res Commun. 2008;373:414–418. doi: 10.1016/j.bbrc.2008.06.051. [DOI] [PubMed] [Google Scholar]
- 51.Manevich Y. Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 52.Matsumura T. Okamoto K. Iwahara S. Hori H. Takahashi Y. Nishino T. Abe Y. Dimer-oligomer interconversion of wild-type and mutant rat 2-Cys peroxiredoxin: disulfide formation at dimer-dimer interfaces is not essential for decamerization. J Biol Chem. 2008;283:284–293. doi: 10.1074/jbc.M705753200. [DOI] [PubMed] [Google Scholar]
- 53.Mizohata E. Sakai H. Fusatomi E. Terada T. Murayama K. Shirouzu M. Yokoyama S. Crystal structure of an archaeal peroxiredoxin from the aerobic hyperthermophilic crenarchaeon Aeropyrum pernix K1. J Mol Biol. 2005;354:317–329. doi: 10.1016/j.jmb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 54.Monteiro G. Horta BB. Pimenta DC. Augusto O. Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci U S A. 2007;104:4886–4891. doi: 10.1073/pnas.0700481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamura T. Kado Y. Yamaguchi T. Matsumura H. Ishikawa K. Inoue T. Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide. J Biochem. 2010;147:109–115. doi: 10.1093/jb/mvp154. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura T. Yamamoto T. Abe M. Matsumura H. Hagihara Y. Goto T. Yamaguchi T. Inoue T. Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate. Proc Natl Acad Sci U S A. 2008;105:6238–6242. doi: 10.1073/pnas.0709822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakamura T. Yamamoto T. Inoue T. Matsumura H. Kobayashi A. Hagihara Y. Uegaki K. Ataka M. Kai Y. Ishikawa K. Crystal structure of thioredoxin peroxidase from aerobic hyperthermophilic archaeon Aeropyrum pernix K1. Proteins. 2006;62:822–826. doi: 10.1002/prot.20796. [DOI] [PubMed] [Google Scholar]
- 58.Nelson JW. Creighton TE. Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry. 1994;33:5974–5983. doi: 10.1021/bi00185a039. [DOI] [PubMed] [Google Scholar]
- 58a.Nelson KJ. Knutson ST. Soito L. Klomsiri C. Poole LB. Fetrow JS. Analysis of the peroxiredoxin family: using active site structure and sequence information for global classification and residue analysis. Proteins. Dec. 2010. [Epub ahead of print]; [DOI] [PMC free article] [PubMed]
- 59.Nelson KJ. Parsonage D. Hall A. Karplus PA. Poole JB. Cysteine pK(a) values for the bacterial peroxiredoxin AhpC. Biochemistry. 2008;47:12860–12868. doi: 10.1021/bi801718d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neumann CA. Cao J. Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8:4072–4078. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nordstrand K. Aslund F. Meunier S. Holmgren A. Otting G. Berndt KD. Direct NMR observation of the Cys-14 thiol proton of reduced Escherichia coli glutaredoxin-3 supports the presence of an active site thiol-thiolate hydrogen bond. FEBS Lett. 1999;449:196–200. doi: 10.1016/s0014-5793(99)00401-9. [DOI] [PubMed] [Google Scholar]
- 62.Ogusucu R. Rettori D. Munhoz DC. Netto LE. Augusto O. Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: rate constants by competitive kinetics. Free Radic Biol Med. 2007;42:326–334. doi: 10.1016/j.freeradbiomed.2006.10.042. [DOI] [PubMed] [Google Scholar]
- 63.Olahova M. Taylor SR. Khazaipoul S. Wang J. Morgan BA. Matsumoto K. Blackwell TK. Veal EA. A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. Proc Natl Acad Sci U S A. 2008;105:19839–19844. doi: 10.1073/pnas.0805507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papinutto E. Windle HJ. Cendron L. Battistutta R. Kelleher D. Zanotti G. Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter pylori. Biochim Biophys Acta. 2005;1753:240–246. doi: 10.1016/j.bbapap.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Parsonage D. Youngblood DS. Sarma GN. Wood ZA. Karplus PA. Poole LB. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pedrajas JR. Miranda-Vizuete A. Javanmardy N. Gustafsson JA. Spyrou G. Mitochondria of Saccharomyces cerevisiae contain one-conserved cysteine type peroxiredoxin with thioredoxin peroxidase activity. J Biol Chem. 2000;275:16296–16301. doi: 10.1074/jbc.275.21.16296. [DOI] [PubMed] [Google Scholar]
- 67.Peskin AV. Low FM. Paton LN. Maghzal GJ. Hampton MB. Winterbourn CC. The high reactivity of peroxiredoxin 2 with H(2)O(2) is not reflected in its reaction with other oxidants and thiol reagents. J Biol Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 68.Pineyro MD. Pizarro JC. Lema F. Pritsch O. Cayota A. Bentley GA. Robello C. Crystal structure of the tryparedoxin peroxidase from the human parasite Trypanosoma cruzi. J Struct Biol. 2005;150:11–22. doi: 10.1016/j.jsb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 69.Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 70.Poole LB. The catalytic mechanism of peroxiredoxins. In: Flohé L, editor; Harris JR, editor. Peroxiredoxin Systems. New York: Springer; 2007. pp. 61–81. [DOI] [PubMed] [Google Scholar]
- 71.Rho BS. Hung LW. Holton JM. Vigil D. Kim SI. Park MS. Terwilliger TC. Pedelacq JD. Functional and structural characterization of a thiol peroxidase from Mycobacterium tuberculosis. J Mol Biol. 2006;361:850–863. doi: 10.1016/j.jmb.2006.05.076. [DOI] [PubMed] [Google Scholar]
- 72.Sarma GN. Nickel C. Rahlfs S. Fischer M. Becker K. Karplus PA. Crystal structure of a novel Plasmodium falciparum 1-Cys peroxiredoxin. J Mol Biol. 2005;346:1021–1034. doi: 10.1016/j.jmb.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 73.Sayed AA. Williams DL. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J Biol Chem. 2004;279:26159–26166. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- 74.Schroder E. Littlechild JA. Lebedev AA. Errington N. Vagin AA. Isupov MN. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 A resolution. Structure Fold Des. 2000;8:605–615. doi: 10.1016/s0969-2126(00)00147-7. [DOI] [PubMed] [Google Scholar]
- 75.Smeets A. Loumaye E. Clippe A. Rees JF. Knoops B. Declercq JP. The crystal structure of the C45S mutant of annelid Arenicola marina peroxiredoxin 6 supports its assignment to the mechanistically typical 2-Cys subfamily without any formation of toroid-shaped decamers. Protein Sci. 2008;17:700–710. doi: 10.1110/ps.073399308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smeets A. Marchand C. Linard D. Knoops B. Declercq JP. The crystal structures of oxidized forms of human peroxiredoxin 5 with an intramolecular disulfide bond confirm the proposed enzymatic mechanism for atypical 2-Cys peroxiredoxins. Arch Biochem Biophys. 2008;477:98–104. doi: 10.1016/j.abb.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 77.Stehr M. Hecht HJ. Jager T. Flohe L. Singh M. Structure of the inactive variant C60S of Mycobacterium tuberculosis thiol peroxidase. Acta Crystallogr D Biol Crystallogr. 2006;62:563–567. doi: 10.1107/S0907444906008249. [DOI] [PubMed] [Google Scholar]
- 78.Trivelli X. Krimm I. Ebel C. Verdoucq L. Prouzet-Mauleon V. Chartier Y. Tsan P. Lauquin G. Meyer Y. Lancelin JM. Characterization of the yeast peroxiredoxin Ahp1 in its reduced active and overoxidized inactive forms using NMR. Biochemistry. 2003;42:14139–14149. doi: 10.1021/bi035551r. [DOI] [PubMed] [Google Scholar]
- 79.Trujillo M. Ferrer-Sueta G. Thomson L. Flohe L. Radi R. Kinetics of peroxiredoxins and their role in the decomposition of peroxynitrite. In: Flohé L, editor; Harris JR, editor. Peroxiredoxin Systems. New York: Springer; 2007. pp. 83–113. [DOI] [PubMed] [Google Scholar]
- 80.Vedadi M. Lew J. Artz J. Amani M. Zhao Y. Dong A. Wasney GA. Gao M. Hills T. Brokx S. Qiu W. Sharma S. Diassiti A. Alam Z. Melone M. Mulichak A. Wernimont A. Bray J. Loppnau P. Plotnikova O. Newberry K. Sundararajan E. Houston S. Walker J. Tempel W. Bochkarev A. Kozieradzki I. Edwards A. Arrowsmith C. Roos D. Kain K. Hui R. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol Biochem Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 81.Wakita M. Masuda S. Motohashi K. Hisabori T. Ohta H. Takamiya K. The significance of type II and PrxQ peroxiredoxins for antioxidative stress response in the purple bacterium Rhodobacter sphaeroides. J Biol Chem. 2007;282:27792–27801. doi: 10.1074/jbc.M702855200. [DOI] [PubMed] [Google Scholar]
- 82.Whitesides GM. Liburn JE. Szajewski RP. Rates of thiol-disulfide interchange reactions between mono- and dithiols and Ellman's reagent. J Org Chem. 1977;42:332–338. [Google Scholar]
- 83.Wilson JM. Bayer RJ. Hupe DJ. Structure-reactivity correlations for the thiol-disulfide interchange reaction. J Am Chem Soc. 1977;99:7922–7926. [Google Scholar]
- 84.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 85.Woo HA. Yim SH. Shin DH. Kang D. Yu DY. Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 86.Wood ZA. Poole LB. Hantgan RR. Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 87.Wood ZA. Poole LB. Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 88.Wood ZA. Schroder E. Harris JR. Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 89.Yamamoto Y. Ritz D. Planson AG. Jonsson TJ. Faulkner MJ. Boyd D. Beckwith J. Poole LB. Mutant AhpC peroxiredoxins suppress thiol-disulfide redox deficiencies and acquire deglutathionylating activity. Mol Cell. 2008;29:36–45. doi: 10.1016/j.molcel.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang KS. Kang SW. Woo HA. Hwang SC. Chae HZ. Kim K. Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 91.Yuan Y. Knaggs MH. Poole LB. Fetrow JS. Salsbury FR., Jr Conformational and oligomeric effects on the cysteine pKa of tryparedoxin peroxidase. J Biomol Struct Dynam. 2003;300:650–653. doi: 10.1080/07391102.2010.10507343. [DOI] [PMC free article] [PubMed] [Google Scholar]