Abstract
Many prokaryotic and eukaryotic dsDNA viruses use a scaffolding protein to assemble their capsid. Assembly of the dsDNA bacteriophage P22 procapsids requires the interaction of 415 molecules of coat protein and 60–300 molecules of scaffolding protein. Although the 303 amino acid scaffolding protein is essential for proper assembly of procapsids, little is known about its structure beyond an NMR structure of the extreme C-terminus, which is known to interact with coat protein. Deletion mutagenesis indicates that other regions of scaffolding protein are involved in interactions with coat protein and other capsid proteins. Single and double cysteine variants of scaffolding protein were generated for use in fluorescence resonance energy transfer and crosslinking experiments designed to probe the conformation of scaffolding protein in solution and within procapsids. We showed that the N- and C-termini are proximate in solution and that the middle of the protein is near the N-terminus but not accessible to the C-terminus. In procapsids, the N-terminus was no longer accessible to the C-terminus, which indicated that there is a conformational change in scaffolding protein upon assembly. In addition, our data are consistent with a model where scaffolding protein dimers are positioned parallel to one another with associated C-termini.
Keywords: crosslinking, fluorescence energy transfer, virus assembly, protein folding, procapsids
Introduction
Scaffolding proteins are required by numerous dsDNA icosahedral viruses to catalyze the proper assembly of their transient precursor capsid, known as a procapsid. These viruses use either an internal or external scaffolding protein, or both, to assemble their procapsids. Some viruses recycle the scaffolding proteins for further rounds of assembly, while others enzymatically degrade them upon DNA packaging.1,2,3 It is clear from studies of many viruses that scaffolding proteins are indispensable for proper assembly, and yet there is no clear structural understanding of their fundamental function.
Bacteriophage P22 provides a paradigm for the assembly of dsDNA viruses. The in vivo morphogenic pathway of the T=7 Salmonella bacteriophage P22 involves the co-assembly of 415 molecules of monomeric coat protein with 60–300 molecules of an internal scaffolding protein, as well as some minor injection proteins and the portal protein complex, which occupies one of the five fold vertices, to form a procapsid.4,5 P22 scaffolding protein is known to direct procapsid assembly. Without scaffolding protein, high concentrations of coat protein will assemble into aberrant forms: T= 4 empty procapsids and spiral structures. The spiral structures appear to have their 5-fold and 6-fold vertices located inappropriately so that closed procapsid structures do not form.6,7 Scaffolding protein is also responsible for incorporation of the ejection proteins and the portal complex.8 The dsDNA is actively packaged into procapsids through the unique portal vertex.8 Concomitant with DNA packaging, scaffolding protein exits from the immature capsids to take part in additional rounds of assembly, and the capsids expand to form a mature virion.9,10 In the processes of folding and assembly of P22 procapsid proteins, none are covalently modified or proteolyzed. Nevertheless, the structure of the procapsid examined by electron cryo-microscopy reveals that coat protein subunits are found in seven quasi-equivalent conformations, six in hexons and one in pentons.10,11 Thus, during assembly coat protein monomers must be ‘switched’ into the necessary conformations to produce a closed icosahedral procapsid. Scaffolding protein has been suggested to control the proper switching of capsid proteins.12 How conformational switching occurs and is controlled by scaffolding proteins during assembly is not understood for any icosahedral virus. In vitro, 420 copies of coat protein and ~300 copies of scaffolding protein assemble into a procapsid-like particle that has the same size and general morphology of in vivo assembled procapsids, though without the portal complex or minor proteins.4,13,14
P22 scaffolding protein is comprised of 303 residues and its functional domains have been mapped through mutagenesis studies. The C-terminus has been identified as the coat-binding domain.15,16,17 Residues 280–294 are the minimum residues required for coat binding and are highly negatively charged.18 The N-terminus is postulated to be involved in autoregulation of scaffolding protein gene expression through interaction with its own mRNA.19,20,21,22 The N-terminus might also function as the signaling domain to control scaffolding protein exit during DNA packaging because N-terminal deletion mutants of scaffolding protein are unable to leave the procapsids upon initiation of DNA packaging.18 Based on functional studies, P22 scaffolding protein is thought to be generally U-shaped with the N-terminus and the C-terminus positioned in an anti-parallel fashion.18 Biophysical studies of P22 scaffolding protein show that it is an elongated ellipsoid mainly composed of α-helices connected by unstructured regions23,24 with dimensions of 22 Å in diameter by 247 Å in length.25 It exists in a monomer-dimer-tetramer equilibrium in solution, but the monomers and the dimers are proposed as the species actively involved in procapsid assembly.25 Internal scaffolding proteins from other viruses such as lambda,T4 and SPP1 are also elongated helical ellipsoids with flexible domains.26,27,28
Phi29 scaffolding protein, which is 100 residues in length, is a dimeric, elongated α-helical structure composed of a four helical bundle with coiled coil tails at the N-terminus.29 The NMR structure of the minimal coat-binding domain at the C-terminus of P22's scaffolding protein also contains a helix-turn-helix similar to phi 29's N-terminus.30,31,32 However, acquiring a high-resolution structure of the P22 scaffolding protein has remained a challenge, likely due to its inherent flexibility and multiple oligomeric states. The arrangement of the scaffolding protein inside procapsids is still ambiguous. Attempts to study scaffolding protein inside procapsids of other dsDNA viruses such as T7 and phi29 have only showed possible network arrangements of inner scaffolding proteins.29,33 Cryo-electron microscopy studies on P22 that imposed icosahedral symmetry on the reconstructions have not been able to visualize the full length scaffolding protein, suggesting a non-icosahedral order inside the procapsids.11 A 22 Å cryo-electron microscopy difference map of procapsids with and without scaffolding protein, revealed that scaffolding protein C-terminus interacts with coat protein trimer tips and are arranged with a distance of 50 Å between adjacent scaffolding proteins.34 A recent reconstruction of P22 procapsids with no symmetry imposed confirms the electrostatic interaction between the C-terminus of scaffolding protein and the N-terminal arm of coat protein. The reconstruction also indicates a second site of interaction of the scaffolding protein C-terminus with the A-loop of the coat protein.35 In total, this reconstruction indicated that each coat protein has an associated scaffolding protein.
In this study we have investigated the general conformation and fold of scaffolding protein in solution, as well as inside procapsids, to increase our understanding of the ability of this protein to direct assembly of procapsids, through the use of fluorescence resonance energy transfer (FRET) and crosslinking. The orientation of the full-length scaffolding protein inside procapsids was also examined by crosslinking. We show that scaffolding protein has a significantly more complex fold than the phi29's scaffolding protein and undergoes conformational changes when bound to the interior of procapsids. We also present evidence that scaffolding proteins are more closely associated inside procapsids than previously suggested.
Results
As noted above, the P22 scaffolding protein is one of the best characterized scaffolding proteins of dsDNA bacteriophages and viruses. Nevertheless, there is little known about its fold or any conformational rearrangements that might occur during assembly. We decided to take an approach that combined site-directed incorporation of cysteine residues with FRET and cysteine-reactive crosslinking between different sites within scaffolding protein to add distance constraints to the existing simple structural models.36,18 To guide us in selection of sites for mutagenesis, we used the secondary structure prediction program, SABLE.37 The prediction by SABLE is consistent with the biophysical data that the P22 scaffolding protein is composed of multiple helical regions, and that the C-terminus is composed of a helix-turn-helix (Figure 1). For this study, we introduced either one or two cysteine residues into an amino-terminal hexa-histidine tagged scaffolding protein. Since scaffolding protein is postulated to be flexible,38 substituting residues with cysteines in predicted unstructured regions (Figure 1) should cause minimal distortion to its inherent fold in solution. We also made a cysteine substitution in a known helical region at K294; although this residue is at the coat binding helix, the K294 residue is not involved in coat binding (Cortines et al., submitted). In addition we substituted S242C, which is found at the end of a predicted α-helix.
Figure 1.
The amino acid sequence of P22 scaffolding protein with the secondary structure predicted by SABLE.37 The residues highlighted in magenta were either singly substituted or made with others in combination. The predicted secondary structure is represented as green arrows for β-sheets, red rods for α-helices and thick blue lines for unstructured flexible regions. Amino acids 269–300 are the coat-binding helix-turn-helix domain.
Procapsid assembly activity of scaffolding protein with cysteine substitutions
Prevelige's group showed that dimeric scaffolding protein, formed by a disulfide bonded R74C/L177I variant, catalyzed assembly better than WT scaffolding protein.25 We wanted to confirm that the scaffolding protein cysteine variants were competent for assembly, and to assess any changes in the ability of the variants to catalyze assembly when they form dimers (Figure 2). Assembly reactions were measured by light scattering, which increases with the formation of procapsids with time. The scaffolding protein variants contain different forms of scaffolding proteins: monomeric (with and without intra-molecular disulfides), dimeric and oligomeric (discussed further below). To determine if the substitutions of residues by cysteines caused any variation in assembly or if changes were due to dimer formation, the mutants were also labeled with N-ethylmaleimide (NEM) to prevent disulfide formation.
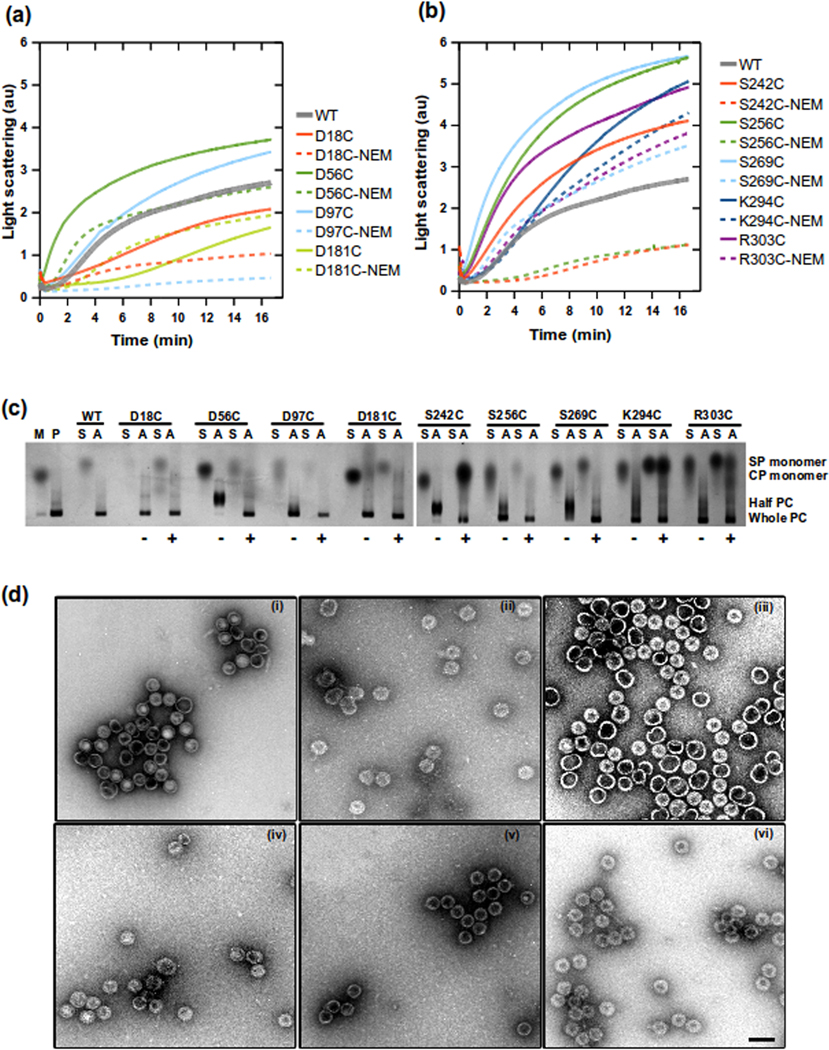
Figure 2.
Assembly reactions catalyzed by scaffolding protein with single cysteine substitutions. (a) and (b) show assembly reactions of all scaffolding protein variants. Reactions were done at 0.5 mg/ml of monomeric coat protein and 0.5 mg/ml scaffolding protein in buffer. The increase in light scattering was monitored at 500 nm at 1 second intervals. Scaffolding protein samples were either oxidized in air or reacted with NEM to prevent disulfide bond formation. The assembly reaction products were visualized with native agarose gels shown in (c). Controls are lane M, monomeric coat protein and lane P, in vivo assembled procapsids. The upper labels indicate which scaffolding protein variants mixed with the monomeric coat protein. S is the scaffolding alone, while A indicates assembly reactions. The symbols at the bottom indicate if the scaffolding protein was reacted (+) or unreacted (−) with NEM. Labels on the right indicate location of scaffolding protein (SP monomer), coat protein (CP monomer) and procapsids (PC). Assembly reactions were analyzed by negative stain electron microscopy at magnification 56700X shown in (d). Micrographs show (i) procapsids assembled in vivo, procapsids assembled in vitro with different scaffolding protein variants: (ii) WT-his, (iii) D18C, (iv) D181C, (v) S242C, (vi) K294C. The scale bar in (vi) represents 100 nm, all micrographs have similar scaling.
Coat protein monomers at 0.5 mg/ml were mixed with scaffolding protein variants at 0.5 mg/ml. Assembly reactions with the oxidized single cysteine variants of scaffolding protein each showed a higher rate of assembly as compared to the NEM-modified proteins, indicating that covalent dimer formation enhanced assembly. The only exception to this was D181C where the unmodified and NEM modified samples had similar assembly rates (Figure 2b). When assembly rates were compared to that catalyzed by wild-type scaffolding protein, the scaffolding proteins with substitutions at D18C or D181C had lower rates of assembly overall, regardless of modification with NEM (Figure 2a). The oxidized D56C mutant had a higher assembly rate and shorter lag time than the wild-type scaffolding protein (Figure 2a) and formed procapsid halves (Figure 2c), indicating a higher affinity to coat protein during assembly.39,40 Scaffolding protein variants: S242C, S256C and S269C had the fastest rate of assembly (Figure 2b), and these mutants formed a mix of whole procapsids and halves at equilibrium (Figure 2c). Even though there were changes in the rates of assembly, all single cysteine scaffolding protein variants, regardless of whether they were labeled with NEM or not, were able to support procapsid assembly. On the native agarose gel (Figure 2c) lane with only monomer coat (M), shows that there is minimal procapsid formation without scaffolding protein. A band corresponding to the procapsid or procapsid halves was observed in the assembly reactions for each scaffolding protein variant. The presence of procapsid halves indicates a higher affinity between coat protein and scaffolding protein, and thereby these reactions should require a slightly higher NaCl concentration to form normal procapsids.40 When extra NaCl was added to these reactions, whole procapsids were generated (data not shown). Electron micrographs of assembly reaction products (Figure 2d) show that whole procapsids similar to morphology of (i) in vivo formed procapsids can be observed for (ii) WT-his, (iii) D18C, (iv) D181C, (v) S242C and (vi) K294C. Other scaffolding variants were also observed to form similar particles (data not shown).
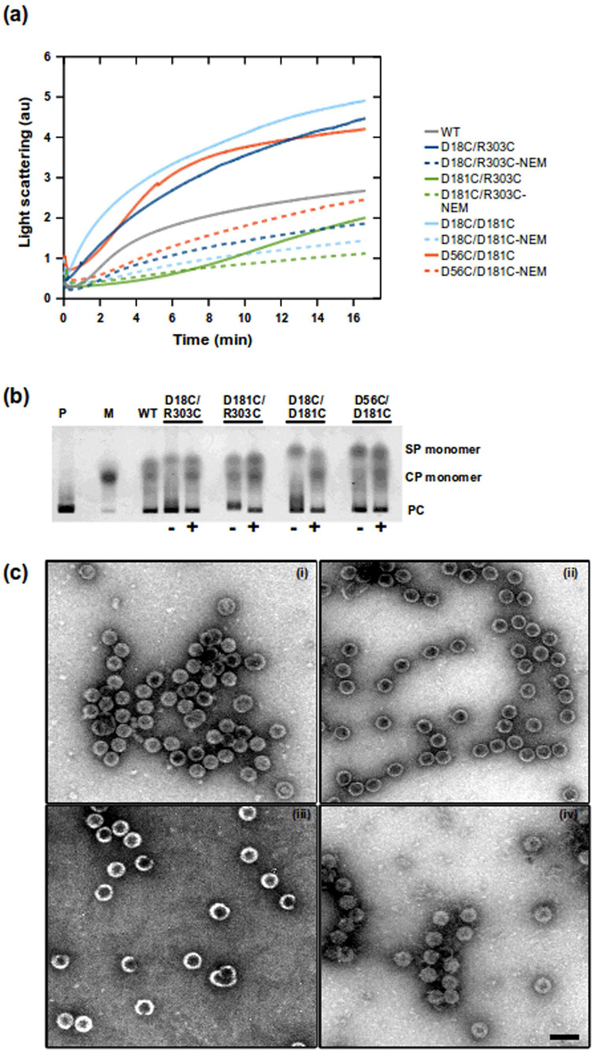
Procapsid assembly with scaffolding proteins with double cysteine substitutions had higher assembly rates when they were not labeled with NEM (Figure 3a). The assembly reaction with D181C/R303C substitution had a longer lag phase than that of the other scaffolding proteins with double cysteine substitutions. Native agarose gels (Figure 3b) of the assembly reactions at equilibrium indicated these double cysteine variants formed procapsids. The micrographs (Figure 3C) of assembly reaction products show that each double cysteine variant of scaffolding protein (without NEM) was capable of producing whole procapsids. Overall, our data indicated that all of the variants were assembly-competent and could be used further in our assays with confidence that we had not significantly affected their activity.
Figure 3.
Assembly reactions of double cysteine variants of scaffolding protein and coat protein. The proteins were mixed at 0.5 mg/ml of each in buffer. Light scattering was monitored at 500 nm wavelength at 1 second intervals. Scaffolding proteins were either oxidized or reacted with NEM. (a) Shows the rate of increase in light scattering of each cysteine variant. (b) The agarose gel of the assembly reactions after overnight incubation at RT. The controls were lane M which is the monomeric coat protein and P which are in vivo assembled procapsids. The gel labels indicate the cysteine variant used in the assembly reactions, which were either unreacted (−) or reacted (+) with NEM. (c) Electron micrographs of negatively stained in vitro assembly reactions. (i) D18C/R303C, (ii) D181C/R303C, (iii) D18C/D181C, (iv) D56V/D181C. All micrographs has similar magnification (56700x) and scale shown by the bar in (iv) which represents 100 nm.
The fold of scaffolding protein in solution determined by FRET
Förster resonance energy transfery (FRET) is a widely used spectroscopic method for determining long-range interactions and the dynamics of biomolecules.41 The rate of energy transfer, kET, between donor and acceptor molecules is proportional to the inverse 6th power of the distance separating them.42 We used FRET as a structural probe to investigate the proximity of the N-terminus and the C-terminus of scaffolding protein. Pyrene is a FRET partner for tryptophan with a Förster distance of 28 Å.43 Pyrene fluorescence emission spectra can give two distance measurements: (1) the spatial distance of the probe from the tryptophan donor and (2) the spatial distance between two pyrene probes44, also known as excimers. The emission of the monomer of pyrene has peaks at 375, 385, 398 and 405 nm. A pyrene excimer has a broad emission band at 480–560 nm and a Förster distance of <10 Å. Another advantage of using pyrene probe is that it has negligible fluorescence in aqueous solutions.45 A single tryptophan residue in scaffolding protein was used as the fluorescence donor and an N-1-pyrene maleimide reacted with a single cysteine residue as the acceptor for the FRET pair.
Scaffolding protein has a single tryptophan residue, W134, which is positioned in the central region of the molecule. However, the tryptophan residue location was not ideal for our desired FRET distance analysis. An N-terminal tryptophan was more useful for this study, so the T10 residue was substituted by a tryptophan and the W134 was changed to a tyrosine. The T10W/W134Y scaffolding protein was assembly competent and formed procapsids similar to procapsids formed with the his-tagged WT scaffolding protein (data not shown). To attach pyrene-1-maleimide, cysteine substitutions were introduced in T10W/W134Y scaffolding protein at the same positions as described above. These proteins were are also found to be assembly competent (data not shown).
Based on previous studies of P22 scaffolding protein, it should have its two termini positioned in an anti-parallel manner.36,18 Therefore, we predicted the FRET measurements would give similar distance measurements if the probe was attached to residues close to the C-terminus or the N-terminus. In Figure 4, the emission spectra of pyrene via FRET from the tryptophan is shown. Observation of pyrene emission due to resonance energy transfer between T10W and K294C-pyrene or R303C-pyrene suggests that the C-terminal and N-terminal domains of scaffolding protein are in close proximity to one another. In addition, the T10W/W134Y/K294C-pyrene had a particularly intense excimer signal even at very low concentrations (0.05 mg/ml) (Figure 4a), pinpointing a possible strong interaction site for dimeric scaffolding protein, which was not identified previously. The excimer formation at 0.05 mg/ml was only seen with the T10W/W134Y/K294C-pyrene variant, confirming that the dimer formation cannot be attributed to stacking of hydrophobic dye molecules in aqueous solution. At 2 mg/ml, where we expect ~25% of scaffolding protein to be dimeric based on analytical ultracentrifugation (AU) experiments,25 other scaffolding protein cysteine variants exhibited only very weak excimer fluorescence, which is consistent with a parallel arrangement of dimeric scaffolding protein (Figure 4b).
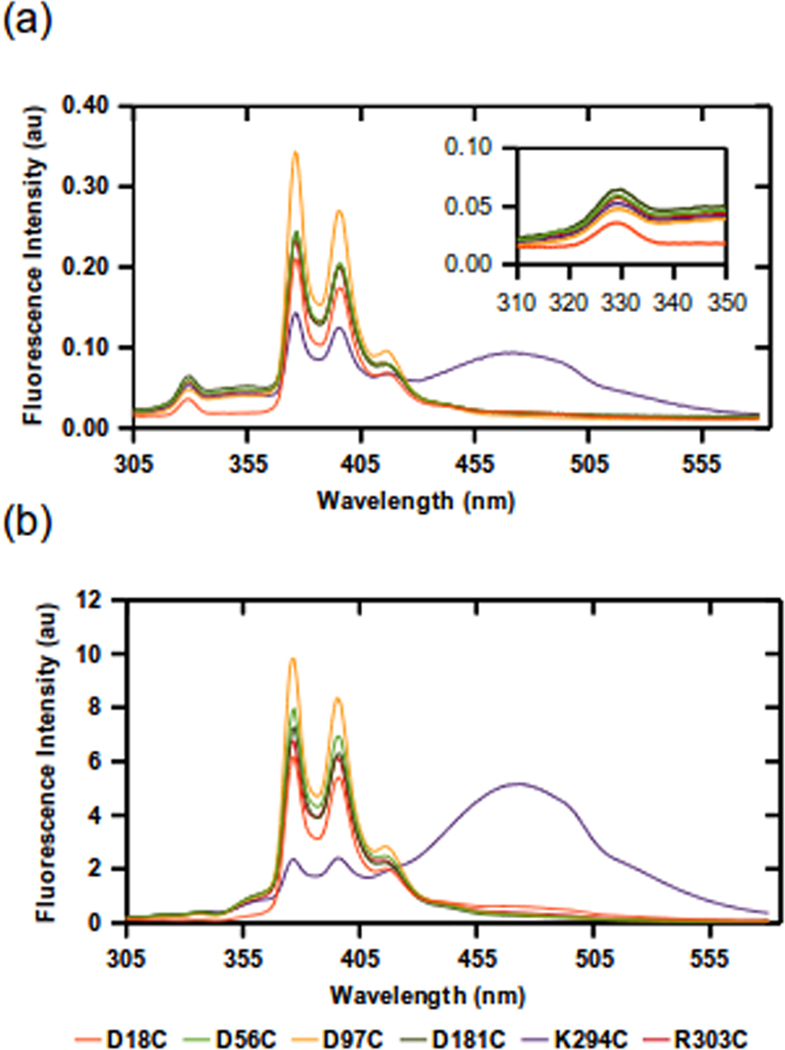
Figure 4.
Emission spectra of scaffolding protein T10W/W134Y with single cysteine variants labeled with N-1-pyrene-maleimide. Solutions of the scaffolding protein variants were excited at 295 nm and the emission was monitored from 305 nm to 580 nm. The measurements were done at (a) 0.05 mg/ml and (b) 2 mg/ml scaffolding protein. Inset in (a) show the tryptophan emission of labeled variants. All variants showed distinct pyrene monomer excitation peaks at 375 nm and 395 nm. The T10W/W134Y/K294C variant showed a prominent excimer band at ~470 nm with both concentrations of protein. The legend indicates location of the N-1-pyrene-maleimide labels.
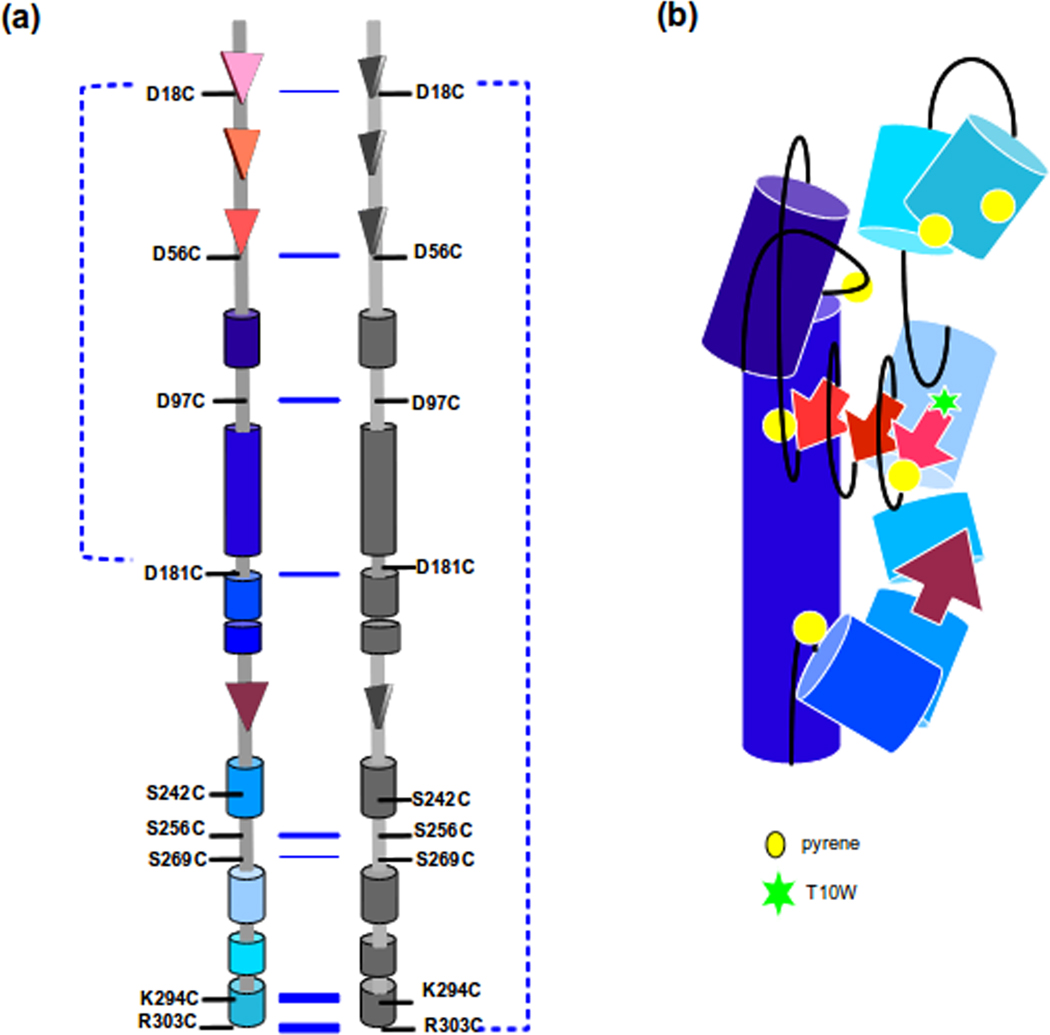
Anisotropy measurements of all 6 labeled variants were similar (data not shown), indicating that each of the pyrenes has similar rotational freedom. Analysis of the emission intensity of the tryptophan donor at ~340 nm show that T10W/W134Y/D18C had the lowest signal, which is expected since it has the shortest distance to the pyrene acceptor compared to other constructs (Figure 4a). Calculation of the efficiency of energy transfer indicated that each cysteine position was within 18–23 Å of T10W, suggesting an overall compact molecule with the C-termini and N-termini in proximate distance. In addition, these data suggest that the middle of the protein may be folded towards the N and C-termini.
Conformation of scaffolding protein in solution determined by crosslinking
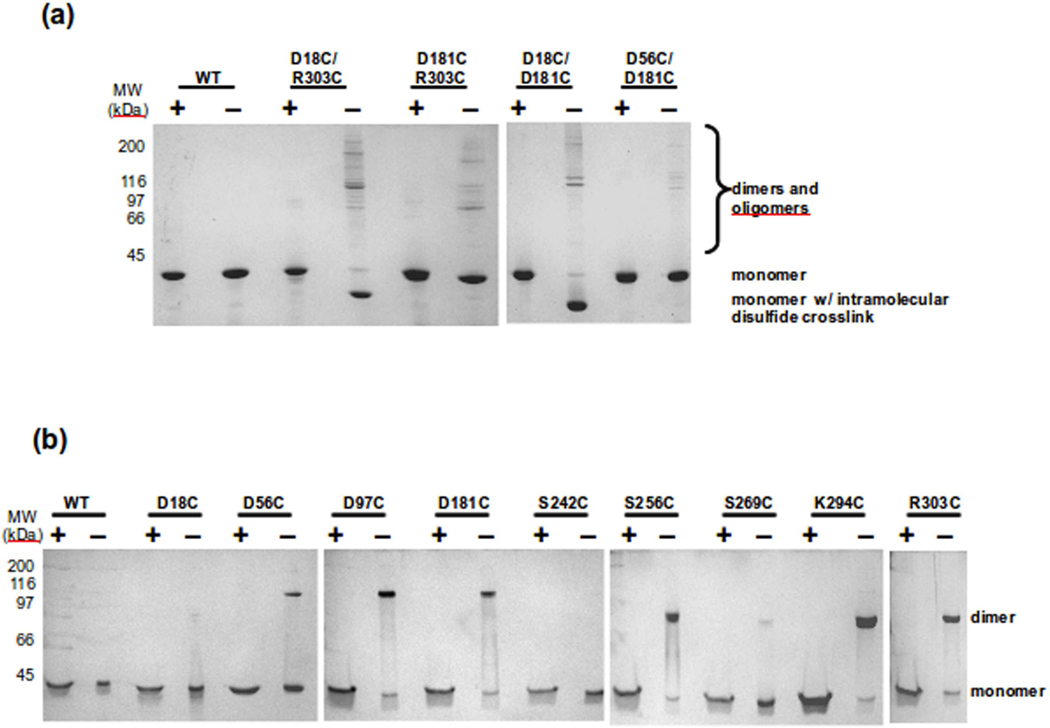
Our FRET data gave surprisingly similar distance results for amino acids that by primary sequence are rather distant. One trivial reason for this result was that because of its flexibility, different regions of scaffolding protein randomly would be close enough for energy transfer to occur. To test this possibility we required a method to determine if two regions were closer than 18–23 Å. Two cysteines can form a disulfide bond if they are less than ~6 Å apart. Therefore, to test if the N-terminus, C-terminus and other regions of scaffolding protein were near each other, double cysteine mutants were generated. We made D18C/R303C, D18C/D181C, D56C/D181C, and D181C/R303C. Each of the single cysteines was accessible to labeling with a maleimide compound (data not shown), indicating that none of these residues are buried. The purified proteins were allowed to oxidize at 0.5 mg/ml, where a solution of scaffolding protein should have 85% monomers and 14% dimers.25 This concentration was chosen for these experiments to be consistent with the sample concentrations used in assembly reactions. The proteins were visualized on SDS gels with and without reducing agent (Figure 5a). Scaffolding protein variants D18C/R303C and D18C/D181C showed strong intramolecular disulfide bonds in solution, seen as bands of faster mobility than monomeric scaffolding protein. Neither D56C/D181C or D181C/R303C were able to form an intramolecular disulfide, despite being accessible to cysteine reagents, suggesting that these regions do not closely interact. These data substantiates our hypothesis that the intramolecular disulfide bonds formed by the other double cysteine variants are specific interactions. The intramolecular disulfide bond observed with D18C/D181C shows that part of the middle domain of scaffolding protein is folded towards the N-terminus, but is somehow constrained from interacting with the C-terminus. These data also show that our FRET results were not simply due to flexibility in the protein, because not all cysteine variants were able to generate disulfide bonds as would be expected if the protein were completely flexible. Our data again suggest an additional complexity to the fold of scaffolding protein beyond the U-shaped model previously suggested.18
Figure 5.
Determination of the oligomeric state of scaffolding protein variants in solution. The scaffolding proteins were allowed to oxidized and samples were mixed with SDS loading buffer with (+) and without (−) β-mercaptoethanol. (a) Shows the double cysteine variants. The intramolecular disulfide crosslinks were found only in D18C/R303C and D18C/D181C. b) Shows the single cysteine mutants of scaffolding protein. D18C, S242C and S269C are the only variants not able to form disulfide bonds.
It can be deduced that scaffolding protein should form dimers in a parallel arrangement since when arranged in this fashion, the C-termini of each protomer would be accessible to interact with coat protein during assembly. However, there is little direct biochemical evidence to support this deduction. In addition, the orientation (i.e., face-to-face vs. side-by-side, etc.) of the protomers relative to one another is not known. To find additional constraints to the dimer model, we determined if single cysteine mutants could form intermolecular disulfide bonds. We generated additional single cysteines at positions S242, S256, S269, to increase the potential interaction sites. We found that 242 and 269 residues were inaccessible to labeling with a maleimide compound, suggesting that they are buried within the folded protein. All other cysteine residues could be completely labeled (data not shown). The scaffolding proteins were allowed to oxidize at 0.5 mg/ml and the ability of the single cysteine residues to form disulfide linked dimers in solution was assessed (Figure 5b). Strong dimer bands formed between all of the single cysteine residues except for positions D18C, S242C and S269C. The inability of the S242C and S269C proteins to form strong dimer bands is consistent with these positions being buried within the scaffolding core and indicates that scaffolding protein may be more tightly folded than previously thought.46 The observation the D18C scaffolding protein does not form a prominent dimer band suggests that the N-termini of the adjacent dimers must be turned away from one another. In addition, this result shows that the formation of disulfide bonded dimers is not simply due to random collisions in solution but is due to specific dimer interactions. The data also indicate that the dimer subunits must be face to face.
Orientation of scaffolding protein inside procapsids
Our understanding of the organization of P22 scaffolding protein inside procapsids is rudimentary. Between 60–300 scaffolding proteins are found within an assembled procapsid; however, only 60 of these are essential for assembly and are tightly bound.47,48,49 The remaining scaffolding proteins are much more loosely associated within the capsid. Wild-type procapsids can be treated with a low concentration of guanidine HCl to produce empty procapsid “shells” that have been stripped of the internal scaffolding protein. The isolated shells retain the same morphology as that of procapsids.7,50 When scaffolding protein is added to the shells in solution, the scaffolding protein re-enters through the holes in the procapsid lattice.15
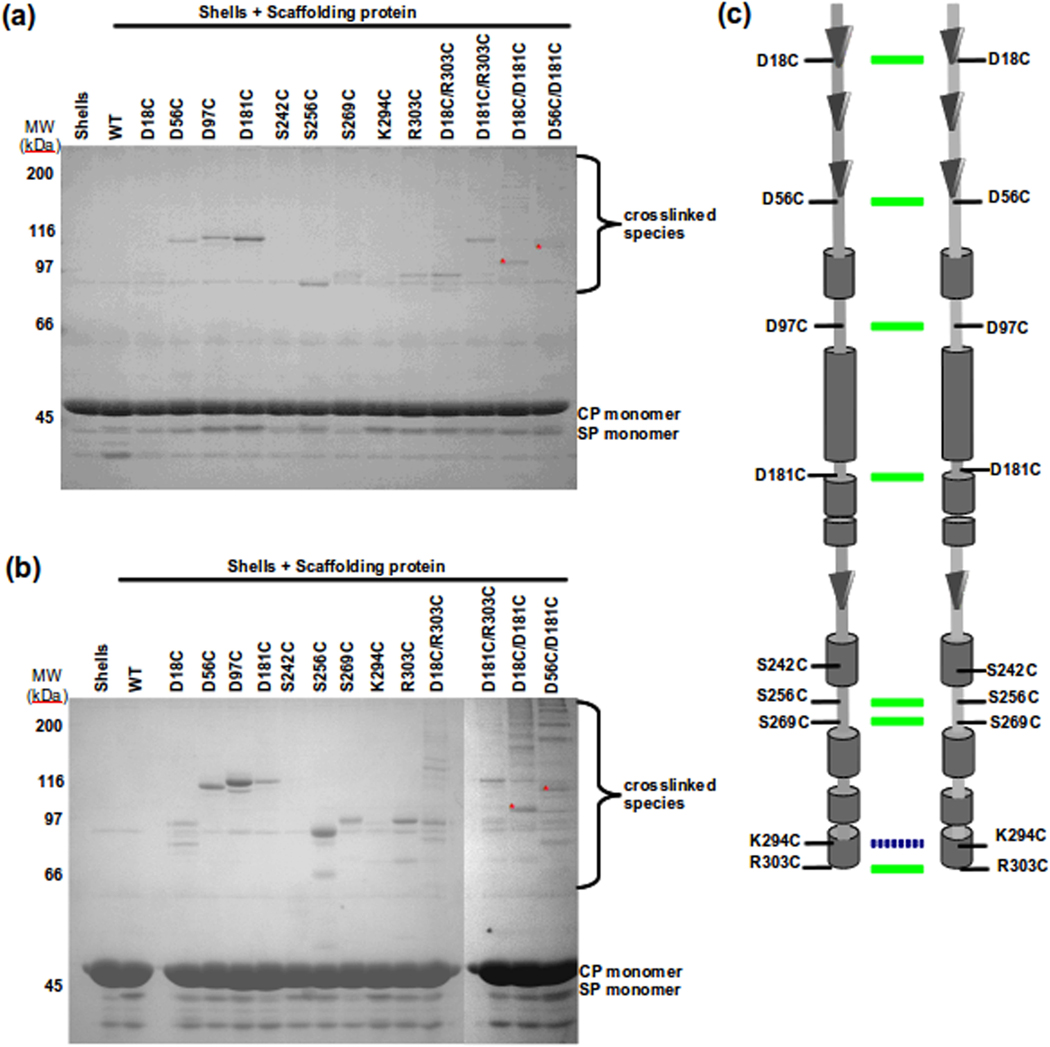
We have investigated the conformation of scaffolding protein within procapsids using the refilling procedure described above, which allows us to strictly control the scaffolding protein concentrations inside procapsids. In our experiments, we used a very low scaffolding protein concentration so that the refilling would result in at most 60 tightly bound scaffolding proteins per shell. We removed any free scaffolding protein from the solution using column chromatography. After re-entry and purification, the proximity of the single cysteine variant scaffolding proteins to each other was first probed using the cysteine reactive crosslinker, BMDB, which has a crosslinking arm length of ~10 Å. One caveat to this experiment would be that P22 coat protein has a single cysteine residue. However, previous studies have shown that the cysteine is not available for crosslinking or labeling.51,52 After crosslinking, the samples were run on SDS-PAGE (Figure 6a). In this gel, bands from ~90 – 125 kDa are due to scaffolding protein crosslinked dimers. The dimer bands were confirmed to be composed of only scaffolding protein by in-gel cleaving of the BMDB crosslinked samples with 30 mM NaIO4 (data not shown). The mobility of these dimers is likely altered from the expected MW of a scaffolding dimer because of the crosslinking. Each of the single cysteines, with the exception of S242C and K294C, showed crosslinking, indicating that the cysteines on two adjacent scaffolding proteins are within ~10 Å of one another. The observation that K294C does not crosslink is consistent with this residue a part of the C-terminal helix involved in interactions with coat protein, and therefore now inaccessible to the reagent. The result with the S242C variant is in agreement with the low crosslinking propensity of this protein in solution, again suggesting it is buried. The same results were observed with BM(PEG)2, which has a crosslinker arm length of 14.7 Å (Figure 6b). D18C and S269C each crosslink with BMBD and BM(PEG)2 even though these variants only generated a low proportion intermolecular disulfide bonded dimers in solution. It is possible that these were now able to form a crosslink with the crosslinking reagents because the crosslinking arm is longer than a disulfide bond. In a control experiment, these proteins were incubated with the BMBD or BM(PEG)2 crosslinkers in solution at 0.5 mg/ml (data not shown). There was still no crosslinking observed for S242C, while D18C and S269C were able to form crosslinked products, suggesting a distance >6 Å between adjacent residues. The data so far are consistent with the parallel face to face arrangement of scaffolding protein observed in solution.
Figure 6.
Assessing the arrangement of scaffolding protein in procapsids. Shells were refilled with scaffolding protein variants and crosslinked with (a) BMDB (arm length 10 Å) or (b) BM(PEG)2 (arm length 14.7 Å). The SDS gel band patterns suggest a parallel arrangement of scaffolding protein inside procapsids. The top labels indicate cysteine variants of scaffolding protein. The lane labeled as shells were without scaffolding protein. WT is wild type scaffolding protein. The position of CP (coat protein), SP (scaffolding protein) is indicated next to the gel. A summary of both gels is shown in (c) with a schematic model of scaffolding protein interactions in procapsids. The green colored lines between scaffolding dimer indicated the ability to form crosslinks in shells with BMDB and BM(PEG)2 while the broken blue colored line indicates ability to form crosslinks in shells only with BM(PEG)2. The secondary structure is based on prediction from SABLE.37
Conformational rearrangement of scaffolding protein in procapsids
To determine if the conformation of scaffolding protein within procapsids remains the same as in solution, the refilling assay was done with the double cysteine variants of scaffolding protein, described above (Figure 6b). With BMDB crosslinker, the D18C/D181C and the D56C/D181C variants showed unique dimeric bands that were not present in the crosslinking pattern of the respective single cysteine variants. In contrast, the bands in the D18C/R303C and D181C/R303C are the simple addition of the bands seen in their single cysteine variants. These data suggest that the scaffolding protein residues that crosslink are aligned in parallel (Figure 6c). However, there is a new intermolecular crosslink formed by D56C/D181C, which is not observed in solution, suggesting a conformational rearrangement of scaffolding protein within procapsids. Intriguingly, no intramolecular disulfide bonds were observed with any of the variants. When BM(PEG)2 was used to probe the double cysteine variants, the SDS-PAGE profile was similar to that generated by BMDB. However, there are additional bands seen for some variants. Even with a longer crosslinking arm, intramolecular crosslinking of scaffolding protein inside procapsids was not observed, even with the D18C/R303C and D18C/D181C variants which form strong intramolecular disulfides in solution, again suggesting conformational rearrangements inside, in particular, movement of the N-terminal domain.
Discussion
Scaffolding proteins are required for most dsDNA viruses to properly form precursor capsids and they also contribute to the stability of the capsid structure. 48,53,54 P22 scaffolding protein is one of the most well characterized. Previous studies on deletion mutants of scaffolding protein suggested a model of two elongated domains composed mostly of helices connected by unstructured regions.24,38 In this study we shed light on the possible orientation of the domains of scaffolding protein, both in solution and within procapsids.
Shape of scaffolding protein in solution
According to Raman spectroscopy studies24 P22 scaffolding protein is a helical protein that is loosely organized without a fixed core. Although scaffolding protein may not have a fixed core, this does not preclude the hypothesis that scaffolding protein maintains a structure and is not an inherently disordered protein. To investigate the arrangement of the different domains of P22 scaffolding protein, we introduced cysteine substitutions in different regions of the P22 scaffolding protein for attachments of probes and crosslinkers. Some cysteine substitutions introduced at different locations in the molecule that were inaccessible to labeling, which suggests that some regions of the protein are not exposed the solvent, inferring a more compact structure than previously proposed.
The specificity of the disulfide bond formation or chemical crosslinking argue that dimer formation occurs via specific interactions. Sedimentation experiments done by Tuma et al 38 showed that the 141–303 scaffolding protein fragment exists in a monomer:dimer equilibrium, suggesting the dimer interface lies within this region. Further details from our FRET and crosslinking data adds that the K294 site contributes to the dimer interface in solution. The excimer data also emphasizes the compact nature of this dimer interface, which must have a distance < 10 Å between molecules. However, crosslinking data of scaffolding protein inside procapsids suggests this residue is not positioned adjacent to another K294C probably because it is located in the C-terminal helical region of the scaffolding protein is involved with coat protein interaction.
Our data indicate a distinct intramolecular fold and a symmetric dimer interface. The scaffolding protein double cysteine mutants suggest a structure wherein part of the central region is bent towards the N-terminus and is constrained from interacting with the C-terminus. The N-terminus and the C-terminus appear to be juxtaposed to each other in solution, as both the FRET and the crosslinking analysis indicated that the N-terminus interacts with the C-terminus. The proximity of the two ends of scaffolding protein supports the hypothesis that one of the functions of the N-terminus is to modulate the affinity of the C-terminus with coat protein. Previous data from Parker and colleagues36 showed that there was an increase in aberrant structures in in vitro assembly with coat and a truncated scaffolding protein 141–303 mutant, which they hypothesized is caused by decreased regulation of coat protein and scaffolding protein assembly. Also, the N-terminus may have the release signal for scaffolding protein during DNA packaging as shown by data from the scaffolding protein deletion mutant 58–303, which is retained inside virions.18
Interaction of scaffolding proteins inside procapsids
Previous studies with cryo-electron microscopy of P22 procapsids have not been able to detect the orientation of scaffolding protein, leading to the hypothesis that scaffolding protein is not icosahedrally arranged.11 The recent cryo-electron microscopy reconstruction by the Chiu group35 showed that the C-terminus of scaffolding protein interacts with every coat protein. In a hexon, scaffolding protein was modeled to interact with the N-arm of coat protein and four of the six scaffolding proteins were shown to also interact with the tip of the A-domain of the adjacent coat protein. Using the model developed by Chiu's group35, the distances between adjacent coat protein N-arms is around 45 Å, and A-domain tip to A-domain tip is about 20 Å. Our data from crosslinking scaffolding protein inside procapsids supports the hypothesis that scaffolding protein could be aligned close to one another. However our data shows a much closer interaction between scaffolding proteins, at a distance ≤10 Å, which are oriented parallel to one another inside the procapsids. Chen et al.35 modeled the C-terminal helix-turn-helix of scaffolding into a density perceived as scaffolding protein, but at their resolution the side chains cannot be assigned and thus the exact sequences are not known. Therefore, we propose that some of the scaffolding density might be due to the interaction of different regions of scaffolding protein, such as the amino-terminus.
Evidence for conformational changes of scaffolding protein with assembly
All scaffolding protein cysteine variants were active in the assembly of procapsids in vitro, albeit each had a different rate of assembly. The rates of assembly of nearly all engineered scaffolding protein variants described here showed a higher rate than that of wild-type scaffolding protein or the mutants modified with NEM to eliminate disulfide crosslinking. These data indicate that the cysteine crosslinking to form covalent dimers is the probable reason for higher assembly rates, and not the introduction of substitutions. A previous study of a P22 scaffolding protein R74C/L177I showed that there is an increase in the rate of assembly of procapsids when the scaffolding protein is disulfide linked compared to the molecules in reducing conditions.25 The variation in the rates of assembly could be attributed to the different ratios of dimer to monomer of the samples under oxidizing conditions. In addition, previous SAX studies showed that the addition of excessive dimeric scaffolding protein leads to incomplete procapsid assembly. However, when monomeric scaffolding protein is added with dimeric scaffolding protein and an excess coat protein is added, the assembly reaction advances to completion. These results suggest that the affinity of coat protein for dimerized scaffolding protein is higher than that for monomeric scaffolding protein.39 Our data is consistent with these prior results, with the exception of D18C and D181C, which were not as good as WT-his scaffolding protein at promoting assembly. In solution, D18C is weakly able to form dimers and promote assembly of procapsids. Conversely, D181C is quite able to make disulfide-linked dimers. We hypothesize that conformational rearrangements may be required during assembly that are precluded by a disulfide between position 181 on adjacent protomers, which is consistent with our chemical crosslinking experiments that show that position 181 undergoes rearrangement upon assembly. The scaffolding proteins with double cysteines under non-reducing conditions also have higher rates of assembly than wild-type scaffolding protein or the NEM treated variants. The exception is D181C/R303C, which is probably caused by an unfavorable conformation or decreased flexibility of the protein in its dimer form that restricts the assembly competence. This result suggests that the flexibility of certain domains of the scaffolding protein is probably requisite for proper assembly.
By combining data from FRET and crosslinking, we have identified key interactions within the monomer of scaffolding protein and provide more details of its structural features. We propose a model that is consistent with a previous model, derived from analysis of deletion mutants, of two elongated helices positioned anti-parallel to each other.18 The current model we are proposing is that scaffolding protein in solution is an elongated protein with its N-terminus and C-terminus in close proximity to each other, while part of the middle domain folds towards N-terminus (Figure 7b). Given that part of the N-terminal region is juxtapositioned between part of the middle domain and the C-terminus, this suggests a flexible arm that moves in proximity of both regions of the scaffolding protein. The importance of the flexibility of certain regions in scaffolding protein is emphasized by the decreased rate of assembly of some of the double cysteine mutants. Our data indicate that inherent flexibility of scaffolding protein is required for it to function in assembly coat protein into procapsids, as well as exit during phage maturation.46 We also propose that the scaffolding protein dimers in solution are formed with the two molecules arranged in parallel with each other and have the C-terminus as part of the dimer interface Conformational rearrangements of scaffolding protein within procapsids are proposed to poise it for release upon DNA packaging. Since scaffolding protein takes part in several rounds of assembly, its ability to change conformation in response to its role in different steps in assembly is critical for its function.
Figure 7.
A schematic model of scaffolding protein in solution. The secondary structure assignment is based on prediction by SABLE.37 The tertiary model is based on FRET and crosslinking. The crosslinking interactions in solution are depicted in (a) with the increasing blue line thickness representing higher tendency to form intermolecular disulfide crosslinks, while the broken lines indicate the observed intramolecular disulfide crosslinks. (b) The model shown is the proposed fold of scaffolding protein in solution as a monomer. The colors of each domain in (b) correspond to the same domain in (a)
Materials and methods
Site directed mutagenesis, expression and purification
The N-terminally hexa histidine-tagged scaffolding protein cysteine mutants were produced by site directed mutagenesis using the primers listed in Table 1. The correct sequences were confirmed by our in house DNA sequencing facility. The template vector used is the pET15b/gp8 which encodes for the 6X his-tag scaffolding protein with a P259H substitution found in some P22 stocks. This construct of scaffolding protein has been previously used and shown to have similar assembly activity to non-his tagged WT scaffolding protein.
Table 1.
Primer table
| 5' | 3' | |
|---|---|---|
| D18C | CTTAACCCTGTCCGGCTGTCATGCAGCGGCATCT | |
| AGATGCCGCTGCATGACAGCCGGACAGGGTTAAG | ||
| D56C | GGACGATGAGACAGCACCAAAACAATGCCCGGCAAAGAAC | |
| GTTCTTTGCCGGGCATTGTTTTGGTGCTGTCTCATCGTCC | ||
| D97C | GTTTACGGGTAAACCCTTGCCTTCCTCCTCAGCCAG | |
| CTGGCTGAGGAGGAAGGCAAGGGTTTACCCGTAAAC | ||
| D181C | GAAAAGCTCAACATCCCTTGCTATCAGGAGAAAGAAG | |
| CTTCTTTCTCCTGATAGCAAGGGATGTTGAGCTTTT | ||
| S242C | GCTGATTGAACTCACTCGACTATGTGAACGCTTAACTCTCAA | |
| CTTGAGAGTTAAGCGTTCACATAGTCGAGTGAGTTCAATCAG | ||
| S256C | CGCGGTAAACAAATCTCTTGCGCTCCCCATG | |
| CATGGGGAGCGCAAGAGATTTGTTTACCGCG | ||
| S269C | GCCTATTACCGGTGATGTCTGTGCAGCAAATAAAGATGCCA | |
| TGGCATCTTTATTTGCTGCACAGACATCACCGGTAATAGGC | ||
| K294C | GGGAGATGTGGAAACCTACCGCTGTCTAAAGGCAAAACTTAAAGG | |
| CCTTTAAGTTTTGCCTTTAGACAGCGGTAGGTTTCCACATCTCCC | ||
| R303C | GCAAAACTTAAAGGAATCTGCTAATAGGATCCGGCTGC | |
| GCAGCCGGATCCTATTAGCAGATTCCTTTAAGTTTTGC | ||
| T10W | CACCGAAATTCAGGCATGGGAAGACTTAACCCTGTC | |
| GACAGGGTTAAGTCTTCCCATGCCTGAATTTCGGTG | ||
| W134Y | CAATGCTGCTAATACCGAATATCTAATGAAAGCGCAGGACG | |
| CGTCCTGCGCTTTCATTAGATATTCGGTATTAGCAGCATTG | ||
All cysteine mutants were expressed in E. coli BL21 cells. A colony from each clone was inoculated in 10 ml LB broth with 100 µg/ml ampicillin and allowed to grow overnight at 30°C. One liter of LB broth with 100 µg/ml ampicillin was inoculated with the overnight cultures and allowed to grow to an O.D. at 600 nm of 0.6 at 37°C, and then induced with 0.1 mM IPTG for 4 hours at 37°C. Cells were then harvested at 4300 rpm for 15 mins, 4 °C and then stored at −20°C. To purify the scaffolding protein, harvested cells were frozen and thawed one time, then suspended in lysis buffer (20 mM Na2HPO4, 300 mM NaCl, 10 mM imidazole and 30 mM phenylmethanesulfonylfluoride). The cell suspension was sonicated for 6 mins, then centrifuged at 10000 rpm, for 10 mins to remove cell debris. The supernatant was loaded onto a Talon® column (Clontech) pre-equilibrated with lysis buffer. The Talon column was washed with lysis buffer, wash buffer (20 mM Na2HPO4, 300 mM NaCl and 25 mM imidazole) and elution buffer (20 mM Na2HPO4, 300 mM NaCl and 250 mM imidazole). The purified protein was concentrated against dry PEG 20000 and then dialyzed against 10 mM Na2HPO4 pH 7.6. The purity was determined by 10% SDS PAGE.
In vitro assembly reactions
Bacteriophage P22 procapsids shells were prepared as previously described.25,55 To make monomeric coat protein, intact P22 procapsid shells were dissociated in buffer containing 6.8 M urea in 10 mM Na2HPO4 pH 7.6 for 30 mins at room temperature, then diluted with same volume 10 mM Na2HPO4 pH 7.6 to 1 mg/ml. The coat protein solution was then dialyzed extensively against 10 mM Na2HPO4 pH 7.6. The samples were centrifuged at 60000 rpm for 20 mins at 4°C, the supernatant was extracted. The concentration of the monomeric coat protein was determined by absorbance at 280 nm.
Procapsid assembly reactions were monitored by mixing 0.5 mg/ml monomeric coat protein with 0.5 mg/ml of scaffolding protein in a SLM Aminco-Bowman 2 spectrofluorometer. The reaction was monitored by the increase in 90° light scattering at 500 nm, with the excitation and emission with bandpass set to 4 nm. The assembly reactions were monitored for 16 mins at 20°C.
The assembly reactions were allowed to reach equilibrium overnight at room temperature, mixed with agarose gel sample buffer (40 mM Tris acetate, 1 mM EDTA, 20% glycerol) and loaded onto 1.0% Seakem LE agarose gel made with the same buffer without glycerol. The gels were run at 100 V for 40 min at room temperature and stained with coomassie blue.47,56
Electron Microscopy
Assembled procapsids were prepared by placing 4 µL of samples on a 400 mesh carbon formvar copper EM grids and allowed to adsorb for 1 min. The grids were then rinsed with 3–4 drops of dH2O followed by staining with 4 µL of 1% uranyl acetate for 1 min. Excess stain was blotted off with filter paper and the grids were allowed to air dry. The prepared grids were viewed in a Philips model 300 TEM with a magnification of 56700X, and operated at an accelerating voltage of 80kV.
Fluorophore labeling and fluorescence measurement
Scaffolding protein cysteine variants were prepared by mixing 150 µM protein with 10 mM Na2HPO4 and 2 mM TCEP for 30 minutes at room temperature. N-(1-pyrene)-maleimide (Molecular Probes) in DMSO was added to the reaction at an approximate 1:2 (mole:mole) ratio of scaffolding protein to the dye and the reaction was incubated for 2 hours. The samples were then filtered through Zeba spin columns (Pierce) to remove unreacted N-(1-pyrene)-maleimide. Samples were then dialyzed with 10 mM Na2HPO4 overnight at 4°C. Protein concentrations were determined by Pierce 660 nm protein assay, using WT scaffolding protein as standard. The degree of labeling with pyrene-maleimide was determined by absorbance measurements at 343 nm with ε = 37500 M−1cm−1 for pyrene57 and absorbance at 280 nm with ε = 17420 M−1cm−1 for scaffolding protein. The degree of labeling was ~100%, and never >100%, which suggests low probability of non-specific labeling.
Fluorescence measurements were performed using a SLM Aminco-Bowman 2 spectrofluorometer. All FRET measurements were made at an excitation of 295 nm and emission scan from 305 nm to 580 nm. The band passes for excitation and emission were 2 nm and 4 nm, respectively. Fluorescence anisotropy was measured at an excitation of 340 nm and emission of 375 nm with both band passes at 4 nm.
Crosslinking scaffolding protein in shells
P22 procapsid shells were re-filled with scaffolding protein variants in buffer with 10 mM Na2HPO4, 45 mM NaCl and 2 mM TCEP-HCl. The reactions were allowed to equilibrate overnight at room temperature. Crosslinkers were then added at an approximate 25:1 (mole:mole) ratio of crosslinker to scaffolding protein. Samples were incubated for 1 hour at room temperature, and the reaction was stopped with 25 mM L-cysteine. To remove the remaining free scaffolding protein, 25 µL of Talon beads were mixed with the reaction mixture. The solution was mixed for 30 minutes, and then centrifuged at 700 xg for 2 mins. Reducing sample loading buffer was added to the supernatant and the samples were then separated in 10% SDS PAGE. The gels were stained with Coomasie blue.
Cleavage of the BMDB crosslink
In gel BMDB cleavage was perfumed as described by manufacturer (Pierce). Briefly, dimer bands were excised from SDS gels and soaked in 20 mM triethanolamine-HCl, pH 7.5 (TEA) for 2 hrs at room temperature with multiple buffer changes. The gel strips were then soaked in TEA buffer with 30 mM NaIO4 for 30 mins at room temperature. The gel strips were incubated in 300 mM NH2OH-HCL, 0.1%SDS (w/v) pH 7.5. The samples were then equilibrated with SDS non-reducing sample buffer for 10 mins. The gel strips were loaded across a 4–15% SDS-PAGE. The gels were stained with SYPRO red (Invitrogen).
Research Highlights.
The conformation of scaffolding protein was studied in solution and inside procapsids.
Our study used FRET and crosslinking to examine the structure.
The N-terminus is modeled in proximity to both the C-terminus and a middle domain.
Inside procapsids, scaffolding protein rearranges to a more open conformation.
Scaffolding proteins are arranged in parallel inside procapsids.
Acknowledgments
We thank Peter Prevelige, Sherwood Casjens and members of the Teschke lab for their helpful comments and suggestions. This work was supported by grant GM076661 to CMT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
G. Pauline Padilla-Meier, Department of Molecular and Cell Biology, University of Connecticut, Storrs, CT, 06269.
Carolyn M Teschke, Department of Molecular and Cell Biology, and Chemistry, University of Connecticut, Storrs, CT, 06269.
References
- 1.Fane BA, Prevelige PEJ. Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 2.Dokland T. Scaffolding proteins and their role in viral assembly. Cell Mol Life Sci. 1999;56:580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King J, Griffin-Shea R, Fuller MT. Scaffolding proteins and the genetic control of virus shell assembly. Q Rev Biol. 1980;55:369–393. doi: 10.1086/411981. [DOI] [PubMed] [Google Scholar]
- 4.King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E. Structure and assembly of the capsid of bacteriophage P22. Philos Trans R Soc Lond B Biol Sci. 1976;276:37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- 5.Prevelige PEJ, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J Mol Biol. 1978;126:721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- 7.Thuman-Commike PA, Greene B, Malinski JA, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazinet C, King J. Initiation of P22 procapsid assembly in vivo. J Mol Biol. 1988;202:77–86. doi: 10.1016/0022-2836(88)90520-7. [DOI] [PubMed] [Google Scholar]
- 9.Earnshaw W, Casjens S, Harrison SC. Assembly of the head of bacteriophage P22: x-ray diffraction from heads, proheads and related structures. J Mol Biol. 1976;104:387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- 10.Prasad BV, Prevelige PE, Marietta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- 11.Thuman-Commike PA, Greene B, Jakana J, Prasad BV, King J, Prevelige PEJ, Chiu W. Three-dimensional structure of scaffolding-containing phage p22 procapsids by electron cryo-microscopy. J Mol Biol. 1996;260:85–98. doi: 10.1006/jmbi.1996.0383. [DOI] [PubMed] [Google Scholar]
- 12.Dokland T, McKenna R, Ilag LL, Bowman BR, Incardona NL, Fane BA, Rossmann MG. Structure of a viral procapsid with molecular scaffolding. Nature. 1997;389:308–313. doi: 10.1038/38537. [DOI] [PubMed] [Google Scholar]
- 13.King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J Mol Biol. 1973;80:697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- 14.Prevelige PEJ, King J. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog Med Virol. 1993;40:206–221. [PubMed] [Google Scholar]
- 15.Greene B, King J. Binding of scaffolding subunits within the P22 procapsid lattice. Virology. 1994;205:188–197. doi: 10.1006/viro.1994.1634. [DOI] [PubMed] [Google Scholar]
- 16.Greene B, King J. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology. 1996;225:82–96. doi: 10.1006/viro.1996.0577. [DOI] [PubMed] [Google Scholar]
- 17.Greene B, King J. In vitro unfolding/refolding of wild type phage P22 scaffolding protein reveals capsid-binding domain. J Biol Chem. 1999;274:16135–16140. doi: 10.1074/jbc.274.23.16135. [DOI] [PubMed] [Google Scholar]
- 18.Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. Molecular genetics of bacteriophage P22 scaffolding protein's functional domains. J Mol Biol. 2005;348:831–844. doi: 10.1016/j.jmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Casjens S, Adams MB, Hall C, King J. Assembly-controlled autogenous modulation of bacteriophage P22 scaffolding protein gene expression. J Virol. 1985;53:174–179. doi: 10.1128/jvi.53.1.174-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casjens S, Adams MB. Posttranscriptional modulation of bacteriophage P22 scaffolding protein gene expression. J Virol. 1985;53:185–191. doi: 10.1128/jvi.53.1.185-191.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyckoff E, Casjens S. Autoregulation of the bacteriophage P22 scaffolding protein gene. J Virol. 1985;53:192–197. doi: 10.1128/jvi.53.1.192-197.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- 23.Thomas GJJ, Li Y, Fuller MT, King J. Structural studies of P22 phage, precursor particles, and proteins by laser Raman spectroscopy. Biochemistry. 1982;21:3866–3878. doi: 10.1021/bi00259a023. [DOI] [PubMed] [Google Scholar]
- 24.Tuma R, Thomas GJJ. Mechanisms of virus assembly probed by Raman spectroscopy: the icosahedral bacteriophage P22. Biophys Chem. 1997;68:17–31. doi: 10.1016/s0301-4622(97)00001-x. [DOI] [PubMed] [Google Scholar]
- 25.Parker MH, Stafford WF, 3, Prevelige PEJ. Bacteriophage P22 scaffolding protein forms oligomers in solution. J Mol Biol. 1997;268:655–665. doi: 10.1006/jmbi.1997.0995. [DOI] [PubMed] [Google Scholar]
- 26.Ziegelhoffer T, Yau P, Chandrasekhar GN, Kochan J, Georgopoulos C, Murialdo H. The purification and properties of the scaffolding protein of bacteriophage lambda. J Biol Chem. 1992;267:455–461. [PubMed] [Google Scholar]
- 27.Mesyanzhinov VV, Sobolev BN, Marusich EI, Prilipov AG, Efimov VP. A proposed structure of bacteriophage T4 gene product 22--a major prohead scaffolding core protein. J Struct Biol. 1990;104:24–31. doi: 10.1016/1047-8477(90)90053-f. [DOI] [PubMed] [Google Scholar]
- 28.Poh SL, el Khadali F, Berrier C, Lurz R, Melki R, Tavares P. Oligomerization of the SPP1 scaffolding protein. J Mol Biol. 2008;378:551–564. doi: 10.1016/j.jmb.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 29.Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BAL, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- 30.Parker MH, Jablonsky M, Casjens S, Sampson L, Krishna NR, Prevelige PEJ. Cloning, purification, and preliminary characterization by circular dichroism and NMR of a carboxyl-terminal domain of the bacteriophage P22 scaffolding protein. Protein Sci. 1997;6:1583–1586. doi: 10.1002/pro.5560060722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Krishna N. 1H and 15N chemical shift assignments of a carboxy-terminal functional domain of the bacteriophage P22 scaffolding protein. Magnetic Resonance in Chemistry. 1999;37:602–604. [Google Scholar]
- 32.Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PEJ, Krishna NR. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J Mol Biol. 2000;297:1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- 33.Agirrezabala X, Martín-Benito J, Castón JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thuman-Commike PA, Greene B, Malinski JA, Burbea M, McGough A, Chiu W, Prevelige PEJ. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys J. 1999;76:3267–3277. doi: 10.1016/S0006-3495(99)77479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker MH, Casjens S, Prevelige PEJ. Functional domains of bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- 37.Adamczak R, Porollo A, Meller J. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins. 2005;59:467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- 38.Tuma R, Parker MH, Weigele P, Sampson L, Sun Y, Krishna NR, Casjens S, Thomas GJJ, Prevelige PEJ. A helical coat protein recognition domain of the bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:81–94. doi: 10.1006/jmbi.1998.1916. [DOI] [PubMed] [Google Scholar]
- 39.Tuma R, Tsuruta H, French KH, Prevelige PE. Detection of intermediates and kinetic control during assembly of bacteriophage P22 procapsid. J Mol Biol. 2008;381:1395–1406. doi: 10.1016/j.jmb.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parent KN, Doyle SM, Anderson E, Teschke CM. Electrostatic interactions govern both nucleation and elongation during phage P22 procapsid assembly. Virology. 2005;340:33–45. doi: 10.1016/j.virol.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 41.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr Opin Struct Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Annalen der Physik. 1948;437:1–112. [Google Scholar]
- 43.Lakowicz JR. Principles of fluorescence spectroscopy. 3rd edition. New York, NY: Springer; 2006. p. 44. [Google Scholar]
- 44.Jung K, Jung H, Wu J, Privé G, Kaback H. Use of site-directed fluorescence labeling to study proximity relationships in the lactose permease of Escherichia coli. Biochemistry. 1993;32:12273–12278. doi: 10.1021/bi00097a001. [DOI] [PubMed] [Google Scholar]
- 45.Patel AB, Khumsupan P, Narayanaswami V. Pyrene fluorescence analysis offers new insights into the conformation of the lipoprotein-binding domain of human apolipoprotein E. Biochemistry. 2010;49:1766–1775. doi: 10.1021/bi901902e. [DOI] [PubMed] [Google Scholar]
- 46.Tuma R, Prevelige PEJ, Thomas GJJ. Structural transitions in the scaffolding and coat proteins of P22 virus during assembly and disassembly. Biochemistry. 1996;35:4619–4627. doi: 10.1021/bi952793l. [DOI] [PubMed] [Google Scholar]
- 47.Teschke CM. Aggregation and assembly of phage P22 temperature-sensitive coat protein mutants in vitro mimic the in vivo phenotype. Biochemistry. 1999;38:2873–2881. doi: 10.1021/bi982739f. [DOI] [PubMed] [Google Scholar]
- 48.Parent KN, Zlotnick A, Teschke CM. Quantitative analysis of multi-component spherical virus assembly: scaffolding protein contributes to the global stability of phage P22 procapsids. J Mol Biol. 2006;359:1097–1106. doi: 10.1016/j.jmb.2006.03.068. [DOI] [PubMed] [Google Scholar]
- 49.Parker MH, Brouillette CG, Prevelige PEJ. Kinetic and calorimetric evidence for two distinct scaffolding protein binding populations within the bacteriophage P22 procapsid. Biochemistry. 2001;40:8962–8970. doi: 10.1021/bi0026167. [DOI] [PubMed] [Google Scholar]
- 50.Fuller MT, King J. Assembly in vitro of bacteriophage P22 procapsids from purified coat and scaffolding subunits. J Mol Biol. 1982;156:633–665. doi: 10.1016/0022-2836(82)90270-4. [DOI] [PubMed] [Google Scholar]
- 51.Kang S, Prevelige PEJ. Domain study of bacteriophage p22 coat protein and characterization of the capsid lattice transformation by hydrogen/deuterium exchange. J Mol Biol. 2005;347:935–948. doi: 10.1016/j.jmb.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 52.Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prevelige PEJ, Thomas D, King J. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J Mol Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 54.Wang S, Chang JR, Dokland T. Assembly of bacteriophage P2 and P4 procapsids with internal scaffolding protein. Virology. 2006;348:133–140. doi: 10.1016/j.virol.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 55.Fuller MT, King J. Purification of the coat and scaffolding proteins from procapsids of bacteriophage P22. Virology. 1981;112:529–547. doi: 10.1016/0042-6822(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 56.Gope R, Serwer P. Bacteriophage P22 in vitro DNA packaging monitored by agarose gel electrophoresis: rate of DNA entry into capsids. J Virol. 1983;47:96–105. doi: 10.1128/jvi.47.1.96-105.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holowka DA, Hammes GG. Chemical modification and fluorescence studies of chloroplast coupling factor. Biochemistry. 1977;16:5538–5545. doi: 10.1021/bi00644a023. [DOI] [PubMed] [Google Scholar]