Abstract
This unit describes the method of following phosphoinositide dynamics in live cells. Inositol phospholipids have emerged as universal signaling molecules present in virtually every membrane of eukaryotic cells. Phosphoinositides are present only in tiny amounts compared to structural lipids but are metabolically very active as they are produced and degraded by the numerous inositide kinase and phosphatase enzymes. Phosphoinositides control the membrane-recruitment and activity of many protein signaling-complexes in specific membrane compartments and have been implicated in the regulation of a variety of signaling and trafficking pathways. It has been a challenge to develop methods that allow detection of phosphoinositides at the single cell level. The only available technique in live cell application is based on the use of the same protein domains selected by evolution to recognize cellular phosphoinositides. Some of these isolated protein modules when fused to fluorescent proteins can follow dynamic changes in phosphoinositides. While this technique can provide information on phosphoinositide dynamics in live cells with subcellular resolution and rapidly gained popularity, it also has several limitations that must be taken into account when interpreting the data. Here, we summarize the design and practical use of these constructs and also review important considerations for the interpretation of the data obtained by this technique.
Keywords: phosphoinositide, green fluorescent protein, pleckstrin homology domain, fluorescence microscopy, live-cell imaging, FRET
INTRODUCTION
The first established role of phosphoinositides was recognized in the mid 80’s as precursors for two important second messengers, the calcium mobilizing inositol 1,4,5-trisphosphate and the protein kinase C activator diacylglycerol (Berridge and Irvine, 1984). However, starting with the discovery of PI 3-kinases (Whitman et al., 1988), several new inositol lipid isomers have been identified and a much broader role of these lipids as organizers of membrane-associated signaling complexes were uncovered (Di Paolo and De Camilli, 2006). The highly compartmentalized nature of inositol lipid signaling processes demanded new methods that allow detection of the presence of the various inositide isomers with subcellular resolution. The idea of how this can be achieved was born out from discoveries that identified protein modules capable of inositol lipids recognition with reasonable specificity in proteins that are regulated by phosphoinositides (Lemmon et al., 1997).
Visualization of phosphoinositide changes in single living cells, was then based on the premise that protein modules that possess high enough affinity and specificity to bind the inositide headgroup of specific phosphoinositides can find the lipids within the cell and visualize it when expressed in the form of a green fluorescent protein (GFP) fusion protein (Balla et al., 2000). As simple as it sounds, the short history of this method has already raised several important technical and theoretical questions that one need to be aware of when attempting to use these methods. We have divided this unit into Protocols that give some practical guidance to users who are not so experienced with live cell microscopy. We also included chapters that deal with theoretical considerations and interpretational issues highly relevant to these measurements. The protocols will not detail common laboratory practices but try to concentrate on aspects that are unique to these applications. We would like to emphasize that the technical comments are intended for less experienced users and not for the experts of this research field.
2. CHOOSING THE FUSION PROTEINS FOR DETECTION OF SELECTED INOSITOL LIPIDS
Table I summarizes protein domains that have been used for imaging purposes. Some of these are better established, while a consensus has yet to be reached for others. These protein domains are widely available and also can be easily duplicated. We are discussing the most important issues related to the individual lipid species for which we have accumulated experience.
Table I.
Phosphoinositide binding modules in use for imaging purposes in live cells.
The usefulness of these domains for imaging purposes is questionable (see (Michell et al., 2005).
The OSH2-PH shows little discrimination between PtdIns4P and PtdIns(4,5)P2 based on in vitro binding (Yu et al., 2004) and it is still not certain whether it actually reports on both of these molecules in some proportions.
PtdIns(4,5)P2
Almost all imaging work for this lipid has used the PLCδ1PH-GFP construct developed independently in the Meyer lab and in our group (see Refs in Table I). This construct expresses very well and decorates the plasma membrane and some vesicular structures but no other organelles. This has raised the question of whether PtdIns(4,5)P2 is only present in detectable amounts in the plasma membrane, or the probe is biased against the plasma membrane pool of the lipid. Few reliable works have compared PLCδ1PH-GFP distribution with staining with PtdIns(4,5)P2 antibodies, and some found the lipid in the Golgi with antibody staining but not with the PLCδ1PH-GFP (Matsuda et al., 2001), while others saw no discrepancy and no Golgi staining with PtdIns(4,5)P2 antibody either (Hammond et al., 2006). Also, EM studies showed some PtdIns(4,5)P2 in the Golgi using GST-fused PLCδ1PH-GFP post fixation (Watt et al., 2002). These data demonstrate one of the possible limitation of this method, namely, that not all pools of the lipids might be equally seen by the domain and this will be even more apparent when imaging other inositol lipid forms (see below).
In spite of this lingering question, the PM pool of PtdIns(4,5)P2 can be monitored by the PLCδ1PH-GFP. This probe very nicely reports changes in this lipid either following PLC activation or after degradation by a phosphoinositide-sensitive 5-phosphatase (Stauffer et al., 1998; Várnai and Balla, 1998; Varnai et al., 2006). However, there is another question that complicates what these changes mean when following PLC-mediated hydrolysis of these lipids. Since PLCδ1PH-GFP recognizes the phosphorylated inositol headgroup within the lipid, the corresponding soluble inositol phosphate (in this case Ins(1,4,5)P3 ) can compete with the membrane PtdIns(4,5)P2 for binding the PH-domain GFP fusion protein. Because of this competing effect, some research groups treat the changes in PLCδ1PH-GFP membrane localization as a faithful index of InsP3 elevations rather than lipid changes within the cell (Nash et al., 2001). We have discussed this topic in detail in a recent review (Varnai and Balla, 2006) and will not elaborate on it further here. The bottom line is that inositol phosphates can compete to the lipid binding of the PH-domain constructs and their effects on changing localization could be quite significant under certain conditions and cannot be ignored. At the same time, it would be just as misleading to treat the PLCδ1PH-GFP translocation response as an index of InsP3 change, and differences between the two have been demonstrated recently by simultaneous measurements of InsP3 and PLCδ1PH-GFP translocation (Matsu-ura et al., 2006). Another PtdIns(4,5)P2 binding module called the Tubby domain was described from the Tubby protein (Field et al., 2005; Santagata et al., 2001; Yaradanakul and Hilgemann, 2007). Two recent studies using a full-length Tubby protein (Nelson et al., 2008) or a mutant form of the Tubby domain has showed that these domains are less sensitive to InsP3 changes than the PLCδ1PH-GFP (Nelson et al., 2008; Quinn et al., 2008).
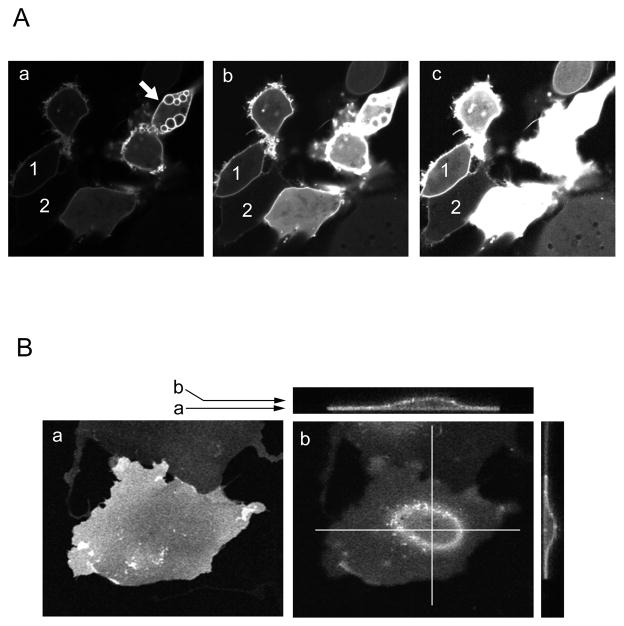
The third complication to remember when using these methods is the inhibitory effect of the expressed domain on cellular responses regulated by the inositol lipids. Binding of the PH (or other) domain-GFP reporters to phosphoinositides should inhibit the lipid-mediated cellular process since it competes with the lipid binding of endogenous effectors. Indeed, high expression of the PLCδ1PH-GFP causes morphological changes that include rounding of the cells, losing attachment and development of intracellular vesicles (Fig. 1). These “toxic” effects are due to the inhibition by the construct of the connections between the cytoskeleton and the plasma membrane (Raucher et al., 2000). This problem can be minimized by choosing cells that express low levels of the protein and using a sensitive microscope that can resolve the weak signals that such cells will have (Fig. 1A).
Figure 1.
Cellular localization of the PLCδ1PH-GFP. (A) Clusters of HEK293 cells transfected for 24h, and expressing the domain at various levels. Note the vesicular structures in the cell pointed to by the arrow that expresses high level of the fusion protein. This is an example of the toxic effects of the protein. Also note that with these illumination settings (panel a), cells 1 and 2 are not even visible, yet these are the cells that one should chose for analysis as shown by the higher illumination that already saturates the signal from the other cells (b and c). (B) Confocal imagaes of a COS-7 cell trasnfected with PLCδ1PH-GFP for 24 h and analyzed by z-sectioning. Panel a shows an image taken close to the bottom of the cell where it attaches to the coverslip. Note that there is no sharp outline of the cell and the signal covers the whole area of the cell. In panel b, the picture is taken at a z-plane higher up in the cell and again, there is no clear outline of the plasma membrane. Compare it with HEK293 cells that are not as flat and show a clear image of the plasma membrane (A). The side views of this COS-7 cell at the cross-sections (top and right side) show better the plasma membrane localization and the shape of the cell.
PtdIns4P
Several PH domains have been used to detect PtdIns4P in living cells. The two most popular ones are the PH domains of the OSBP (oxysterol binding protein) and FAPP1 (four-phosphate-adaptor protein) that were first identified as specific binders to PtdIns4P by fat blots (Dowler et al., 2000). Based on lipid vesicle binding assays, however, these domains not only bind PtdIns4P but also PtdIns(4,5)P2 (Levine and Munro, 1998; Roy and Levine, 2004; Yu et al., 2004). These PH domains, as well as their close relative found in the CERT (ceramide transfer) protein, were indeed localized to the Golgi pool of PtdIns4P both in yeast and in mammalian cells (Levine and Munro, 2002), but not to the plasma membrane, where PtdIns(4,5)P2 is abundant. These data suggest that within the cells these PH domains do not recognize PtdIns(4,5)P2 efficiently. Moreover, it was also found that both the OSBP – and FAPP1-PH domains require active (GTP-bound) Arf1 for Golgi localization and their Golgi targeting requires binding to both PtdIns4P and Arf1-GTP (Levine and Munro, 2002). Neither interaction alone is sufficient for efficient membrane recruitment. This also means that brefeldin A treatment that prevents the formation of Arf1-GTP in the Golgi causes release of these PH domains from these locations. The limited lipid binding specificity and the need for additional protein interaction for membrane targeting make these probes less than optimal for lipid imaging in live cells. However, because of the lack of better ones they have been used in many laboratories including ours (se Table 1 for references).
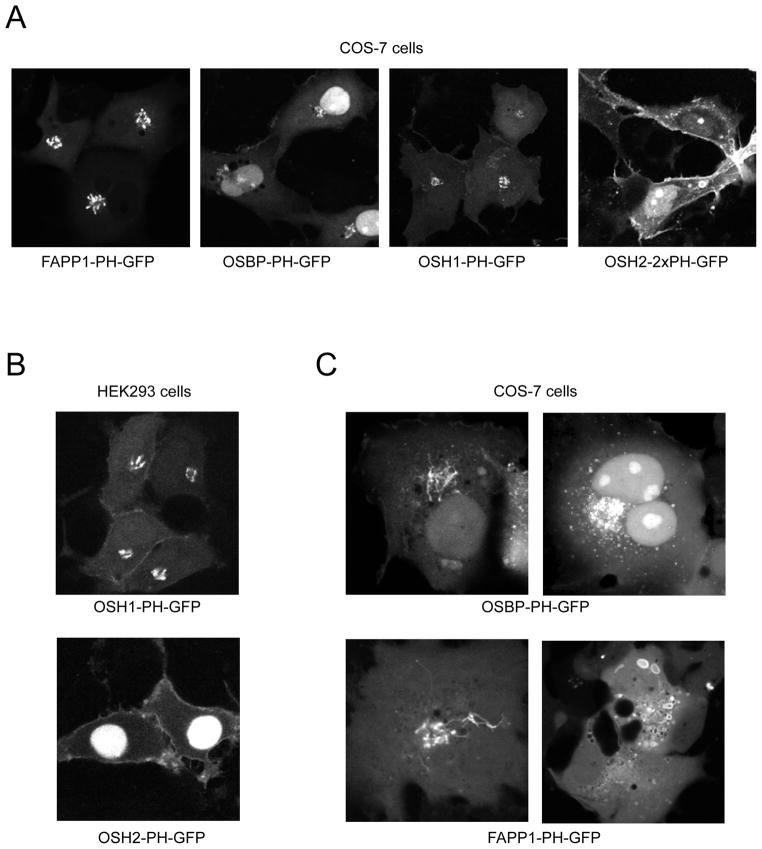
Two additional PH domains, namely those of OSH1 and OSH2 (oxysterol binding protein homologues of S. cerevisiae) were found to show cellular localization consistent with binding to PtdIns4P in yeast (Roy and Levine, 2004; Yu et al., 2004). Remarkably, the OSH2-PH (used in tandem to increase its apparent affinity) showed both the plasma membrane and the Golgi pool of PtdIns4P in yeast, while the OSH1 PH domain only detected the pool in the Golgi (Roy and Levine, 2004; Yu et al., 2004). This spatial discrimination was the more surprising as these PH domains showed very little discrimination between PtdIns4P and several other inositides including PtdIns(4,5)P2 in various in vitro lipid binding assays (Yu et al., 2004). Studies in our laboratory on mammalian cells with these two PH domains yielded somewhat different results: while the OSH1-PH is found as a very good marker for PtdIns4P in the Golgi (as it was in the yeast), the OSH2-2xPH (or the single PH domain) construct only localizes to the plasma membrane but does not show Golgi localization (Fig. 2C). In spite of its very limited in vitro lipid binding specificity, the OSH2-2xPH appears to be biased toward PtdIns4P over PtdIns(4,5)P2 in the plasma membrane, based on the resistance of its membrane localization to phosphoinositide 5-phosphatases that eliminate PtdIns(4,5)P2 (Balla et al., 2007). The extent of this discrimination as well as the mechanism underlying it (interaction with other proteins that would restrict access to PtdIns(4,5)P2 but not PtdIns4P), however, needs further investigations.
Figure 2.
Localization of the various domains used for imaging PtdIns4P in COS-7 and HEK293 cells. (A) COS-7 cells transfected with the indicated domains for 24 h. Note the sharp contrast and prominent recruitment of the FAPP1-PH domain and the higher nuclear staining of the OSBP-PH domain. The yeast OSH1-PH-GFP also shows the Golgi but is also localized to a small extent to the plasma membrane (better seen on panel B). The OSH2-2xPH-GFP prominently labels the plasma membrane but does not show Golgi localization. For this picture cells were selected that are not so flat, do demonstrate better the plasma membrane localization. (B) Localization of the OSH1-PH-GFP and OSH2-PH-GFP constructs in HEK293 cells. Note the Golgi and moderate plasma membrane localization of the OSH1-PH and the lack of Golgi localization and high nuclear signal with the OSH2-PH domain. The nuclear localization is less prominent with the OSH2-2xPH-GFP construct. (C) Examples for interference of the domains with cellular functions. Both the OSBP- and FAPP1-PH domains cause tubulation of the Golgi. Remarkably, this always occurs at moderate level of expression and never at high expression levels and only in a fraction of cells. This indicates that this effect is conditional and requires a certain functional state of the Golgi. At high expression levels, the two constructs have very different effects: the OSBP-PH brakes the Golgi to small vesicles that eventually cover the whole cytoplasm. These are completely resistant to brefeldin A, a treatment that rapidly eliminates the Golgi localization of the construct indicating the Arf1-GTP requirement of the localization (not shown). In contrast, the FAPP1-PH domain shows no Golgi localization at high expression levels and instead produces large vacuoles in the cell with FAPP1-PH domain attached to their limiting membranes. (C)
Taken all these data together, it is clear that only specific pools of PtdIns4P can be monitored with these domains and there is not a single domain identified as yet that would detect all PtdIns4P pools within a cell. In fact, we have not found a domain that recognizes the PtdIns4P produced by type-II PI 4-kinases on endosomes. Even within the Golgi, PtdIns4P is produced by different PI 4-kinases (De Matteis et al., 2005) and it is possible that the different PH domains do not detect these pools equally. Moreover, there is an effect of the overexpression of the domains on the Golgi itself. For example, the FAPP1-PH localizes primarily in the trans-side of the Golgi (Godi et al., 2004), but its localization between the cis- and trans- side depends on the expression level (Weixel et al., 2005). In COS-7 cells, increased level of expression of FAPP1 and OSBP causes quite distinct morphological changes (Fig. 2B) suggesting that they interact with distinct proteins (in addition to PtdIns4P and Arf1) indicating that even though they may appear in the same Golgi compartment at low expression level, they still detect functionally distinct pools of the lipids. These are all important signs to indicate that not all PtdIns4P are created equal and cannot simply be imaged by a single probe.
PtdIns(3,4,5)P3
There are a large number of studies imaging PtdIns(3,4,5)P3 dynamics due to the high interest in PI 3-kinase signaling and its role in polarized cell movements such as chemotaxis. In Dictyostelium, a widely used model for polarization migration, the PH domain of the CRAC protein (cytosolic regulator of adenyl cyclase, not to be mistaken with calcium release activated channels as the same acronym is often used in mammalian cells) has been a very good reporter of PtdIns(3,4,5)P3 distribution (Dormann et al., 2002; Huang et al., 2003). In mammalian cells, the Akt-PH domain has served best for following polarized PtdIns(3,4,5)P3 production (Servant et al., 2000), even though this PH domain also recognizes PtdIns(3,4)P2. Curiously, the Btk-PH domain that is more specific for PtdIns(3,4,5)P3 and detects the lipid increases after PDGF or insulin stimulation in the plasma membrane (Varnai et al., 1999) has not seen similar popularity presumably because it does not show this polarization so effectively. This already suggests that in addition to PtdIns(3,4,5)P3, protein-protein interactions probably also play a role in the effective recruitment of these PH domains to the plasma membrane Because of its good in vitro specificity, the Grp1-PH domain (or its close relative, ARNO-PH) is considered to be the most specific probe to detect PtdIns(3,4,5)P3. However, our experience is not so positive with this construct. In many cells it shows a relatively poor response, it shows high nuclear localization (independent of PtdIns(3,4,5)P3) and its membrane recruitment is largely dependent on active (GTP-bound) Arf6 (Cohen et al., 2007). Probably because of this latter feature, the Grp-PH is also relatively toxic to the cells and inhibits several Arf6-dependent functions including attachment and spreading of cells (Varnai et al., 2005). A recent study compared several PtdIns(3,4,5)P3 binding PH domains for their in vitro binding specificity and cellular localization response (Manna et al., 2007). This study found strikingly different membrane recruitment kinetics between the various domains in PDGF-stimulated NIH3T3 cells also suggesting that in addition to inositide lipid binding, membrane penetration and possibly protein-protein interactions have a role in membrane association of the domains. Based on all of the measurements presented in that study, the Btk-PH appeared to be the best probe for the detection of PtdIns(3,4,5)P3 in intact cells.
PtdIns3P
This was one of the first inositol lipid for which a recognition domain other than a PH domain was found. In fact, no PH domain has been reported as of to date, that recognize PtdIns3P specifically. It was the FYVE domain (the acronym originates from the first letter of the first four proteins in which the domain was described: Fab1p, YOTB, Vac1p and EEA1) that was found to be responsible for PtdIns3P recognition and shown to recognize this lipid in its isolated form fused to GFP (Burd and Emr, 1998). Since the strictly defined FYVE domain will recognize PtdIns3P very specifically in vitro, but poorly localizes without some adjacent residues that help its dimerization (and partially may also bind Rab5), the most widely used construct from the Stenmark lab is based on a tandem FYVE domain of the Hrs protein (Gillooly et al., 2000). The construct we have made from the EEA1 (Early Endosome-Associated Antigen) FYVE domain is the slightly longer version (Hunyady et al., 2002). Both of these constructs decorate an early endosomal compartment and will fall off from the vesicles upon inhibition of PI 3-kinases consistent with the view that the Class III PI 3-kinase (the mammalian Vps34p homologue) constitutively generates PtdIns3P on early endosomes.
The other domain that recognizes PtdIns3P is the PX domain of several proteins, but most prominently that of p40phox (Kanai et al., 2001). Expression of this PX domain as a GFP fusion protein also labels the early endosomes. However, in contrast to the FYVE domains, this construct leads to the accumulation of very brightly stained aggregated vesicles in many cells that express higher amounts of the protein. Moreover, these bright vesicles do not lose their signal after addition of PI 3-kinase inhibitors (while the solitary vesicles labeled in cells with low expression do) suggesting that the PX domain is part of a more stable protein complex that cannot disassemble and is probably responsible for the aggregation of vesicles. This again is an indication of protein-protein interactions that clearly differ between the FYVE and PX domains, and a good example of why cells with low expression levels should be used in these studies.
PtdIns(3,4)P2, PtdIns(3,5)P2 and PtdIns5P
We have not had much experience with visualizing any of these lipids, therefore, we can only summarize what is known to us from the literature. The only PH domains that showed in vitro specificity to binding PtdIns(3,4)P2 was those of the TAPP1 and TAPP2 proteins (Dowler et al., 2000). The crystal structure of the TAPP1 PH domain clearly revealed the structural features responsible for this specificity (Thomas et al., 2001). GFP-tagged TAPP1 PH domain has been shown to detect the lipid in live cells in the plasma membrane under conditions where PtdIns(3,4)P2 but not PtdIns(3,4,5)P3 was elevated (Kimber et al., 2002). In addition, a GST-fused TAPP1 PH domain labeled the membrane of internal vesicles and the multivesicular body in fixed cells processed for EM analysis (Watt et al., 2004). Moreover, this domain does not show membrane association when only PtdIns(3,4,5)P3 is elevated, indicating that it, indeed, can discriminate between these two otherwise closely related lipid products within the cell. Other studies also found this domain useful in detecting the formation of PtdIns(3,4)P2 in phagocytic cups in macrophages (Horan et al., 2007). The PX domain of p47phox has also been claimed as a PtdIns(3,4)P2 recognizing module (Zhan et al., 2002) and used to detect the lipid as a GFP fusion protein (Stahelin et al., 2003). However, in our hand the p47phox PX domain-GFP chimera does not show any indication of binding to membranes in a lipid-dependent manner (our unpublished observations) and its binding to other phospholipids such as phosphatidic acid (Karathanassis et al., 2002) and proteins (Zhan et al., 2002) may limit the usefulness of this PX domain as an imaging tool.
The formation of PtdIns(3,5)P3 and its role in trafficking to the vacuole in yeast and to the multivesicular body in higher eukaryotes (Gary et al., 1998; Ikonomov et al., 2003; Jefferies et al., 2008) has made this lipid a highly interesting target for imaging studies. Unfortunately, in spite of several attempts and claims there are no established tools to accomplish this task reliably. Two proteins, the yeast Ent3p (Friant et al., 2003) and Svp1p (Dove et al., 2004) have been found to be targets of PtdIns(3,4)P2. The inositide binding site was located within the ENTH domain of Ent3p, while it was attributed to a cluster of basic residues on a beta propeller within Svp1p. To our knowledge there have been no systematic analysis on the intracellular distribution of any isolated domains extracted from these molecules with the aim of localizing PtdIns(3,5)P2 in live cells.
PtdIns5P is a lipid that was first identified as a substrate of the type II PIP kinases (PI5P 4-kinase) (Rameh et al., 1997b). The main route(s) of its production in cells is highly debated, but most likely is the result of dephosphorylation of polyphosphoinositides (Coronas et al., 2007; Zou et al., 2007) rather than direct phosphorylation of PtdIns. The only domain that recognizes PtdIns5P has been identified in the nuclear protein ING2 within a domain (PHD as for plant homeodomain) that binds PtdIns5P, and to a lesser degree PtdIns3P, in vitro (Gozani et al., 2003). A GFP fusion protein made of the 3xPHD domain of ING2 detected PtdIns5P in the plasma membrane in response to overexpressed bacterial 4-phosphatase, IpgD (Gozani et al., 2003; Pendaries et al., 2006) and did not show endosomal localization suggesting that it does not bind PtdIsn3P in the cells. Nevrtheless, more experience is needed with this domain to determine where PtdIns5P is found in the cells and whether its role in the nucleus is associated with detectable changes in its nuclear level.
The common message from these examples is that none of these tools are useful as general lipid reporters without specific constraints that vary from probe to probe. However, if treated with caution and after performing proper controls, these reporters are very useful to answer questions that no other techniques can provide.
3. CHOOSING THE FLUORESCENT PROTEIN TO BE FUSED
Most of these fluorescent reporters have been originally created as fusion proteins with the enhanced green fluorescent protein (EGFP). However, over the past few years, a large number of fluorescent proteins were introduced now offering a wide variety of colors and other unique features. In addition to the spectral variants that allow multicolor imaging of several probes in parallel, new features include photoactivation, photoswitching, spectral change during maturation, pH stability, resistance to photobleaching or dimerization etc. These have been summarized in several recent reviews and will not be detailed in this unit(Giepmans et al., 2006; Shaner et al., 2005; Wiedenmann and Nienhaus, 2006). These proteins are often very useful for a particular application but there is considerable confusion as to the source of these proteins. In addition to the GFP variants based on the pioneering work of the Tsien laboratory, many new proteins originate from the Miyawaki laboratory, and the company, Evrogen. These proteins have been cloned from a variety of species and are not derivatives of the jellyfish Aequorea Victoria GFP. A good summary of these proteins and their features are found in (Muller-Taubenberger and Anderson, 2007). However, it would be a mistake to believe that these proteins only differ in their optical behavior and they can be replaced with one another without any problem. Our experience is that from time to time a probe behaves quite differently depending on whether an EGFP, an mRFP, or EosFP molecule is attached to it. Therefore, it is better to start with a few colors that are proven to work similarly with a particular lipid binding domains than to generate a whole series of colors assuming that they will behave identically within the cells. As most studies were done with the pEGFP plasmids and their initial color versions it is wise to stay with these for initial experiments. Unfortunately, the original pEGFP-N and –C series plasmids are no more available since Clontech has become part of Takara and these companies now offer their own fluorescent proteins that originate from a different species. The original EGFP and its color variants are now sold by Invitrogen in a different plasmid backbone. This often causes confusion among users who just begin collecting their fluorescent proteins and do not realize that a green fluorescent molecule now can mean different entities depending on the company that sells it.
BASIC PROTOCOL 1. EXPRESSION OF FLUORESCENT PROTEINS IN MAMMALIAN CELLS
The following procedures describe general methods for preparing to analyze cells expressing EGFP or GFP-fusion proteins by microscopic techniques.
Materials
Cultured cells of interest
6-well culture plates
Coverslips (PG Science, cat: 60–4884–25) or (Warner Instruments, Cat: 64–0705) for TIRF applications
Poly-L- lysine 0.1% (w/v) in H2O (Sigma, P-8920)
Plasmid DNA (usually a Midiprep)
Transfection reagents (individual choice depending on the cells in use)
Culture Medium with serum and antibiotics (depending on the cells)
-
2% paraformaldehyde
Paraformaldehyde should be made up freshly by dissolving the appropriate amount of EM grade paraformaldehyde in PBS and heating (in a chemical hood) until the aldehyde goes into solution. Keep the bottle cap loosened so that pressure does not build up. Cool down to 20°C and pH to 7.4. Aqua Poly/Mount (Polysciences Inc.)
Clear Nail polish
Prepare poly-lysine-coated cover slips
Because most plasticware used for cell culture work has high autofluorescence, we recommend that cells be cultured and transfected on poly-lysine-coated glass cover slips so that they can be viewed in the microscope after transfection without replating. For certain cell types and transfections, it is more advantageous to transfect the cells in culture dishes and re-plate the transfected cells on the poly-lysine-coated coverslips before microscopy.
-
1
Rinse 25-mm coverslips with 98% ethanol in the cell culture hood and air dry.
-
2
Place one coverslip in each well of 6-well culture dish.
-
3
Add 1 ml of 1:50 or 1:100 dilution of Poly-lysine in deionized water (sterile filtered) to cover the entire surface and allow it to stand at room temperature for 1 h.
Plate cells on coverslips coated at each concentration to determine which is best for the cells being studied. -
4
Aspirate the solution from the coverslip and air dry before plating cells.
Transfect cells with plasmid DNA
Transfection protocols are available for the different reagents, and for each cell type the reagent and the procedure that gives the best result can differ considerably. Therefore, we refer the user to the manufacturer’s instructions as far as specifics for cell transfection are concerned. The optimal level of expression has to be determined for each cell type and expression construct. It is important to remember that the expressed proteins often interfere with the functions of the lipids that they recognize, and in high concentrations the lipid-binding fusion proteins are often toxic to the cells.
We recommend that initially cells are also transfected with a mutant version of the lipid-binding domain that does not bind lipids. For example, many constructs localize to the nucleus, but this localization is not dependent on lipid binding; thus, lipid-mediated localization can be confirmed by comparing the distribution of the native lipid-binding domain to that of the non-lipid binding mutant. We also recommend transfecting cells with the GFP protein alone without the lipid-binding domain. These two controls help to track phenotypic changes and potential cellular toxicity associated with overexpression of the lipid-binding domain, as well as to serve as controls for monitoring localization that accurately reflects lipid binding.
-
5
Plate ~50,000 cells directly onto the poly-lysine-coated coverslips in 2 ml of the appropriate culture medium and grow to the density best suited for transfection (usually 2 days).
-
6
Transfect the cells with the desired plasmid DNA construct using a method that is most appropriate for the cells. We usually use Lipofectamine 2000 and 0.5 μg plasmid DNA per coverslip.
-
7
Grow the cells for 24 h to allow expression of the transfected protein.
-
8
Incubate cells in serum-free culture medium for 6 to 10 h to render them quiescent before microscopy.
We do not recommend growing the cells longer than 34 hours total after transfection, because of the potential toxicity of the lipid-binding fusion protein and the ability of the expressed protein to interfere with the functions of the lipids they bind. Not every experiment requires serum deprivation of cells.
Immunostain cells to detect expressed GFP fusion protein
This part can be skipped when doing live cell microscopy (see below). However, it may be necessary to confirm expression of the fusion protein, especially if the GFP signal is not bright enough. This confirmation can be accomplished by immunocytochemistry with antibodies to the GFP portion of the fusion protein. Co-localization of the GFP-fused domain with other molecular markers may also require immunostaining.
-
9
Rinse the transfected cells with 2 ml of PBS.
-
10
Add 2 ml of 2% paraformaldehyde and incubate for 10 min at room temperature.
-
11
Wash the cells three times with 2 ml of PBS for 10 min each.
-
12
Cover the cells with 2 ml of blocking solution (10% FBS in PBS made freshly) to block nonspecific antibody binding. Incubate for 10 min at room temperature.
-
13
Add the primary antibody diluted appropriately in blocking solution complemented with 0.2 % saponin. Incubate for 1 h at room temperature.
100 μl of diluted antibody should be sufficient for a 25-mm circular or 22 mm × 22 mm square coverslip, if it is inverted on a glass slide and incubated in a humidified Petri dish. -
14
Wash the cells three times with 2 ml of blocking solution for 5 min each wash.
-
15
Add the fluorescent secondary antibody diluted in the blocking solution containing 0.2 % saponin. Incubate for 1 h at room temperature protected from light.
-
16
Wash the cells three times with 2 ml of PBS.
-
17
Air-dry until the coverslips are only damp.
-
18
Mount the coverslips with the cells down on a glass slide using Aqua Poly/Mount.
-
19
Seal the coverslips on the side with clear nail polish to prevent drying.
BASIC PROTOCOL 2
In this protocol we describe how to determine whether the fluorescent protein construct remains intact when expressed in the cells. This is an important control in addition to sequencing the DNA construct before performing any microscopy work. Often the fusion protein is cleaved within the cell so that the green fluorescence is not coming from the molecule that was designed. Our experience is that free EGFP is more often present when using the pEGFP-N1 plasmids than with the pEGFP-C1-variant. (This can happen of an internal ribosomal entry site allows the translation of GFP itself, which can be prevented by removing the start codon of the original GFP). In one case we found that a protein expressed in COS-7 cells was cut in half when placed COOH-terminal to EGFP. We recommend analyzing cell lysates by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) either using a phosphorimager equipped with a blue laser line (described below) or by conventional Western blotting techniques using antibodies against the GFP portion of the fusion protein. We find that in many cases, samples that are boiled and analyzed by Western blotting show more degradation product than those analyzed directly from the gel. Curiously, the electrophoretic mobility of the EGFP molecule is clearly different when expressed from pEGFP-C1 versus pEGFP-N1. This may be due to altered migration of the non-denatured protein by extra amino acids encoded within the multiple cloning site of the pEGFP-C1 plasmid.
Materials
Cultured cells of individual choice
12-well culture plates
Plasmid DNA (usually a Midiprep made by the Qiagen kit)
Transfection reagents (individual choice depending of the cells in use)
Culture Medium with serum and antibiotics (depending on the cells)
1.5 ml Eppendorf tubes
SDS gel apparatus
Laemmli buffer
Phosphorimager (or reagents and apparatus for Western Blotting)
Confirm the integrity of the expressed protein
Seed COS-7 (or other cells to be studied) 2 × 105 cells/2ml onto 12-well culture plates and incubate for 24 h.
Transfect cells with the method of choice and incubate for 1 days (or for the time the experiments would be done).
Wash cells with 2 ml PBS and aspirate the PBS.
Dissolve cells in 100 μl Laemmli buffer and transfer to Eppendorf tubes. Do not boil, but briefly sonicate to disrupt DNA.
Load 40 μl of the sample per lane of a small (10 cm) SDS-PAGE gel with an acrylamide concentration that will resolve proteins in the size range of interest.
Run the gel with 100 V until the front reaches the bottom of the gel.
Remove gel from cassette and place in running buffer.
Place the wet gel directly on a phosphorimager and scan using the appropriate laser line (blue for GFP and Red for mRFP or similar colors)
If no PhosphorImager is available, the fluorescent proteins can be detected with Western Blotting using an anti-GFP (or other appropriate) antibody. We provide no protocol for this standard procedure. In this case, we do boil the samples before the SDS Gel analysis. It is also important to remember that the anti GFP antibody does not recognize proteins that are derived from other species than Aequorea Victoria.
4. OBSERVE THE GFP SIGNAL BY MICROSCOPY
In this part we give a few practical advice on what to pay attention to when observing cells by microscopy. These are not intended for experienced users but we often find that researchers who want to observe their proteins as GFP-fusion constructs in live cells has limited practical knowledge of microscopes. This protocol is not a substitute for training on any specific microscope system but rather an aid to help common-sense practices.
Wide field or confocal microscopy?
The first question to be answered is whether to view the cells with a conventional fluorescence microscope or with a confocal microscope. We suggest that the distribution of GFP fluorescence and other initial experiments assessing transfection efficiency etc. should be performed with a conventional fluorescence microscope using filters suitable for fluorescein isothiocyanate (FITC) detection (excitation at 470–490 nm, emission at 500–550 nm). It is not necessary to examine the cells immediately with a confocal microscope, and it is more difficult to get a general impression of what the cells look like when expressing the construct in a confocal microscope. Individual cells, especially COS cells, show enormous variability in their shape, size, and general appearance, and often the level of expression changes their appearance. Conventional fluorescence microscopy is a significantly more efficient way to browse through many cells and notice trends in cellular morphology. In addition, many cells are flat in culture (especially COS cells), so there is little benefit from analyzing the cells in a confocal microscope. Confocal microscopy can be saved for recording cells and changes in fluorescence distribution once the conditions have been optimized with a fluorescence microscope. A further advantage of viewing cells in a fluorescence microscope is that autofluorescence often can be distinguished from the GFP signal because its color is different from the green color of GFP. It is important to remember that confocal microscopes detect light intensity without colors and the “color” given is artificial. Therefore, in each case, the autofluorescence has to be determined so that the GFP signal can be reliably used. For this, observation of untransfected cells is a very useful control.
Fixed or live cells?
The next question is whether to analyze cells live or fixed. EGFP fluorescence can persist in fixed cells under proper fixation conditions described above (Basic Protocol 1, steps 9–19). Fixed cells can be processed for immunostaining, which is often necessary to determine co-localization with organelle markers for which antibodies are available. Also, fixed cells can be stored and studied whenever convenient. On the other hand, changes cannot be followed as they happen in real time when using fixed cells. Moreover, fixation and permeabilization procedures may distort cellular morphology. For example, we find that vesicular structures shrink during fixation, and long canaliculi can turn into small vesicles. The use of live cells definitely is the most reliable way of assessing undistorted morphology, but it is also the most time-consuming and least efficient. For live cell imaging, it is best to use an inverted microscope. New water-immersion objectives make it possible to look at cells in upright microscopes, but the objective has to be in the culture medium. In addition, live cell imaging would generally require some form of temperature control with all the complications associated with it while fixed cells do not.
When analyzing live cells, one major advantage is to record time-lapsed images sequentially after a stimulus is applied to the cells. In confocal microscopes the speed of scanning determines how fast one can record an image, and generally the faster is the scan, the poorer is the quality of the individual pictures. Finally, one of the greatest difficulties is to keep the cell in focus after stimulation, because of shape changes that often occur in response to the stimulus (HEK 293 cells are especially lively). This change in position of the originally imaged plane can make the whole recording unsuitable. We are aware of software programs being developed to compensate for “focal-drift” due to cell movements. These new developments may already be available for some confocal imaging systems.
BASIC PROTOCOL 3
In this protocol we describe steps that can help guide a less experienced user to navigate the live cell imaging process.
Materials and ecquipments
Chambers that can hold the coverslips. We use the metal Atto chambers from Invitrogen
Wide-field fluorescence microscope equipped with sensitive camera and appropriate software for data acquisition or confocal microscope.
Objective heater and heated stage or a complete temperature control enclosure.
Computer controlled valve-system and perifusion (optional)
-
Medium appropriate for the experiments. The modified Krebs-Ringer solution we use has the following composition:
NaCl, 120 mM;
KCl, 3.7 mM;
Na2HPO4, 1.2 mM;
CaCl2, 1.2 mM;
MgSO4, 0.7 mM;
Glucose, 10 mM;
Na-Hepes, pH 7.4, 20 mM;
Bovine serum albumin (BSA,) 0.1%
Forceps
Kim-Wipe paper
Lens cleaning paper
Immersion oil
Mount transfected cells
-
1
Place coverslip with transfected cells on upper side into metal chambers. Tighten the upper part with the O-ring in place gently but firmly so no leak occurs. Wash cells with medium and add 1 or 2 ml medium to the cells. Clean the bottom of the coverslip with a clean Kim-Wipe.
Make sure that the coverslip is in the grooved area otherwise it will brake during tightening. Test for leakage using a clean paper where the metal meats the glass coverslip. Leakage can get worse as the temperature changes on the stage. In any case the use of a lens protector against leakage is advised when using live cell imaging. -
2
Dust off the objective and clean with a cleaning solution provided by the manufacturer using a lens cleaning paper. Alternatively, use isopropanol for this purpose.
Cleaning the lens is important but it can do more damage than good if not done properly. Consult the expert before doing it. Make sure not to use Kim-Wipe or any other paper or cotton swab for this purpose. Never clean the lens with dry lens paper and do not use solutions containing ammonia (e.g. windex) or organic solvents other than recommended. -
3
Add one small drop of immersion oil to the lens without allowing it to flow down along the surface.
-
4
Place the metal chamber on the stage and slowly elevate the objective with eye control until it touches the coverslip.
Choose cells
Looking into the fluorescence microscope, one can usually see cells with a wide range of fluorescence intensities. Depending on the quality of the microscope and the intensity of the light source, sometimes only the cells with the highest expression levels are visible and these are the cells one should avoid. It is our rule to study cells in which the GFP signal is as low as possible but clearly distinguishable from the autofluorescence of untransfected control cells. Since microscopes are very different, there are no arbitrary rules but we recommend the following steps:
-
5
Observe the transfected cells and, if necessary, the control non-transfected cells using the fluorescence setting of the confocal microscope fitted with filters suitable for the chosen fluorophore. (470–490 nm excitation and 500 to 550 nm emission for EGFP).
Often a separate nontransfected cell control is not necessary, because even in the transfected samples, not all cells express the fusion proteins. The nontransfected cells in the population can usually be distinguished by their autofluorescence, allowing easy identification of transfected and nontransfected cells from the same sample when viewed using the fluorescent setting. -
6
Choose cells in which expression levels are just high enough to be resolved above the background autofluorescence, and that are not obviously unhealthy. A good quality image should be obtained in confocal microscopy using 3 to 5% of the maximum laser power (assuming a 30 mW 488 nm laser) (see Fig. 1A).
Analysis of cells that are unhealthy and suffer from toxicity induced by the expression of the fusion protein should be avoided. For example, cells that round up and are about to detach (a common phenomenon with cells expressing very high levels of a fusion protein consisting of the PH domain of PLC and GFP (PLCδ1PH-GFP) are not likely to behave normally. Another indication of toxicity is the appearance of large intracellular vesicles.
Time lapse analysis of live cells
An important and often unappreciated problem in live cell imaging is the proper temperature of the cells that are being observed. It should be remembered that even with a heated stage, an objective acts as a heat sink, keeping the cells in the observation field at a temperature near that of the objective. This temperature is closer to room temperature especially when rooms are kept on the cold side because of the lasers. Many processes in the cells slow down or do not work properly below 34 °C. Therefore, we recommend the use of an objective heater available from Bioptechs (http://www.bioptechs.com). Unfortunately, the heater collar does not fit all objectives, and heating may be damaging to the objective if it is warmed very fast from a cold temperature. Alternative methods of maintaining the proper temperature include perfusion of cells with a high flow of warm medium or the use of a hair dryer to keep the objective at the proper temperature. Complete incubator enclosures are also available from various companies that can keep both the temperature and CO2 concentration and humidity of cells on the stage at the desired levels. Unfortunately, they make manipulations of the cells often difficult.
-
7
Record and store an image before applying any stimulus.
-
8
Set the software to record time-lapsed images.
The speed of recording has to be decided based on the expected speed of the response and the scan speed that is required to generate an image. Phospholipase C activation by receptors is a fast event but it can be followed by obtaining an image every 5 sec. Depending on the software, the scanning time (1.0 to 2.5 s/image for one color) is taken into account when setting up the speed of image acquisition. It is important to remember that frequent scanning leads to fast photobleaching but including a brake between scans helps to counter the bleaching problem. -
9
Start data acquisition and add stimuli or inhibitors after a control period (5–10 images depending on the speed).
Addition of a treatment is a more complicated question than it seems. The best method is to use a computer-controlled valve-system linked to constant perfusion of the cells with prewarmed media. This ensures proper mixing, timing and keeping the composition of the medium not affected by evaporation. However, our experience is that many of the lipophylic compounds (ionomycin, thapsigargin, rapamycin, etc) stick to the plastic tubing and to the valve components to such an extent that it is almost impossible to clean them even with organic solvents. This is also true for the metal chambers including the plastic oring seal. This may cause very significant problems, especially in multi-user settings. Because of this potential problem, we simply use a pipet to stimulate the cells. For this, we remove 200 μl warm medium from the cells and add it to an Eppendorf tube that already contains the desired amount of drug in 1–5 μl, and after mixing, add back the medium to the cells. With some practice, one can pipette the medium back at an angle that will yield proper mixing that is further helped with one up and down pipetting. This takes about 2 seconds, which is acceptable for many applications but may be too slow for others. It is up to the users whether they want to follow our practices or the more sophisticated valve-controllers. -
10
Once the scanning sequence is done do not forget to save the results. Analysis of the data can be done post acquisition using appropriate softwares.
-
11
Quantify the data
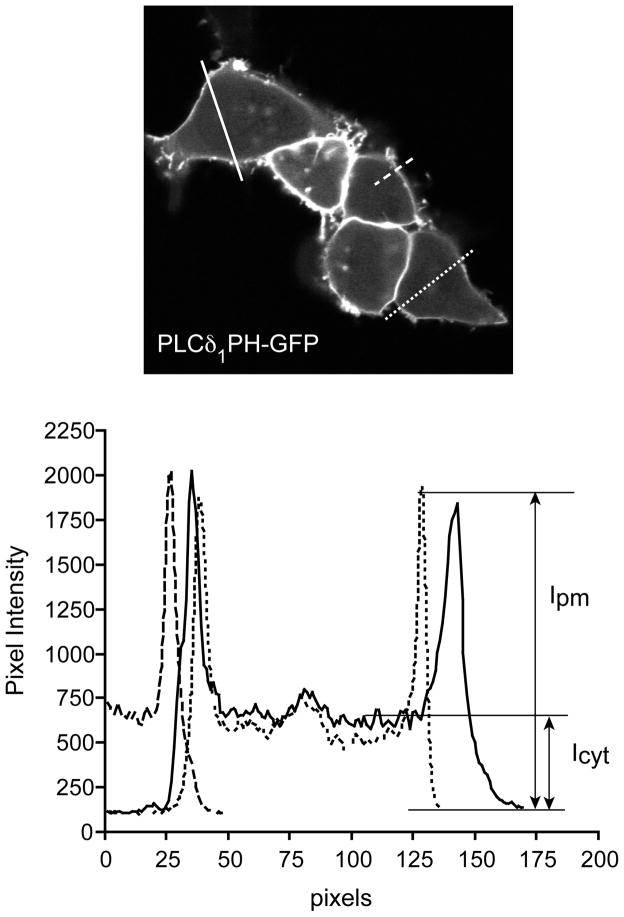
The most demanding part of the analysis of time-lapsed sequences is the quantification of data. Obvious changes can be simply documented in a series of pictures or movies that describe what is happening. However, as our imaging tools are improving, some of the changes are not so obvious to the eye, especially when the number of vesicles, their moving patterns or intensities all show some changes. Quantification is necessary when determining a dose-response relationship, comparing the relative effectiveness of two stimuli, or investigating the efficacy or potency of an inhibitor. It is difficult to give recommendations that would cover all of these areas. In most of the applications dealing with plasma membrane phosphoinositides it is necessary to assess the extent of plasma membrane recruitment so we concentrate on methods that determine plasma membrane association. The simplest way is to monitor the cytosolic intensity since it will significantly increase as a domain translocates from the membrane to the cytosol. It is more demanding to calculate the ratio of membrane to cytosolic intensities. This can be done after creating a line-intensity histogram through a selected line across the cell. It is important that the highest intensities should not be in saturation, which requires a fine optimization of the dynamic range before recording in 8-bit systems (256 levels of gray intensity) (Fig. 3). This is less of a problem with the newer 12-bit systems (4096 levels of intensity) or with even higher dynamic range. A more accurate but also more demanding way of quantification of the extent of membrane association of fluorescent proteins is to use fluorescence resonance energy transfer (FRET) or total internal reflection fluorescence (TIRF) microscopy as detailed in (Balla, 2007; Varnai and Balla, 2006). The former is able to detect the radiationless energy transfer between two appropriate fluorophore pairs when they are within an optimal distance, while the latter only detects fluorescence originating from the plane of the membrane attached to the coverslips. The variability of the cell population and the requirement for analysis of a large number of cells to obtain quantitative estimates of the fluorescence changes remain the most laborious part of obtaining reliable, reproducible results.
Figure 3.
Quantification of the plasma membrane localization of an inositide binding domain. HEK293 cells are shown expressing the PLCδ1PH-GFP. The pixel intensity histograms are calculated for the three lines placed on this recording. Note that the scale shows that this is a 12 bit image. An 8 bit image would only have only 256 levels of intensities. Also note that the peak intensities are not at saturation. The lower panel shows how the intensity values from the membrane and the cytosol are calculated. Their ratio is than a good measure of localization. Also note that no lines are placed over areas where two cells are joined. These calculations have to be made for each picture from a sequence to obtain a full time-course of change. It is advised that more than one line is placed on a cell to get a more accurate value as the intensities even vary along the perimeter.
COMMENTARY
Background Information
The classical methods to study phosphoinositides relied upon metabolic labeling of phosphoinositides. Labeling cells with myo-[3H]inositol or 32P-phosphate followed by lipid extraction and separation by TLC (or other methods) has been widely used to measure phosphoinositide changes (e.g.(Christy et al., 1998). However, these techniques require millions of cells to obtain a sufficient signal. Moreover, depending on the labeling time and the turnover rate of the metabolically distinct inositide pools, it is not certain whether isotopic equilibrium is reached. Determination of the subcellular location of inositides requires even more cells and cell fractionation procedures, and by the end it is still questionable whether the distribution really reflects what was present in the intact cell. Total cellular mass of inositides have also been measured based on quantitation of the inositide headgroup that is liberated from the extracted lipid species (Pearson et al., 2000). Detection of phosphoinositides or inositol phosphate mass separated on HPLC has been done with metal dye-detection (Pittet et al., 1989) or suppressed conductivity detection (Nasuhoglu et al., 2002). Excellent collections of the conventional methods have been published (Irvine, 1990; Shears, 1997). In the meantime, newer methods have been introduced for mass measurement of inositides without extensive purification with the aid of protein domains that specifically recognize the inositide headgroup (Guillou et al., 2007; Luo et al., 2003). However, even in their most simple forms these methods are cumbersome and are unable to resolve the small changes that occur in subcellular compartments, especially against the higher background of non-responsive inositide pools. The realization that inositides are rapidly changing in restricted cellular compartments brought about the desire to detect them at the single cell level preferably in live cells where the dynamics can be followed in real time.
Since the method employing protein domain-fluorescent protein (FP) chimeras in live cells has certain limitation, there is a legitimate need for alternative methods in which the lipids are detected post-fixation, hence, without interfering with the biological process. Moreover, none of the live cell imaging can compete with the resolving power of electron-microscopy. Post-fixation detection of the lipids have been achieved with anti-phosphoinositide antibodies or recombinant GST-fused inositol lipid binding domains in immuno-cytochemical or –electron microscopy applications (Watt et al., 2004; Watt et al., 2002). The value of these techniques is clear; theoretically they would tell us where the lipids are in an intact cell without any distortion caused by the detection process and many cells can be analyzed with no time constrains. EM studies revealed the presence of lipids in compartments where in vivo PH domain imaging failed to do so (for example PtdIns(4,5)P2 in the Golgi (Watt et al., 2002)). However, these techniques also have their drawbacks. First, the fixation process has a major influence on what inositide pools are “visible” to the antibodies or the GST-fused inositide-binding module. Second, the sensitivity of these methods is hard to evaluate. The fixation process is even more critical than in proteins in preserving the lipids and yet to make them accessible to the antibodies or protein modules. This technical difficulty explains the variability of the results obtained in different laboratories with the antibodies and their failure to work in some cases. The specificity of the antibodies should be a rigorous criterion and the data should be consistent with already existing knowledge on the distribution of phosphoinositides.
Related methods for quantitative analysis
Assessing membrane localization by FRET
A quantitative assessment of the membrane localization of fluorescent probes is not always easy based on confocal images. An increase in the fluorescent intensity of the membrane can reflect a change in membrane volume or shape and not a real recruitment. Similarly, the cytosolic intensity of the probe can increase due to shrinkage instead of its release from the membranes. To overcome these problems, the FRET principle was used in several studies to obtain a signal that reflects true binding of the inositide binding domain to membranes. The method developed in the Jalink laboratory and also used in our studies is based on co-expression of the CFP- and YFP-tagged versions of the same PH domain (e.g. PLCδ1-PH). These fluorophores are the most widely used pairs for FRET studies although the more pH resistant Venus replaces YFP and cerulean replaces CFP in newer applications. Newer fluorophore pairs are also available now for FRET studies but they are not as well established as the GFP derivatives (see under point 3. above). Nevertheless, the principle is the same; when the donor (CFP) and the acceptor (YFP) are within FRET distance (<8 nm) there will be energy transfer from CFP to YFP causing a decrease in CFP emission (475 nm) and an increase in YFP emission (525 nm) when using CFP excitation only (430 nm). In the so-called sensitized emission method, the efficiency of the energy transfer is numerically calculated after making all necessary corrections (such as bleed through of the CFP and YFP signals into the other pair’s emission channels). However, for all practical purposes the simple fluorescence ratio of 525/475 can be used if the two wavelengths show opposite changes. This principle is applicable to the lipid binding PH domains when expressed both as a CFP or YFP fusion proteins. The two fluorophores will show efficient energy transfer when their attached PH domains are bound to the lipids at the membrane. However, upon PLC activation the PtdIns(4,5)P2 molecules are hydrolyzed and the PH domains leave the membrane decreasing the FRET signal (van Der Wal et al., 2001). This method is quite sensitive and even small PLC activation can be detected and quantified. The method does not require a confocal microscope (in fact it is not the best way to use it) and can be used either in individual cells or in cell suspensions (Balla et al., 2005). A disadvantage of the method is that the FRET efficiency is not very high at low expression levels (which would be desirable to minimize the ill effects of the presence of the probes) and even at high probe concentrations there could be low FRET signal if the density of the lipids is below a certain level. Although we have not used this application, FRET can also be assessed by FLIM (fluorescence life time imaging microscopy) that calculates the half life of the excited state of the fluorophore that is very different when the energy is emitted in the form of photons or is transferred to an acceptor by FRET (Bastiaens and Squire, 1999). FLIM has many advantages over sensitized emission to evaluate FRET but it requires a separate special instrumentation that is not as easily available as fluorescence microscopes.
In the above examples two separate molecules are used to detect FRET between neighboring molecules (intermolecular FRET). A better solution would be to have inositol lipid probes based on intramolecular FRET in which case both fluorophores are attached to the same inositide-recognizing domain. Lipid binding then induces a molecular rearrangement that changes the distance (or more likely the dipole orientation) of the two fluorophores and, hence, the FRET signal. Such a probe based on the Grp1-PH domain was targeted to different membranes for PtdIns(3,4,5)P3 detection (Sato et al., 2003) and a similar principle was utilized to generate FRET probes for monitoring InsP3 concentrations in the cytoplasm (Matsu-ura et al., 2006). The difficulty here is to ensure a big enough conformational change upon lipid binding to substantially change the FRET efficiency. Construction of a useful probe requires lots of experimentation with the domains themselves as well as with the linkers to connect the fluorophores. In a recent study, the AktPH domain was used to detect PtdIns(3,4,5)P3/PtdIns(3,4)P2 changes using a clever molecular design. The conformational change between the lipid-bound and unbound stages was achieved by inserting a negatively charged “pseudoligand” in the probe that binds to the PH domain (presumably to the lipid binding site) when lipids are not present. Binding of the appropriate lipids abolishes this intramolecular interaction amplifying the conformational change and amplifying the change in the FRET signal (Ananthanarayanan et al., 2005). These single molecule FRET probes do not require co-expression, their readout does not depend on their expression level (once above reliable detection limits) or on lipid density in the membrane. We expect that more efforts will be made to generate similar probes for the detection of lipid production in the various cellular compartments.
More detailed technical and theoretical background on FRET measurements either with sensitized emission or with fluorescence lifetime imaging microscopy (FLIM) are covered in recent publications elsewhere (Sekar and Periasamy, 2003; Thaler et al., 2005; van Rheenen et al., 2004).
Membrane localization based on TIRF analysis
TIRF (total internal reflection fluorescence) analysis has also been used to monitor plasma membrane association of inositol lipid binding domains (Tengholm et al., 2003). The basis of this technique is a special form of illumination where the light is shot at the sample at a shallow angle in a way that the photons do not illuminate the specimen beyond a ~ 200 nm thickness above the coverslip. This way the excitation is limited to the fluorescence molecules that are found close to the membrane of the cell attached to the coverslips. This makes this method quite suitable to detect membrane-associated events and also useful to study association and dissociation of molecules form the membrane. It should be noted, though, that if the footprint of the cell changes that, itself, could cause a change in fluorescence intensity unrelated to the actual amount of fluorescent molecules at the membrane. For this reason, it is advised that a fluorescent membrane marker is used as a control and the fluorescence changes evaluated against this reference signal.
6. TROUBLE SHOOTING
This part describes the most common problems that one encounters starting from plasmid propagation to the final microscopy steps.
No Bacterial Colonies
The pEGFP plasmids have the kanamycin selection marker so transformed bacteria must be grown on kanamycin-containing plates and not ampicillin-containing plates. However, as now many companies sell the various plasmids in different plasmids one cannot assume that all GFP constructs are Kanamycin selectable. Moreover, some labs add the GFP to their chosen domain in Ampicillin resistance plasmids and we often find that people make wrong assumptions when selecting colonies of transformed bacteria. This should be the first thought when no colonies are found after transformation of bacteria during the subcloning or propagation of plasmids. Problems with subcloning, ligation reaction, or bacterial transformation can also result in no bacterial gowth, but discussion of these variables is beyond the scope of this article.
No Green Fluorescence
It may sound trivial but the most common reason for not seeing fluorescence is due to inappropriate microscope settings. Confocal microscopes used for fluorescence imaging can be intimidating for newcomers, with several filters and light directions that must be set properly to see the fluorescent signal. It is a good practice to find the right focus of the cells with transmitted light. Switching to fluorescence one has to see the blue (or other color) light coming through the objective to be sure that the illumination and the light pass is properly set. It is very useful to have a slide of fixed GFP-expressing cells to use as a control. If the microscope settings are right and there are still no GFP positive cells (yet the autofluorescence is visible with a 40× or higher objective), then the problem is with the transfection. Transfection problems can be caused by several factors, including inaccurate subcloning, impure plasmid for transfection, inappropriate transfection conditions for the cells. Even transfected primary cultured cells should have a few positive cells. Using cells transfected with the original pEGFP plasmid as control should help to determine whether the transfection procedure or the DNA construct is the source of the problem. If these cells are positive for GFP fluorescence, then the most likely source of the problem is the construct itself. The DNA construct could be defective by having an unwanted stop codon or a frame-shift either because of wrong design or by a mutation. The plasmids always should be confirmed by DNA sequencing and the expression of the full-length protein should be confirmed by SDS-PAGE as described above.
Weak fluorescence
If there are positive cells but their fluorescence is weak, the microscope may not be set properly. Again, comparison with cells transfected with EGFP alone will help to determine if the problem is with the microscope or the construct to be studied. If the EGFP-expressing cells are bright, but the fusion protein-expressing cells are weakly fluorescent, then the problem is inherent to the fusion protein. Our experience is that the larger the protein fused to GFP, the weaker its fluorescence. This could be because of the difficulty in the folding of GFP as part of a larger protein or that the translation of longer proteins is less efficient. Since the constructs listed in Table 1 express very well, low levels of fluorescence usually indicates a transfection problems that is also corroborated by few cells showing fluorescence. Many cells showing weak fluorescence indicates good transfection efficiency but low yield of the fluorescent protein.
Large “aggregates” are in the cytoplasm
The localizations of the lipid-binding domains listed in Table 1 are well documented. However, from time to time, one will encounter a problem that we describe here, when making a new probe or introducing mutations or deletions in any of these probes. This is a “localization” that is not related to interactions with phospholipids but is due to technical problems. High concentrations or more often folding problems of the expressed fusion proteins are usually responsible for this phenomenon. This results in the formation of large fluorescent “aggregates” that are found in various parts of the cell but mostly associated with the perinuclear area and could overlap with the Golgi. Very often this localization corresponds to the ER-associated protein degradation and represent the proteosome (Hitchcock et al., 2003). We experienced such problems with the EBFP-fused PLCδ1PH and with some chimeric constructs. In such cases a fraction of the expressed protein still could fold properly and functions as expected and it also happens that there are cells that express less of the protein or just for a shorter time that do not show this problem. It is important to recognize this aggregation artifact and not to take it as a true localization. Sometimes it helps to reduce expressing less protein by using a less active promoter or to lower the temperature by 5 to 7°C during transfection and culture. However, we did not have great success with these manipulations once the construct showed this aggregation behavior.
No localization is observed on the membrane where the lipid is expected to be
If GFP fluorescence is observed, but it is not associated with the membrane, it should be confirmed that the expression construct contains the sequence for the lipid-binding domain. Plasmid preparations of pEGFP-based plasmids often contain a ~500 bp DNA piece that is present without digestion by restriction enzymes. This band is usually faint but can be mistaken for an insert when confirming the DNA construct by restriction enzyme digestion analysis. This is especially problematic because many PH domains are encoded by DNA sequences of ~500 bp.
It also has to be realized that the amount of lipid produced and available for binding by a fluorescent probe is limited. Therefore, the membrane-bound fraction may not be distinguishable from the high cytosolic bacground of the unlocalized fusion protein when a cell expresses high amounts of the fusion protein. This is another reason to study cells that express low concentrations of the fusion protein. Curiously, when PtdIns(4,5)P2 is monitored with PLCδ1PH-GFP we did not observe saturation within a wide range of expression level. We assume that there is a compensatory increase in the amount of PtdIns(4,5)P2 in the cells expressing larger amounts of the lipid-binding protein. Compensatory increases are less likely to occur with lipids that are only formed in response to acute stimulation, such as PtdIns(3,4,5)P3.
Nuclear localization
All of the domains listed in Table 1 can penetrate the nuclear pore and enter the nucleus, although their movement in and out is clearly limited. Many of the PH domains (Grp1-PH, ARNO-PH, Btk-PH, OSBP-PH, OSH2-PH) show very prominent nuclear localization. With the increased interest in the nuclear phosphoinositides (Irvine, 2006) and the clear presence of these lipids in the nuclear matrix (in a still poorly understood physical form) this localization of the PH domains makes many researchers very excited. However, nuclear localization of these domains is also observed with mutants that do not bind the inositol lipids. This does not rule out that the wild-type constructs do have some nuclear binding components but it shows that the major force of nuclear enrichment is not a reflection of lipid binding. It is very likely that the numerous basic residues characteristic of these domains are responsible for their nuclear localization; this is supported by the fact that algorithms based on the primary sequences of these proteins accurately predict their nuclear localization. However, if the protein binds to a lipid that is already present and abundant during expression, such as PtdIns(4,5)P2, it will keep the domain out of the nucleus, provided that the amount of the expressed protein does not saturate the lipid available for binding. Again, choosing cells in which the expression levels are low also ensures less localization in the nucleus.
Plasma membrane localization true or false?
It is our experience that many investigators have wrong ideas about how plasma membrane localization can be recognized especially in a confocal microscope. They expect that cells will be outlined with a strong fluorescence signal and no signal over the cytoplasm. This certainly is the case in cells that are round shaped or arranged as cobblestones such as MDCK cells. However, plasma membrane localization can be more difficult to recognize in cells that are solitary and very flat such as COS cells. In such flat cell one could see even fluorescence when imaging the glass-attached membrane surface giving the impression that the probe is also present in the cytosplasm. Moving up on the cell this can change to a very fuzzy ring with some worm like intensities (representing ruffles and microfilopodia) as the membrane covering the nucleus appears in the z-plane (Fig. 1B). It requires a trained eye to differentiate between plasma membrane and cytosolic constructs in such cells. A good guide is to see if the fluorescence clearly outlines the nucleus and its intensity is diminishing toward the periphery of the cells, both being signs of cytosolic localization.
In an opposite way, cytosolic proteins can show a phenomenon that we call “pseudo membrane localization” that gives the impression that the protein is in the membrane. In very flat cells the edge of the cell and the membrane ruffles often appear as high-intensity lines or wrinkles because of the way the membrane appears in the imaged plane. Since membrane ruffling is a common response of cells to stimuli that activate PI 3-kinases and can be reversed with PI3K inhibitors this “apparent” localization will show the same PI 3-kinase dependence as the real plasma membrane recruitment of PtdIns(3,4,5)P3 recognizing domains. Therefore it can give the impression that the construct localizes to the membrane ruffles in stimulated cells even when it does not actually bind to PtdIns(3,4,5)P3. Comparing the fusion protein-expressing cells with control cells expressing only GFP is essential to resolving true versus false membrane localization. A useful measure of true plasma membrane recruitment is to monitor the decrease in intensity of the probe in the cytoplasm. We find that often this decrease can be best judged by the increased contrast between the cytoplasm and the nucleus, which appears sharper as the cytoplasmic fluorescence decreases. True membrane localization also has to fulfill several criteria. The localization of the fusion protein should follow the lipid changes that are evoked by physiological or pharmacological means. For example, PI 3-kinase inhibitors, such as wortmannin or LY294002 prevent the formation of 3-phosphorylated lipid products, so treatment of the transfected cells with these inhibitors should eliminate localization of fluorescent probes that recognize 3-phosphorylated lipids. Also, robust activation of PLC should cause a decrease in the localization of probes detecting PtdIns(4,5)P2. However, the extent of change often is smaller than what can be detected by the eye or even with more sensitive quantification. For example, many G protein-coupled receptors evoke a Ca2+ signal (indicating PLC activation), yet no detectable change in PLCδ1PH-GFP distribution. This does not mean that the probe does not work. It only means that the PLC activation is not robust enough, or the PIP 5-kinase activity is so active that there is little change in the concentration of the PtdIns(4,5)P2 in the membrane.
A further observation of practical importance is that live cells lose localization of the PLCδ1PH-GFP (and some other domains) when they are kept at room temperature for more than 15 to 20 min. The reason for this has not been explored in detail, but it may be that lipid synthesis is slowed because of either ATP depletion or the physicochemical properties of the membrane change. Importantly, warming the cells to 37°C does not correct the situation within 30 min. Thus, live cell imaging should be performed using a temperature controlled microscope system within a short period of time after placing the cells in the observation medium.
Concluding remarks
Live cell imaging of inositol lipids has become a standard approach in many laboratories. It has generated some controversies with examples for its uncritical use as well as for its complete dismissal as an unreliable method. The truth is that this method – as every other one – has its unique benefits as well as its limits. We would like to emphasize repeatedly that we treat phosphoinositides imaged with these molecular tools as only representative of a pool associated with certain processes and not necessarily being representative of all of that particular inositide within the cell. This is especially true and obvious for PtdIns4P and may be less notable but still true for the other inositides. We also caution about high expression levels that inhibit and distort cellular processes. Generating quantitative data with these methods is an added challenge but a very important necessity. As more domains that interact with inositides are identified in proteins it is important to remember that many of these will not work as reporters because their cellular localization is more dependent on protein-protein interactions than on lipid binding. However, some other probes should be compared for their imaging properties, especially for PtdIns(4,5)P2 and PtdIns4P. Development of FRET-based probes relying on intramolecular rearrangements should also be facilitated. These tasks will make this research area very busy for the near future, and the high interest in phosphoinositides also demands that we constantly try to improve our tools to further the knowledge on these exciting molecules.
Anticipated results
Expression of GFP-fused inositol lipid binding domains should yield a fluorescent signal that is easy to recognize and distinguish from the background autofluorescence. In fact, the bigger problem often is too high expression especially in COS-7 cells. In these cells the biology is greatly distorted and therefore these cells should not be used for analysis (see Figure 1 for an example). HEK293 or HeLa cells will express more moderate levels of the same constructs since the do not express the large T antigen that generates many copies of the plasmid in COS cells. The distribution of the individual domains will reflect the localization of the lipid to which it binds except that additional interactions of the domain with other membrane components or proteins may significantly alter the localization of the probe. This should always be kept in mind. Lastly, the lipid changes are not always large enough to be appreciated with the eye or even by quantification of the data. Many cells generate clearly detectable Ca2+ signals by PLC activation without a noticeable change in PLCδ1PH-GFP localization. These negative data do not necessarily mean that the probe is not working. A positive control is always a useful way to check a newly studied construct.
Time Considerations
Performing a live cell imaging experiment is not particularly time consuming. A typical experiment starts out by preparing the coverslips (~ 2 h with drying) and seeding cells for the experiment. Cells are transfected the following day with the plasmid DNA (~ 1 h depending on the number of coverslips). For most phosphoinositide-binding domains we start the transfection around 2 pm and change the transfection medium to one with serum around 7 pm for experiments that are planned the following day. It is not advised to use cells after longer than 24–30 h after transfection with these domains. The actual experiment requires setting up the microscope and the heated stage (~ 30 min) and wait for equilibration (~ 30 min). Depending on the type of experiment a sample is rarely longer on the stage than 30 min. Data analysis is done off-line and can be more time consuming than the actual experiment depending on the application. Design and generation of a new construct is a more time-consuming process that can take from a week (optimal) to several weeks.
It is important to remember that the time consuming part of live cell imaging is the many times an experiment has to be repeated. Many recordings cannot be used because of technical problems such as moving cells or focus-drift occurring during lapse imaging. Also, one has to repeat experiments many times before being confident that the chosen pictures actually represent a reproducible biological process. Due to individual variations among cells this is often not appreciated by users who are used to methods that give averaged cell responses.
Acknowledgments
We would like to thank Dr. Mark Lemmon (Univ. Pennsylvania, Philadelphia) for the OSH1-PH-GFP and OSH2-PH-GFP constructs. The confocal imaging was performed at the Microscopy & Imaging Core of the National Institute of Child Health and Human Development, NIH with the kind assistance of Drs. Vincent Schram and James T. Russell. This research was supported in part by the Intramural Research Program of the National Institute of Child Health and Human Development of the National Institutes of Health. P.V. is also a Bolyai Fellow of the Hungarian Academy of Science and was also supported by the Hungarian Scientific Research fund (OTKA NF-68563) and the Medical Research Council (ETT 440/2006).
Literature cited
- Ananthanarayanan B, Ni Q, Zhang J. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proc Natl Acad Sci U S A. 2005;102:15081–6. doi: 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Ju Kim Y, Varnai P, Szentpetery Z, Knight Z, Shokat KM, Balla T. Maintenance of Hormone-sensitive Phosphoinositide Pools in the Plasma Membrane Requires Phosphatidylinositol 4-Kinase III{alpha} Mol Biol Cell. 2007;19:711–721. doi: 10.1091/mbc.E07-07-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Imaging and Manipulating Phosphoinositides in Living Cells. J Physiol. 2007;582:927–937. doi: 10.1113/jphysiol.2007.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Bondeva T, Varnai P. How accurately can we image inositol lipids in live cells? Trends Pharmacol Sci. 2000;21:238–241. doi: 10.1016/s0165-6147(00)01500-5. [DOI] [PubMed] [Google Scholar]
- Bastiaens PI, Squire A. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999;9:48–52. doi: 10.1016/s0962-8924(98)01410-x. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Irvine RF. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Christy AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy Jg, Gascard P, Takakuwa Y, Huang SC, Benz EJ, Jr, Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Arpin M, Louvard D, Tonks NK, Anderson JM, Fanning AS, Bryant PJ, Woods DF, Hoover KB. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998;23:281–282. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FB, Balla T, Donaldson JG. Active Arf6 recruits ARNO/Cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 2007 doi: 10.1091/mbc.E06-11-0998. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronas S, Ramel D, Pendaries C, Gaits-Iacovoni F, Tronchere H, Payrastre B. PtdIns5P: a little phosphoinositide with big functions? Biochem Soc Symp. 2007:117–28. doi: 10.1042/BSS0740117. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Di Campli A, Godi A. The role of the phosphoinositides at the Golgi complex. Biochim Biophys Acta. 2005;1744:396–405. doi: 10.1016/j.bbamcr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–7. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dormann D, Weijer G, Parent CA, Devreotes PN, Weijer CJ. Visualizing PI3 kinase-mediated cell-cell signaling during Dictyostelium development. Curr Biol. 2002;12:1178–88. doi: 10.1016/s0960-9822(02)00950-8. [DOI] [PubMed] [Google Scholar]
- Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, Parker PJ, Lemmon MA. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. Embo J. 2004;23:1922–33. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PRJ, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40phox. Nat Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- Field SJ, Madson N, Kerr ML, Galbraith KA, Kennedy CE, Tahiliani M, Wilkins A, Cantley LC. PtdIns(4,5)P2 functions at the cleavage furrow during cytokinesis. Curr Biol. 2005;15:1407–12. doi: 10.1016/j.cub.2005.06.059. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt protooncogene product by PI3,4P2. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Friant S, Pecheur EI, Eugster A, Michel F, Lefkir Y, Nourrisson D, Letourneur F. Ent3p Is a PtdIns(3,5)P2 effector required for protein sorting to the multivesicular body. Dev Cell. 2003;5:499–511. doi: 10.1016/s1534-5807(03)00238-7. [DOI] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–24. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Gillooly DJ, I, Morrow C, Lindsay M, Gould R, Bryant NJ, Gaullier LM, Parton GP, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Di Campi A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Logovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenov J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Guillou H, Stephens LR, Hawkins PT. Quantitative measurement of phosphatidylinositol 3,4,5-trisphosphate. Methods Enzymol. 2007;434:117–30. doi: 10.1016/S0076-6879(07)34007-X. [DOI] [PubMed] [Google Scholar]
- Hammond GR, Dove SK, Nicol A, Pinxteren JA, Zicha D, Schiavo G. Elimination of plasma membrane phosphatidylinositol (4,5)-bisphosphate is required for exocytosis from mast cells. J Cell Sci. 2006;119:2084–94. doi: 10.1242/jcs.02912. [DOI] [PubMed] [Google Scholar]
- Hitchcock AL, Auld K, Gygi SP, Silver PA. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc Natl Acad Sci U S A. 2003;100:12735–40. doi: 10.1073/pnas.2135500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan KA, Watanabe K, Kong AM, Bailey CG, Rasko JE, Sasaki T, Mitchell CA. Regulation of FcgammaR-stimulated phagocytosis by the 72-kDa inositol polyphosphate 5-phosphatase: SHIP1, but not the 72-kDa 5-phosphatase, regulates complement receptor 3 mediated phagocytosis by differential recruitment of these 5-phosphatases to the phagocytic cup. Blood. 2007;110:4480–91. doi: 10.1182/blood-2007-02-073874. [DOI] [PubMed] [Google Scholar]
- Huang YE, Iijima M, Parent CA, Funamoto S, Firtel RA, Devreotes P. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P3 to the leading edge of chemotaxing cells. Mol Biol Cell. 2003;14:1913–22. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunyady L, Baukal AJ, Gaborik Z, Olivares-Reyes JA, Bor M, Szaszak M, Lodge R, Catt KJ, Balla T. Differential PI 3-kinase dependence of early and late phases of recycling of the internalized AT1 angiotensin receptor. J Cell Biol. 2002;157:1211–1222. doi: 10.1083/jcb.200111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Foti M, Carpentier JL, Shisheva A. PIKfyve controls fluid phase endocytosis but not recycling/degradation of endocytosed receptors or sorting of procathepsin D by regulating multivesicular body morphogenesis. Mol Biol Cell. 2003;14:4581–91. doi: 10.1091/mbc.E03-04-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF. Methods in Inositide Research. Raven Presss; New York: 1990. [Google Scholar]
- Irvine RF. Nuclear inositide signalling -- expansion, structures and clarification. Biochim Biophys Acta. 2006;1761:505–8. doi: 10.1016/j.bbalip.2006.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Cooke FT, Jat P, Boucheron C, Koizumi T, Hayakawa M, Kaizawa H, Ohishi T, Workman P, Waterfield MD, Parker PJ. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–70. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL. Binding of the PX domain of p47phox to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimber WA, Trinkle-Mulcahy L, Cheung PC, Deak M, Marsden LJ, Kieloch A, Watt S, Javier RT, Gray A, Downes CP, Lucocq JM, Alessi DR. Evidence that the tandem-pleckstrin-homology-domain-containing protein TAPP1 interacts with Ptd(3,4)P2 and the multi-PDZ-domain-containing protein MUPP1 in vivo. Biochem J. 2002;361:525–36. doi: 10.1042/0264-6021:3610525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing plekstrin and Sec7 homology domains. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Tsiaras W, Holik JJ, Chawla A, Czech MP. Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J Biol Chem. 2000;275:32816–32821. doi: 10.1074/jbc.M002435200. [DOI] [PubMed] [Google Scholar]
- Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Peter Downes C, Safrany ST, Alessi DR, van Aalten DM. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. Embo J. 2004;23:3918–28. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Falasca M, Ferguson KM, Schlessinger J. Regulatory recruitment of signalling molecules to the cell membrane by plekstrin-homology domains. Trends Cell Biol. 1997;7:237–242. doi: 10.1016/S0962-8924(97)01065-9. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, O’Brian R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated plekstrin homology domain. Proc Natl Acad Sci U S A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S. The pleckstrin-homology domain of oxysterol-binding protein recognizes a determinant specific to Golgi membranes. Curr Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, Snyder SH. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–72. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Manna D, Albanese A, Park WS, Cho W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J Biol Chem. 2007;282:32093–105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- Matsu-ura T, Michikawa T, Inoue T, Miyawaki A, Yoshida M, Mikoshiba K. Cytosolic inositol 1,4,5-trisphosphate dynamics during intracellular calcium oscillations in living cells. J Cell Biol. 2006;173:755–65. doi: 10.1083/jcb.200512141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Paterson HF, Rodriguez R, Fensome AC, Ellis MV, Swann K, Katan M. Real time fluorescence imaging of PLC gamma translocation and its interaction with the epidermal growth factor receptor. J Cell Biol. 2001;153:599–612. doi: 10.1083/jcb.153.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell RH, V, Heath L, Lemmon MA, Dove SK. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci. 2005;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Muller-Taubenberger A, Anderson KI. Recent advances using green and red fluorescent protein variants. Appl Microbiol Biotechnol. 2007;77:1–12. doi: 10.1007/s00253-007-1131-5. [DOI] [PubMed] [Google Scholar]
- Nagel W, Schilcher P, Zeitlmann L, Kolanus W. The PH domain and the polybasic c domain of cytohesin-1 cooperate specifically in plasma membrane-association and cellular function. Mol Biol Cell. 1998;9:1981–1994. doi: 10.1091/mbc.9.8.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash MS, Young KW, Willars GB, Challiss RA, Nahorski SR. Single-cell imaging of graded Ins(1,4,5)P3 production following G-protein-coupled-receptor activation. Biochem J. 2001;356:137–42. doi: 10.1042/0264-6021:3560137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasuhoglu C, Feng S, Mao J, Yamamoto M, Yin HL, Earnest S, Barylko B, Albanesi JP, Hilgemann DW. Nonradioactive analysis of phosphatidylinositides and other anionic phospholipids by anion-exchange high-performance liquid chromatography with suppressed conductivity detection. Anal Biochem. 2002;301:243–54. doi: 10.1006/abio.2001.5489. [DOI] [PubMed] [Google Scholar]
- Nelson CP, Nahorski SR, Challiss RA. Temporal profiling of changes in phosphatidylinositol 4,5-bisphosphate, inositol 1,4,5-trisphosphate and diacylglycerol allows comprehensive analysis of phospholipase C-initiated signalling in single neurons. J Neurochem. 2008;107:602–15. doi: 10.1111/j.1471-4159.2008.05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold and extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- Pendaries C, Tronchere H, Arbibe L, Mounier J, Gozani O, Cantley L, Fry MJ, Gaits-Iacovoni F, Sansonetti PJ, Payrastre B. PtdIns5P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. Embo J. 2006;25:1024–34. doi: 10.1038/sj.emboj.7601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet D, Schlegel W, Lew DP, Monod A, Mayr GW. Mass changes in inositol tetrakis- and pentakisphosphate isomers induced by chemotactic peptide stimulation in HL-60 cells. J Biol Chem. 1989;264:18489–18493. [PubMed] [Google Scholar]
- Quinn KV, Behe P, Tinker A. Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J Physiol. 2008;586:2855–71. doi: 10.1113/jphysiol.2008.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh LE, Arvidsson A, Carraway Iii KL, Couvillon AD, Rathbun G, Crompton A, VanRentherghem B, Czech MP, Ravichandran KS, Burakoff SJ, Wang DS, Chen CS, Cantley LC. A comparative analysis of the phosphoinositide binding specificity of plekstrin homology domains. J Biol Chem. 1997a;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997b;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- Roy A, Levine TP. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J Biol Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CIE, Driscoll PC, Waterfield MD, Panayotou G. Distinct specificity in the recognition of phosphoinositides by the plekstrin homology domains of dynamin and Bruton’s tyrosine kinase. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- Sato M, Ueda Y, Takagi T, Umezawa Y. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat Cell Biol. 2003;5:1016–22. doi: 10.1038/ncb1054. [DOI] [PubMed] [Google Scholar]
- Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol. 2003;160:629–33. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shears SB. Signalling by Inositides. Oxford University Press; Oxford, New York, Tokyo: 1997. [Google Scholar]
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Burian A, Bruzik KS, Murray D, Cho W. Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J Biol Chem. 2003;278:14469–79. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- Tengholm A, Teruel MN, Meyer T. Single cell imaging of PI3K activity and glucose transporter insertion into the plasma membrane by dual color evanescent wave microscopy. Sci STKE. 2003:PL4. doi: 10.1126/stke.2003.169.pl4. [DOI] [PubMed] [Google Scholar]
- Thaler C, Koushik SV, Blank PS, Vogel SS. Quantitative multiphoton spectral imaging and its use for measuring resonance energy transfer. Biophys J. 2005;89:2736–49. doi: 10.1529/biophysj.105.061853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CC, Dowler S, Deak M, Alessi DR, van Aalten DM. Crystal structure of the phosphatidylinositol 3,4-bisphosphate-binding pleckstrin homology (PH) domain of tandem PH-domain-containing protein 1 (TAPP1): molecular basis of lipid specificity. Biochem J. 2001;358:287–94. doi: 10.1042/0264-6021:3580287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Wal J, Habets R, Varnai P, Balla T, Jalink K. Monitoring Phospholipase C activation kinetics in live cells by FRET. J Biol Chem. 2001;276:15337–15344. doi: 10.1074/jbc.M007194200. [DOI] [PubMed] [Google Scholar]
- van Rheenen J, Langeslag M, Jalink K. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys J. 2004;86:2517–29. doi: 10.1016/S0006-3495(04)74307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim Biophys Acta. 2006;1761:957–67. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Várnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium-and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Bondeva T, Tamas P, Toth B, Buday L, Hunyady L, Balla T. Selective cellular effects of overexpressed pleckstrin-homology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J Cell Sci. 2005;118:4879–88. doi: 10.1242/jcs.02606. [DOI] [PubMed] [Google Scholar]
- Varnai P, Rother KI, Balla T. Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton’s tyrosine kinase pleckstrin homology domain visualized in single living cells. J Biol Chem. 1999;274:10983–10989. doi: 10.1074/jbc.274.16.10983. [DOI] [PubMed] [Google Scholar]
- Varnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol. 2006;175:377–82. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K, Gunn-Moore F, Oatey PB, Tavare JM, Cullen PJ. Nerve growth factor- and epidermal growth factor-stimulated translocation of the ADP-ribosylation factor-exchange factor GRP1 to the plasma membrane of PC12 cells requires activation of phosphatidylinositol 3-kinase and the GRP1 pleckstrin homology domain. Biochem J. 1998a;335(Pt 1):139–46. doi: 10.1042/bj3350139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K, Gunn-Moore F, Tavare JM, Cullen PJ. EGF-and NGF-stimulated translocation of cytohesin-1 to the plasma membrane of PC12 cells requires PI 3-kinase activation and a functional cytohesin-1 PH domain. J Cell Sci. 1999;112:1957–1965. doi: 10.1242/jcs.112.12.1957. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Oatey PB, Tavare JM, Cullen PJ. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol. 1998b;8:463–466. doi: 10.1016/s0960-9822(98)70181-2. [DOI] [PubMed] [Google Scholar]
- Watt SA, Kimber WA, Fleming IN, Leslie NR, Downes CP, Lucocq JM. Detection of novel intracellular agonist responsive pools of phosphatidylinositol 3,4-bisphosphate using the TAPP1 pleckstrin homology domain in immunoelectron microscopy. Biochem J. 2004;377:653–63. doi: 10.1042/BJ20031397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt SA, Kular G, Fleming IN, Downes CP, Lucocq JM. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem J. 2002;363:657–666. doi: 10.1042/0264-6021:3630657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watton J, Downward J. Akt/PKB localisation and 3′ phosphoinositide generation at sites of epithelial cell-matrix and cell-cell interaction. Curr Biol. 1999;9:433–436. doi: 10.1016/s0960-9822(99)80192-4. [DOI] [PubMed] [Google Scholar]
- Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem. 2005;280:10501–8. doi: 10.1074/jbc.M414304200. [DOI] [PubMed] [Google Scholar]
- Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type-I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- Wiedenmann J, Nienhaus GU. Live-cell imaging with EosFP and other photoactivatable marker proteins of the GFP family. Expert Rev Proteomics. 2006;3:361–74. doi: 10.1586/14789450.3.3.361. [DOI] [PubMed] [Google Scholar]
- Yaradanakul A, Hilgemann DW. Unrestricted diffusion of exogenous and endogenous PIP(2 )in baby hamster kidney and Chinese hamster ovary cell plasmalemma. J Membr Biol. 2007;220:53–67. doi: 10.1007/s00232-007-9074-4. [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Virbasius JV, Song X, Pomerleau DP, Zhou GW. The p40phox and p47phox PX domains of NADPH oxydase target cell membranes via direct and indirect recruitment by phosphoinositides. J Biol Chem. 2002;277:4512–4518. doi: 10.1074/jbc.M109520200. [DOI] [PubMed] [Google Scholar]
- Zou J, Marjanovic J, Kisseleva MV, Wilson M, Majerus PW. Type I phosphatidylinositol-4,5-bisphosphate 4-phosphatase regulates stress-induced apoptosis. Proc Natl Acad Sci U S A. 2007;104:16834–9. doi: 10.1073/pnas.0708189104. [DOI] [PMC free article] [PubMed] [Google Scholar]