Abstract
The unfolded protein response (UPR), which is activated when unfolded or misfolded proteins accumulate in the endoplasmic reticulum, has been implicated in the normal physiology of immune defense and in several human diseases including diabetes, cancer, neurodegenerative disease, and inflammatory disease. In this study, we found that the nervous system controlled the activity of a non-canonical UPR pathway required for innate immunity in Caenorhabditis elegans. OCTR-1, a putative octopamine G protein-coupled catecholamine receptor (GPCR, G protein–coupled receptor), functioned in sensory neurons designated ASH and ASI to actively suppress innate immune responses by down-regulating the expression of non-canonical UPR genes pqn/abu in non-neuronal tissues. Our findings suggest a novel molecular mechanism by which the nervous system may sense inflammatory responses and respond by controlling stress-response pathways at the organismal level.
Endoplasmic reticulum (ER) stress has been linked to several human diseases, including diabetes, cancer, neurodegenerative disease, and inflammatory disease (1-3). The ER has developed specific signaling pathways, known as the unfolded protein response (UPR), to cope with ER stress and restore ER homeostasis. Recent studies indicate that increased demand on protein folding in the ER, which may occur during bacterial infections, must be alleviated by UPR pathways for a complete immune response to be mounted (4-8).
We took advantage of the simple nervous and immune systems of the nematode Caenorhabditis elegans to investigate the role of the nervous system in the organismal control of pathways involved in innate immune responses. Three sensory neurons (AQR, PQR and URX) are known to regulate resistance to pathogen infections by controlling the activation of a p38 mitogen-activated protein kinase (MAPK) pathway and the C. elegans avoidance to certain pathogens (9, 10). In addition, a range of chemosensory neurons that penetrate the cuticle and directly detect and respond to different environmental cues have the potential to control innate immune responses.
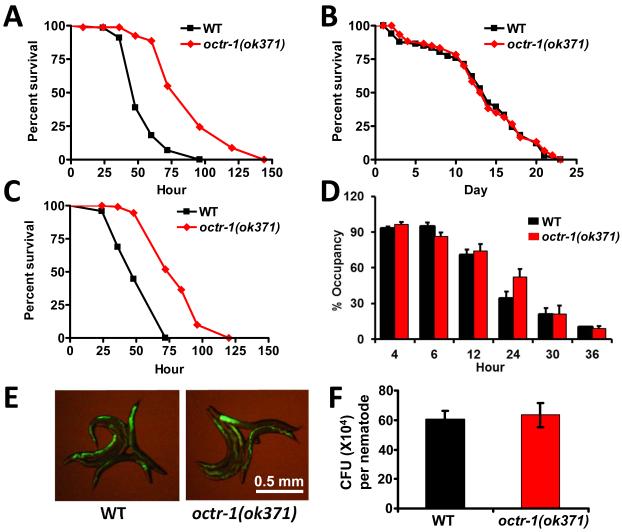
To elucidate the molecular mechanism by which these neurons regulate innate immunity in response to pathogen infection, we first determined the susceptibility to the human opportunistic pathogen Pseudomonas aeruginosa of octr-1(ok371) animals, which lack a G protein-coupled catecholamine receptor normally expressed in the cilia of neurons located in sensory openings, including ASH, ASI, AIY, and dopaminergic ADE/CEP neurons (11). octr-1(ok371) animals exhibited enhanced resistance to killing by P. aeruginosa (Fig. 1A), which suggests that the loss of OCTR-1 signaling increases the general immune function of the nematodes. We observed no difference in survival between octr-1(ok371) and wild-type animals that were fed heat-killed P. aeruginosa or Escherichia coli (Figs. 1B and S1). Thus, octr-1 mutation affects the immune response to living pathogenic bacteria without altering the basic lifespan of the nematodes.
Fig.1.
C. elegans G-protein coupled receptor OCTR-1 is involved in immunity to P. aeruginosa. (A) Wild-type and octr-1(ok371) animals were exposed to P. aeruginosa. (B) Wild-type and octr-1(ok371) animals were exposed to heat-killed P. aeruginosa. (C) Wild-type and octr-1(ok371) animals were exposed to a full lawn of P. aeruginosa. The survival graphs represent combined results of two independent experiments, N=90 adult animals per strain. (D) Animals were placed on a small spot of P. aeruginosa in a 3.5cm plate and monitored over time for their presence or absence on the lawn. The graph represents combined results of three independent experiments, N=60 adult animals per strain. (E) Wild-type and octr-1(ok371) animals were exposed to P. aeruginosa expressing GFP for 48 hours and then visualized using a MZ FLIII Leica stereomicroscope. (F) Wild-type and octr-1(ok371) animals were exposed to P. aeruginosa expressing GFP for 48 hours and the colony forming units were quantified. Ten animals were used for each condition. The graph represents combined results of three independent experiments. Bars represent mean ± SEM.
Because pathogen avoidance is part of the C. elegans defense response to P. aeruginosa (9), we examined the susceptibility to P. aeruginosa-mediated killing of octr-1(ok371) animals on agar plates that were completely covered in bacteria, a condition that eliminates pathogen avoidance. octr-1(ok371) animals died at a slower rate than did wild-type animals (Fig. 1C), indicating that pathogen avoidance does not play a role in octr-1(ok371)-enhanced resistance to P. aeruginosa. In addition, the magnitude of pathogen avoidance of octr-1(ok371) and wild-type animals was similar (Fig. 1D). The enhanced resistance of octr-1(ok371) to S. enterica (Fig. S2), a pathogen that does not elicit an avoidance behavior (12), further supports the function of OCTR-1 in the regulation of immune responses.
Certain mutants resistant to pathogen infection exhibit resistance to pathogen accumulation (13). Therefore, we examined whether octr-1 mutation affected bacterial accumulation in the intestine. Compared to wild-type animals, octr-1(ok371) animals exhibited a similar accumulation pattern of P. aeruginosa/green fluorescent protein (GFP) or S. enterica/GFP (Figs. 1E and S3). In addition, the number of bacterial cells in octr-1(ok371) animals was similar to that in wild-type animals (Figs. 1F and S4). Thus, the bacterial load to which the animals were exposed was comparable, and reduced bacterial accumulation levels in the intestine did not contribute to the enhanced immunity of octr-1(ok371) animals, indicating that octr-1(ok371) animals exhibit enhanced endurance to P. aeruginosa infection.
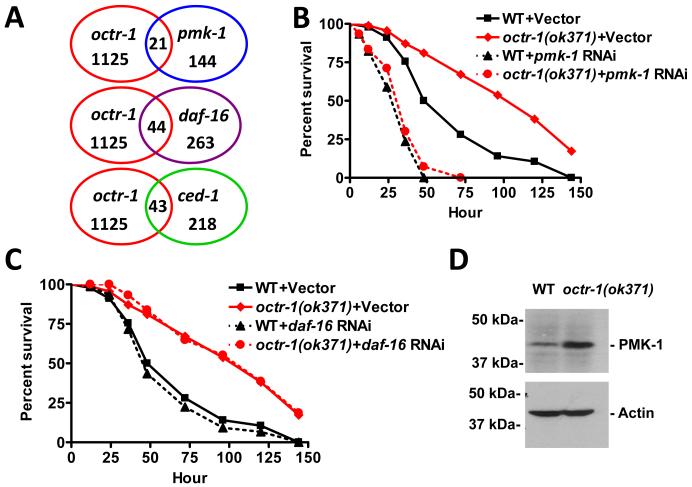
To provide insights into the mechanism underlying the enhanced immunity of octr-1(ok371) animals, we used genome microarrays to find clusters of genes commonly misregulated in octr-1(ok371) relative to wild-type animals exposed to live P. aeruginosa. OCTR-1-regulated genes previously known to be regulated by pathways involved in C. elegans innate immunity, including a DAF-2/DAF-16 insulin pathway, a p38/PMK-1 MAPK pathway, and a CED-1 pathway were enriched (Fig. 2A, Table S1-S2). Unexpectedly, the enhanced resistance to P. aeruginosa-mediated killing of octr-1(ok371) animals was suppressed by pmk-1 RNA interference (RNAi) or mutation, but not by daf-16 RNAi or mutation (Figs. 2B, 2C, and S5). Mutation in sek-1, which encodes for a MAPK kinase essential for PMK-1 activation, also suppressed the enhanced resistance of octr-1(ok371) animals (Fig. S6). Consistent with the idea that derepression of PMK-1 transcriptional activity by lack of OCTR-1 signaling activates innate immunity, octr-1(ok371) animals exhibited higher levels of active PMK-1 than wild-type animals (Fig. 2D). Thus, in contrast to AQR, PQR, and URX neurons, which control not only PMK-1 activity but also pathogen avoidance, OCTR-1 expressing neurons control PMK-1 activity without affecting pathogen avoidance.
Fig.2.
OCTR-1 controls pathways required for C. elegans innate immunity. (A) Venn diagrams of genes that are up-regulated in octr-1(ok371) animals and positively regulated by daf-16, pmk-1, and ced-1. (B) Wild-type and octr-1(ok371) animals grown on double-stranded RNA (dsRNA) for vector control or dsRNA for pmk-1 were exposed to P. aeruginosa. (C) Wild-type and octr-1(ok371) animals grown on dsRNA for vector control or dsRNA for daf-16 were exposed to P. aeruginosa. The survival graphs represent combined results of two independent experiments, N=90 adult animals per strain. (D) Active PMK-1 was detected in wild-type and octr-1(ok371) animals. Animals were grown at 20°C until 1-day-old adult and whole-worm lysates were used to detect active PMK-1 with an antibody against human p38 from Promega (Madison, WI). Actin was used as loading control.
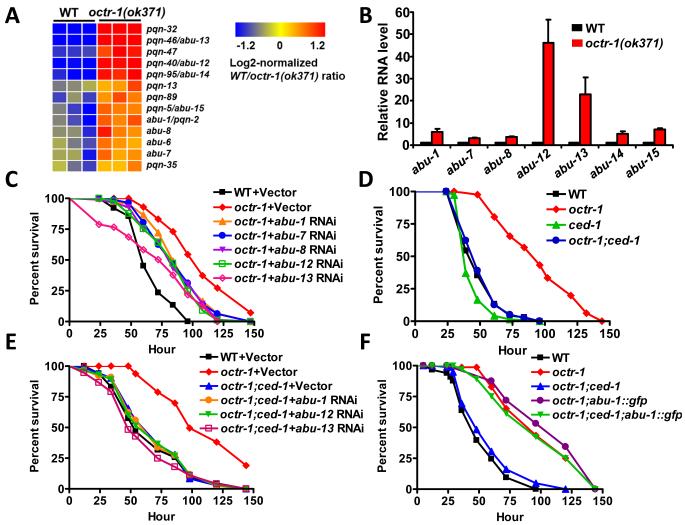
Among genes up-regulated in octr-1(ok371) animals, a group of 43 genes transcriptionally regulated by the phagocyte receptor CED-1 (4) was significantly enriched (Fig. 2A and Table S2). Indeed, 7 out of 14 genes (Fig. 3A) that were up-regulated greater than twofold in octr-1(ok371) animals correspond to a family of pqn (prion-like glutamine[Q]/asparagine[N]-rich domain-bearing protein) genes that are controlled by CED-1 (4). Eleven genes in the pqn family are classified as abu (activated in blocked unfolded protein response) (14), because they are activated in xbp-1 mutant animals when ER stress is induced by tunicamycin treatment. The abu genes encode UPR proteins that function in parallel with the canonical UPR pathway (14). We studied whether the most highly up-regulated pqn genes in octr-1(ok371) mutants could be induced by tunicamycin in an xbp-1 mutant background. Indeed, pqn-40, pqn-46, pqn-95, and pqn-5 genes were found to be activated by ER stress in animals with blocked UPR (Table S3) and were named abu-12, abu-13, abu-14, and abu-15. abu-1, -7, -8, -12, -13, -14, and -15 were significantly up-regulated in octr-1(ok371) animals (Fig. 3B), suggesting that the non-canonical UPR pathway is inhibited by OCTR-1. Consistent with this idea that abu genes encode non-canonical UPR proteins that may interact with abnormal ER client proteins, ABU-1 is retained in the ER by its transmembrane domain (14). Furthermore, with the exception of pqn-46, all pqn/abu genes that were up-regulated in octr-1(ok371) animals encode proteins that have both an ER signal sequence and a transmembrane domain (Table S4).
Fig. 3.
pqn/abu genes are required for the enhanced immunity of octr-1(ok371) animals to P. aeruginosa. (A) Cluster of pqn/abu genes that are up-regulated in octr-1(ok371) mutants. (B) Quantitative reverse transcription-PCR analysis of abu-1, abu-7, abu-8, abu-12, abu-13, abu-14 and abu-15 expression in octr-1(ok371) relative to wild-type animals exposed to P. aeruginosa. N=3; bars represent mean ± SEM. (C) Wild-type animals grown on dsRNA for vector control and octr-1(ok371) animals grown on dsRNA for vector control or dsRNA for abu genes were exposed to P. aeruginosa. (D) Wild-type, octr-1(ok371), ced-1(e1735) and octr-1(ok371);ced-1(e1735) animals were exposed to P. aeruginosa. (E) Wild-type and octr-1(ok371) animals grown on dsRNA for vector control and octr-1(ok371);ced-1(e1735) animals grown on dsRNA for vector control or dsRNA for abu genes were exposed to P. aeruginosa. (F) Wild-type, octr-1(ok371), octr-1(ok371);ced-1(e1735), octr-1(ok371);abu-1::gfp(zcEx8), and octr-1(ok371);ced-1(e1735);abu-1::gfp(zcEx8) animals were exposed to P. aeruginosa. The survival graphs represent combined results of two independent experiments, N=90 adult animals per strain.
Nine out of thirteen pqn/abu genes up-regulated in octr-1(ok371) animals are strongly expressed in the pharynx and the intestine, primary interfaces between host cells involved in immune responses and bacterial pathogens. Transgenic animals show expression of ABU-1::GFP in vesicular structures corresponding to the pharyngeal cells (14). In situ hybridizations completed as part of NEXTDB (The Nematode Expression Pattern Database) reveal expression of eight pqn/abu genes in the pharynx and the intestine from L3 to adult stages (http://nematode.lab.nig.ac.jp). To investigate the relation between the non-canonical UPR pathway and host responses to P. aeruginosa infection, we tested whether exposure to this pathogen would up-regulate abu genes. Indeed, P. aeruginosa exposure up-regulated abu genes (Table S5 and Fig. S7), indicating that the non-canonical UPR pathway may be required for C. elegans immune response against this pathogen. We then studied the resistance to P. aeruginosa-mediated killing of wild-type and octr-1(ok371) animals where abu genes were inhibited by RNAi. Inhibition of abu-1, -7, -8, -12, and -13, partially suppressed the enhanced immunity of octr-1(ok371) animals (Fig. 3C). RNAi-mediated inhibition of abu genes also resulted in enhanced susceptibility to P. aeruginosa-mediated killing (Fig. S8).
Because ced-1 is part of a pathway that transcriptionally controls the expression of pqn/abu genes (4), we asked whether ced-1 mutation suppresses the enhanced immunity of octr-1(ok371) animals. The survival of octr-1(ok371);ced-1(e1735) animals was comparable to that of wild-type animals (Fig. 3D). We observed no additive effect of abu RNAi ablation on octr-1(ok371);ced-1(e1735) animals (Fig. 3E), confirming that ced-1 and abu genes are part of the same pathway responsible for the enhanced resistance to pathogen infection of octr-1(ok371) animals. Furthermore, abu-1 overexpression protected octr-1(ok371);ced-1(e1735) animals from P. aeruginosa-mediated killing (Fig. 3F). Thus, higher CED-1-mediated signaling is responsible for the enhanced resistance to P. aeruginosa-mediated killing in animals with impaired neural function due to mutation in octr-1.
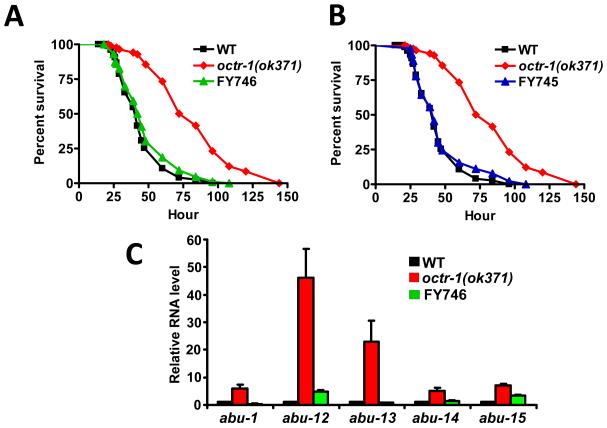
To determine the specific foci of OCTR-1 activity involved in the control of innate immunity, we studied the pathogen susceptibility of octr-1(ok371) animals expressing octr-1 under the control of the sra-6 promoter, which drives the expression of octr-1 to the ASH, ASI, and PVQ neurons (15). Expression of octr-1 under the control of the sra-6 promoter fully rescued the mutant phenotype of octr-1(ok371) animals (Fig. 4A), indicating that OCTR-1-expressing neurons regulate innate immunity in C. elegans. Consistent with this idea, OCTR-1 expression under the regulation of its own promoter also fully rescued the mutant phenotype of octr-1(ok371) animals (Fig. 4B). Because PVQ neurons do not express octr-1, our results suggest that ASH and ASI neurons negatively regulate innate immunity. Thus, we studied the susceptibility to P. aeruginosa of a strain in which mutation in polyQ enhancer-1 promotes early onset degeneration of ASH neurons caused by polyQ toxicity (16). The strain lacking ASH neurons exhibited a significantly increased survival on P. aeruginosa (Fig. S9), indicating that ASH neurons suppress innate immunity.
Fig.4.
OCTR-1 expressing neurons ASH and ASI suppress innate immunity by controlling the expression of pqn/abu genes. (A) Wild-type, octr-1(ok371) and FY746 octr-1(ok371);grEx158[Psra-6::octr-1] animals were exposed to P. aeruginosa. (B) Wild-type, octr-1(ok371) and FY745 octr-1(ok371);grEx157 [Poctr-1::octr-1::gfp] animals were exposed to P. aeruginosa. The survival graphs represent combined results of two independent experiments, N=90 adult animals per strain. (C) Quantitative reverse transcription-PCR analysis of abu-1, abu-12, abu-13, abu-14, and abu-15 expression in octr-1(ok371) and FY746 octr-1(ok371);grEx158[Psra-6::octr-1] relative to wild-type animals exposed to P. aeruginosa. N=3; bars represent mean ± SEM.
We confirmed that ASH and ASI neurons negatively regulate abu genes required for appropriate innate immunity by studying the gene expression levels of abu-1, -12, -13, -14, and -15 in wild-type, octr-1(ok371), and octr-1(ok371) animals expressing octr-1 under the control of the sra-6 promoter. Whereas the expression levels of abu genes in octr-1(ok371) animals are higher than those in wild-type animals, their expression levels are close to those of the wild-type in octr-1(ok371) animals where octr-1 expression in ASH and ASI was restored (Fig. 4C).
We have shown that OCTR-1 expressing neurons ASH and ASI negatively regulate innate immunity, at least in part, by controlling the expression of a family of genes that are part of a non-canonical UPR pathway regulated by the apoptotic receptor CED-1. Furthermore, the conserved p38 MAPK pathway is under the control of OCTR-1 expressing neurons. Recent studies indicate that the murine nervous system, through the vomeronasal organ, has the potential to “smell” molecules related to disease or inflammation in the outside world (17). Even though the mammalian nervous system plays a key role in the regulation of inflammation (18), the specific role of neurons in contact with the outside world in the control of immune responses remains unclear.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center (University of Minnesota) for strains used in this study, R. Komuniecki (University of Toledo) for providing FY745 and FY746, and D. Ron for providing the abu-1 transgenic strain. A.A. is funded by the Dana Foundation and the NIH (grant GM070977).
References
- 1.Hotamisligil GS. Cell. 2010;140:900. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd DJ, Lee AH, Glimcher LH. Nat Rev Immunol. 2008;8:663. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 3.Kim I, Xu W, Reed JC. Nat Rev Drug Discov. 2008;7:1013. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 4.Haskins KA, Russell JF, Gaddis N, Dressman HK, Aballay A. Dev Cell. 2008;15:87. doi: 10.1016/j.devcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richardson CE, Kooistra T, Kim DH. Nature. 2010;463:1092. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischof LJ, et al. PLoS Pathog. 2008;4:e1000176. doi: 10.1371/journal.ppat.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinon F, Chen X, Lee AH, Glimcher LH. Nat Immunol. 2010;11:411. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo CW, et al. Nat Cell Biol. 2009;11:1473. doi: 10.1038/ncb1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Styer KL, et al. Science. 2008;322:460. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aballay A. Cell Cycle. 2009;8:966. doi: 10.4161/cc.8.7.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wragg RT, et al. J Neurosci. 2007;27:13402. doi: 10.1523/JNEUROSCI.3495-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenor JL, Aballay A. EMBO Rep. 2008;9:103. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrman LE, Goel AK, Smith J, Shianna KV, Aballay A. PLoS Genet. 2009;5:e1000657. doi: 10.1371/journal.pgen.1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urano F, et al. J Cell Biol. 2002;158:639. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Cell. 1995;83:207. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 16.Faber PW, Voisine C, King DC, Bates EA, Hart AC. Proc Natl Acad Sci U S A. 2002;99:17131. doi: 10.1073/pnas.262544899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riviere S, Challet L, Fluegge D, Spehr M, Rodriguez I. Nature. 2009;459:574. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- 18.Tracey KJ. Nature immunology. 2010;11:561. doi: 10.1038/ni0710-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.