Abstract
The role of metabolic compartmentation in spatially organizing metabolic enzymes into pathways, regulating flux through metabolic pathways, and controlling the partitioning of metabolic intermediates among pathways is appreciated, but our understanding of the mechanisms that establish metabolic architecture and mediate communication and regulation among interconnected metabolic pathways and networks is still incomplete. This review discusses recent advancements in our understanding of metabolic compartmentation within the pathways that constitute the folate-mediated one-carbon metabolic network and emerging evidence for a need to regulate the trafficking of folates among compartmentalized metabolic pathways.
Introduction
The role of metabolic compartmentation in spatially organizing metabolic enzymes into pathways, regulating flux through metabolic pathways, and controlling the partitioning of metabolic intermediates among pathways has been appreciated for decades (1–3). However, our understanding of the underlying mechanisms that establish metabolic architecture and mediate communication and regulation among interconnected metabolic pathways and networks is still incomplete. Cellular metabolic compartmentation can be achieved by isolating pathways within physical barriers (such as the mitochondrion, lysosome, or nucleus) and also through multienzyme complexes that transiently and reversibly assemble, referred to as metabolons, which can exist within the cytoplasm or cellular organelles (4, 5). The formation of metabolic complexes provides another layer of regulation at the level of complex assembly, which can be largely independent of the kinetic properties of the individual enzymes that constitute the pathways. Moreover, metabolon formation can enable metabolic channeling of substrates, which refers to the direct transfer of substrates and products among enzymes along a pathway. Metabolic channeling circumvents the need for diffusion-mediated delivery of substrates among enzymes and thereby can accelerate metabolic flux through a pathway. Furthermore, channeling permits regulated partitioning of substrates at metabolic branch-points within metabolic networks and stabilizes labile metabolic intermediates. This review discusses recent advancements in our understanding of metabolic compartmentation within the pathways that constitute the folate-mediated one-carbon metabolic network and emerging evidence for a need to regulate the trafficking of folates among compartmentalized metabolic pathways.
Current status of knowledge
Chemical and physical properties of folate cofactors and metabolic channeling
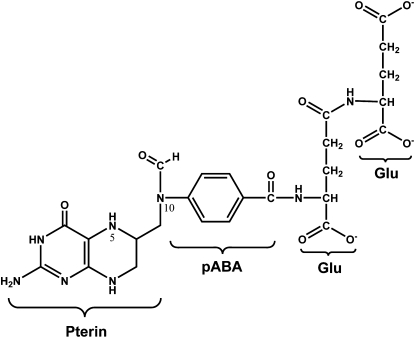
The necessity for metabolic compartmentation of the pathways that constitute folate-mediated one-carbon metabolism and the potential for metabolic channeling of folate cofactors among folate-dependent enzymes has been inferred from knowledge of the physical and chemical properties of folate cofactors and is supported by both theoretical and empirical evidence. Tetrahydrofolates (THF)3 are water-soluble B vitamins that contain 3 distinct chemically linked moieties: 1) a fully reduced pterin ring; 2) a p-aminobenzoyl group; and 3) the amino acid glutamate. They are the form of the vitamin that is absorbed in the intestine from natural food (Fig. 1). Folic acid, an oxidized, chemically stable, and synthetic provitamin present in fortified foods and vitamin supplements, is also absorbed across the intestinal epithelium by the proton-coupled folate transporter (6) and then reduced to THF. Serum folates circulate as monoglutamate derivatives and are imported into cells through either the reduced folate carrier or through a receptor-mediated endocytosis of the folate receptors (7, 8). Once transported into cells, the vitamin is processed to a metabolic cofactor by the addition of a polyglutamate peptide comprised of up to 9 glutamate residues linked by unusual γ-peptide linkages and is substituted with a one-carbon moiety at the N5 and/or N10 position at the oxidation level of formate (e.g. 10-formylTHF), formaldehyde (5,10-methyleneTHF) or methanol (5-methylTHF) (9). The glutamate polypeptide serves to retain the vitamin within cells and to increase its affinity for folate-dependent enzymes.
Figure 1.
Structure of 10-formyl-tetrahydrofolate diglutamate. pABA, para-aminobenzoate.
Early elucidation of the chemical properties of reduced folate cofactors indicated that metabolic channeling may be necessary to prevent the oxidative degradation of folate cofactors (10). Many folate cofactors, including THF, dihydrofolate (DHF), and 10-formylTHF, are chemically labile and susceptible to irreversible oxidative degradation (10). In vitro, THF in solution has a half-life of minutes in the absence of reduced thiols and ascorbate (11), yet the mean residency time of whole-body folate in humans can exceed 100 d (12). Folates have been shown to be stabilized and protected from oxidative degradation when bound to protein (10), indicating that the elimination of diffusion-mediated transfer of folate cofactors among enzymes may be necessary to decrease random oxidation events. Second, metabolic channeling of folate coenzymes is predicted by estimates of cellular folate concentrations compared to total cellular folate binding capacity. Several studies indicate that the cellular concentration of folate-binding enzymes and proteins exceeds, by severalfold, the cellular concentration of folate cofactors (13). Considering that most cellular folate-binding proteins bind folate cofactors with dissociation constants in the nanomolar range, and folate enzymes are present in the micromolar range in liver (14, 15), it is apparent that cellular folates are protein bound with limited opportunity to accumulate in the cellular milieu. Last, there is evidence that folate-dependent enzymes within metabolic pathways physically associate. Several folate-binding proteins are multifunctional proteins with contain 2 or more folate-dependent activities and active sites on a single polypeptide (16), and in vitro kinetic studies have demonstrated direct transfer of the folate cofactor between these active sites without the need for dissociation of the enzyme-folate complex (17). Furthermore, there is also kinetic evidence of folate cofactor channeling between enzyme active sites from distinct proteins in metabolic pathway reconstitution experiments (18). It has been suggested that direct transfer of the cofactor among enzyme active sites is facilitated by the γ-glutamyl polypeptide moiety of the cofactor (18).
Folate-mediated one-carbon metabolism
Folate cofactors have been identified in all subcellular organelles (19), but the vast majority of folate cofactors are distributed between the cytoplasm and mitochondria (20). Because folate cofactors are not known to function in any other cellular process other than metabolism, it has been assumed that metabolic processes occur wherever these cofactors are present, with the exception of the lysosome, where folate polyglutamates can be converted to folate monoglutamates through the activity of γ-glutamyl hydrolase (21).
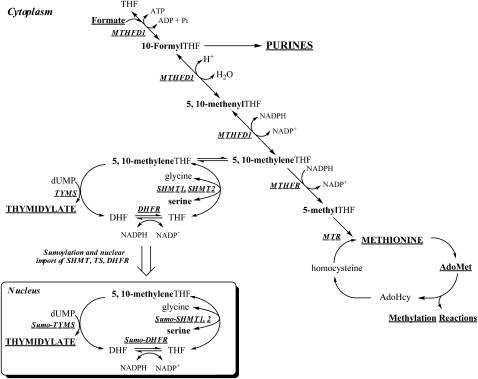
Folate polyglutamates function in cells as a family of metabolic cofactors that chemically activate and accept or donate single carbons. Folate-mediated one-carbon metabolism in the cytoplasm includes 3 interdependent biosynthetic pathways that catalyze the de novo synthesis of purine nucleotides, thymidylate (dTMP), and the remethylation of homocysteine to methionine (Fig. 2). Methionine is a precursor for the synthesis of S-adenosylmethionine (AdoMet or SAM), a cofactor and methyl group donor for numerous methylation reactions, including the methylation of cytosine bases in DNA, histones, RNA, neurotransmitters, and other small molecules, phospholipids, and other proteins. S-adenosylmethionine–dependent methylation reactions serve to regulate fundamental biological processes, including gene transcription, mRNA translation, cell signaling (22), protein localization (23), and the degradation of small molecules (24). Although one-carbons can be derived directly in the cytoplasm from the catabolism of histidine, purines, and serine (9), the primary source of one-carbons for cytoplasmic one-carbon metabolism comes from formate that is derived from mitochondrial one-carbon metabolism (25–27). Serine is a primary source of one-carbon units for cytoplasmic one-carbon metabolism (9), mainly through its conversion to formate in mitochondria (25, 27, 28).
Figure 2.
Compartmentation of folate-mediated one-carbon metabolism in the cytoplasm and nucleus. One-carbon metabolism in the cytoplasm is required for the de novo synthesis of purines and thymidylate and for the remethylation of homocysteine to methionine. One-carbon metabolism in the mitochondria is required to generate formate for one-carbon metabolism in the cytoplasm. One-carbon metabolism in the nucleus synthesizes thymidylate from uridylate and serine. MTR, methionine synthase; TYMS, thymidylate synthase; Pi, phosphate.
Compartmentation of one-carbon metabolism in the mitochondrion
Mitochondria contain as much as 40% of total cellular folate (19, 20). Folates are transported into mitochondria in the monoglutamate form through the activity of the mitochondrial folate transporter SLC25A32 (29). Folate polyglutamates in mitochondria are a distinct pool that are not in equilibrium with folate polyglutamates in the cytoplasm (20). Genetic disruption of folate metabolism in mitochondria of Chinese hamster ovary cells results in a glycine auxotrophy, indicating glycine synthesis from serine is an essential role of one-carbon metabolism in this compartment (30, 31). One-carbon metabolism is also necessary to form fMet-tRNA from Met-tRNA using 10-formylTHF as a cofactor for the initiation of mitochondrial protein synthesis and to synthesize formate from 10-formylTHF for cytoplasmic one-carbon metabolism. Formate is derived from the catabolism of serine, glycine, dimethylglycine, and sarcosine (32) (Fig. 2). The THF-dependent catabolism of the these amino acids each generates 5,10-methyleneTHF, which is subsequently oxidized to 5,10-methenylTHF and then to 10-formylTHF in a reaction catalyzed by the bifunctional enzymes MTHFD2 (33, 34) and MTHFD2L (35). MTHFD1L then liberates formate from 10-formylTHF (32, 36). Formate derived in mitochondria traverses to the cytoplasm to support cytoplasmic one-carbon metabolism (25). A comprehensive review of mitochondrial folate metabolism recently has been published (37).
Evidence for metabolic compartmentation in the cytoplasm and nucleus and need for intracellular trafficking folate among compartmentalized pathways
The 3 cytoplasmic pathways that comprise folate-mediated one-carbon metabolism have been suggested to function in a metabolic network that interconnects the 3 biosynthetic pathways, namely de novo purine biosynthesis, de novo dTMP biosynthesis, and homocysteine remethylation. In this model, the 3 pathways compete for a limiting pool of folate cofactors as illustrated in Figure 2. Mathematical models have been generated to predict metabolic flux through the network based on the kinetic properties of the individual enzymes (38, 39). This concept of metabolic competition within the folate network has been supported in part through kinetic isotope tracer as well as empirical evidence through stable isotope studies (9). The competition for folate cofactors is viewed as being especially keen for 5,10-methyleneTHF between de novo dTMP biosynthesis and homocysteine remethylation (27, 40), and this includes human carriers of a common genetic variant in the MTHFR gene (677 C→ T) who exhibit less MTHFR enzymatic activity due to thermolability of the enzyme (41). Because MTHFR generates 5-methylTHF from 5,10-methyleneTHF for homocysteine remethylation, homozygous carriers of the less active MTHFR variant therefore exhibit higher mean rates of de novo dTMP biosynthesis (42). However, the model of direct substrate competition between folate-dependent pathways is being challenged by more recent studies, which are demonstrating that both de novo purine biosynthesis and de novo dTMP biosynthesis pathways are transiently isolated from the other folate-dependent pathways through both dynamic physical compartmentation and metabolon formation. The transient compartmentation of these pathways adds additional dimensions and complexity to regulation of these pathways, including the requirement to colocalize enzymes, as well as the necessity for trafficking mechanisms to mobilize folate cofactors among compartmentalized pathways.
De novo dTMP biosynthesis
The folate-dependent pathway for the de novo synthesis of dTMP from deoxyuridylate involves the enzymes thymidylate synthase (TYMS), serine hydroxymethyltransferase (SHMT), the trifunctional enzyme methylenetetrahydrofolate dehydrogense (MTHFD1) and DHF reductase (DHFR). 5,10-MethyleneTHF is the one-carbon donor for this reaction in which TYMS converts deoxyuridylate to dTMP and DHF (9). This is the only folate-dependent reaction whereby the folate cofactors serve both as one-carbon donors and sources of 2 electrons through the oxidation of THF to DHR. The THF cofactor is regenerated from DHF in a reaction catalyzed by the NADPH-dependent enzyme DHFR. The 3rd step required to complete the de novo dTMP synthesis cycle is the generation of 5,10-methyleneTHF, which can be derived from formate, ATP, NADPH, and THF catalyzed by the trifunctional enzyme MTHFD1 or by vitamin B-6–dependent SHMT, which catalyzes the transfer of the hydroxymethyl group of serine to THF to generate glycine and 5,10-methyleneTHF (Fig. 2). Isotope tracer studies have demonstrated that 5,10-methyleneTHF generated by SHMT is preferentially incorporated into dTMP compared to 5,10-methyleneTHF generated by MTHFD1 (27). This preferential enrichment of SHMT-derived 5,10-methyleneTHF into dTMP is consistent with compartmentation of SHMT with TYMS, which was subsequently demonstrated to involve cell cycle-dependent nuclear localization of SHMT, TYMS, and DHFR in the nucleus (43–45).
The concept that one-carbon metabolism occurs in the nucleus was suggested as early as 1976 when Shin et al. (19) at the University of California at Berkeley determined that 10% of total cellular folate was present in nuclei of rat liver following injection of [3H] folic acid. In this study, >95% of nuclear folate was present in the polyglutamate form, indicating that nuclear folate could serve as a metabolic cofactor. In 1980, Prem veer Reddy and Pardee (46) reported the isolation of a nuclear-localized multienzyme complex involved in DNA synthesis in Chinese hamster embryo fibroblast cells. The enzyme activities were present in nuclear fractions from S-phase cells, whereas the enzyme activities were localized to the cytoplasm in G1 phase cells. The authors proposed that DNA precursor synthesis occurs within a multienzyme complex termed a replitase, which assembles in proximity to the DNA replication machinery during the S-phase of the cell cycle. Nuclear TYMS was shown to form part of a putative replitase complex along with DNA polymerase α, ribonucleotide reductase, thymidylate kinase, nucleotide diphosphate (NTP) kinase, and the folate-dependent enzyme DHFR (46–48). More recently it has been shown that ribonucleotide reductase localizes to the cytoplasm in mammalian cells (49). This study was based on earlier work by Mathews et al. (50) investigating bacteriophage T4-infected cells. In a series of studies, multienzyme complexes involved in the synthesis of deoxyribonucleotides were shown to be physically associated with the DNA replication machinery, indicating that DNA precursor synthesis occurs at the site of DNA synthesis in prokaryotes.
The identification of the replitase suggested the possibility that deoxyribonucleotide synthesis and folate-mediated one-carbon metabolism occurred in the nucleus of eukaryotic cells. In support of the replitase concept, TYMS was physically localized within the nucleus in several mammalian cell types, indicating that folate-dependent de novo biosynthesis could occur in the nuclear compartment (44, 46, 51–54). However, localization studies in Saccharomyces cerevisiae indicated that although TYMS was associated with purified nuclei, it was present only on the nuclear envelope and/or endoplasmic reticulum membrane exterior surface but was not within the lumen (55). More recent studies have directly demonstrated that nuclei isolated from mouse liver can synthesize [3H] dTMP from [3H] serine, indicating that nuclei contain the entire folate-dependent de novo dTMP pathway (45). SHMT, TYMS, and DHFR were also found to be present in nuclei isolated from mouse liver (44, 45). In human Michigan Cancer Foundation-7 (MCF-7) cells, nuclear localization was restricted to the S and G2M phases of the cell cycle (45). Interestingly, both SHMT isozymes function in nuclear thymidylate biosynthesis. SHMT1 encodes a protein that localizes to the cytoplasm and nucleus, whereas SHMT2 encodes 2 transcripts that serve as templates for the synthesis of a mitochondrial isozyme (SHMT2) and a cytoplasmic/nuclear isozyme (SHMT2α). Nuclear localization of the enzymes involved in nuclear thymidylate biosynthesis occurs through the UBC9-mediated modification with the small ubiquitin-like modifier (SUMO), which targets proteins for nuclear localization during S-phase (43, 44). A common SHMT1 variant, C1420T, results in an amino acid substitution, L474F, that prevents SHMT SUMOylation and modifies risk for cardiovascular disease and acute lymphocytic leukemia (43). Collectively, these studies provide direct evidence for cell cycle–dependent nuclear dTMP biosynthesis in the nucleus.
There are many unanswered questions regarding the role and regulation of nuclear de novo dTMP biosynthesis. Nothing is known about the transport, processing, and accumulation of folates into the nucleus, the one-carbon forms of folate present in the nucleus, and the relationship between cell cycle dependency of de novo dTMP biosynthesis and cell cycle-dependent accumulation of nuclear folate. Active transport of folates into the nucleus may not be required, because the nuclear membrane breaks down and reassembles with each cell division, offering the opportunity to capture folate cofactors from the cytoplasm into the nucleus during each mitotic event. Such a passive accumulation of folates negates the need for transport of folate monoglutamates and subsequent polyglutamylation into the nucleus, but lacks specificity. This mechanism would result in the sequestering of all folate one-carbon forms in the nucleus, including 5-methylTHF, which is the predominate form of folate in the cytoplasm. However, only THF, DHF, and methyleneTHF are involved in the dTMP biosynthesis pathway, and 5-methylTHF is a potent inhibitor of SHMT1 and SHMT2 (56). Alternatively, folates may traverse the nuclear membrane bound to TYMS, SHMT, or DHFR, a mechanism that does not preclude the transport of inhibitory 5-methylTHF into the nucleus.
The biological rationale for dTMP biosynthesis in the nucleus, but not purine deoxyribonucleotide biosynthesis, remains unexplained. Recently, the salvage pathway for dTMP synthesis has also been localized to the nucleus (57). Thymidine is unique from other deoxyribonucleotides required for DNA replication in that its metabolic precursor, deoxyuradine nucleotide, can be incorporated into DNA during replication. Decreased rates of deoxythymidine triphosphate synthesis (58) results in incorporation of dUTP into DNA, because DNA polymerases do not discriminate between dUTP and deoxythymidine triphosphate (58). It remains to be established if the presence of the entire de novo dTMP synthesis is located directly at the replication fork during DNA synthesis to limit the accumulation of uracil into DNA or rather serves other purposes.
De novo purine biosynthesis
Nucleotide biosynthesis can occur either through salvage pathways or through de novo biosynthesis. De novo purine nucleotide biosynthesis is a 10-step pathway that is folate dependent, with 10-formylTHF supplying the number 2 and number 8 carbon of the purine ring through the activities of glycinamide ribonucleotide transformylase (GAR Tfase) and aminoimidazolecarboxamide ribonucleotide transformylase (AICAR Tfase). Alternatively, purine nucleotides can be synthesized through the single-step salvage pathway in which hypoxanthine and phosphoribosyl pyrophosphate are condensed to form inosine monophosphate, a precursor for AMP and GMP synthesis. The purine salvage pathway strongly inhibits the de novo salvage pathway through feedback inhibition of the de novo pathway by purine nucleotides synthesized through the salvage pathway (59, 60), and this has been the presumed mechanism whereby de novo purine biosynthesis is inhibited when the salvage pathway is active.
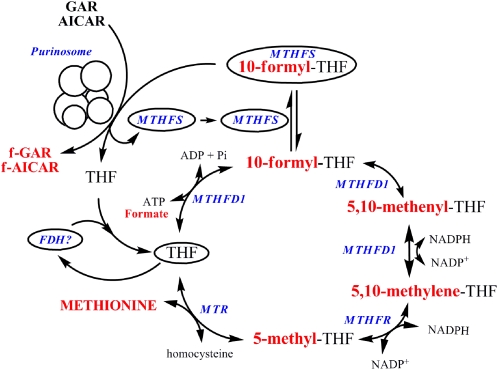
More recent studies have indicated another mechanism, whereby the salvage pathway regulates the de novo purine nucleotide biosynthesis pathway. The 6 enzymes that constitute the de novo synthesis pathway have been shown to form a multicomplex or metabolon in the cytoplasm referred to as the purinosome (Fig. 3) (61). The complex forms reversibly and once formed, it dissociates upon exposure to exogenous purines nucleotides or casein kinase II inhibitors (62), thereby indicating another potential mechanism whereby the salvage pathway regulates de novo purine nucleotide biosynthesis in addition to feedback inhibition (61).
Figure 3.
Putative trafficking of folate in the cytoplasm. The one-carbon unit is shown in red. f-GAR, formyl-GAR; f-AICAR, formyl-AICAR; Pi, phosphate.
The identification of the purinosome and knowledge that de novo purine biosynthesis only occurs when the salvage pathway is not functional indicate that although the enzymes that constitute the de novo synthesis pathway are present in the cytoplasm, the pathway is not functional until the multienzyme complex is established. The compartmentation of the purinosome within the cytoplasm requires a delivery mechanism to supply the complex with chemically labile 10-formylTHF cofactors and, similarly, a mechanism to remove THF following catalysis. MTHFD1, which synthesizes 10-formylTHF from formate and THF, is the most obvious candidate to deliver 10-formylTHF to the purinosome; however, this enzyme was shown not to colocalize with purinosome (61). Methenyltetrahydrofolate synthetase (MTHFS) has been proposed to be another candidate enzyme to deliver 10-formylTHF to GAR Tfase and AICAR Tfase within the purinosome (63). MTHFS catalyzes the ATP-dependent conversion of 5-formylTHF to 5,10-methenylTHF in a reaction that regulates the levels of 5-formylTHF, but also binds 10-formylTHF tightly as a tight-binding inhibitor. Increased expression of MTHFS in cells enhances rates of de novo purine biosynthesis and also protects de novo purine biosynthesis from antifolate inhibitors that target GAR Tfase and AICAR Tfase (63–65). Recently, we have validated experimentally that MTHFS colocalizes with the purinosome (Fig. 3).
10-Formyltetrahydrofolate dehydrogenase (FDH) is a candidate protein to remove THF from the purinosome and deliver the unsubstituted cofactor to either SHMT or MTHFD1 to regenerate 5,10-methyleneTHF or 10-formylTHF, respectively. FDH catalyzes the conversion of 10-formylTHF and NADP+ to THF and CO2. FDH exhibits strong product inhibition by THF, which binds tightly with a dissociation constant of 15 nmol/L (13). FDH product inhibition by THF is eliminated in vitro in the presence of either SHMT or MTHFD1, suggesting the potential for rapid transfer of THF between these enzymes potentially through substrate channeling. However, there is no evidence to date for a physical interaction between FDH and the purinosome, SHMT, or MTHFD1.
The homocysteine remethylation pathway
The folate-dependent remethylation of homocysteine to methionine in the cytoplasm requires the reduction of methyleneTHF to 5-methylTHF in an NADPH-dependent reaction catalyzed by MTHFR (Fig. 3). The 5-methylTHF cofactor is the one-carbon donor for folate-dependent homocysteine remethylation, which is catalyzed by methionine synthase. This reaction requires vitamin B-12 in the form of cobalamin to convert 5-methylTHF and homocysteine to methionine and THF. Because the MTHFR-catalyzed generation of 5-methyl-THF is essentially irreversible in vivo, accumulation of 5-methyl-THF can impair purine and thymidylate de novo biosynthesis, which occurs in severe vitamin B-12 deficiency. The enzymes MTHFD1, MTHFR, and methionine synthase constitute a metabolic cycle capable of converting formate and homocysteine to form methionine, but there is no evidence in the literature that these enzymes associate or are compartmentalized in the cytoplasm. Further research is needed to determine if and how the homocysteine remethylation cycle is compartmentalized.
Conclusions
There is increasing evidence that individual folate-dependent pathways are compartmentalized both within organelles and/or through the formation of multi-enzyme metabolons. These recent observations support early studies that inferred metabolic compartmentation and substrate channeling from the chemical and physical properties of folate cofactors, as well as kinetic and in vitro reconstitution studies of metabolic pathways (18). These studies are leading to a new appreciation of the relationships among folate-dependent pathways, which may not be regulated by direct competition of folate cofactors by folate-dependent enzymes but rather mediated by the partitioning and trafficking of folate cofactors among these pathways. The elucidation of the trafficking of folate cofactors among these complexes, their regulation, and impact of metabolic flux will be the next challenge to understanding folate-dependent one-carbon metabolism and its relationship to human health and disease.
Acknowledgments
P.J.S. and M.S.F. wrote the paper. P.J.S. had primary responsibility for final content. Both authors read and approved the final manuscript.
Footnotes
Supported by Public Health Service grant DK58144 to P.J.S.
Author disclosures: P. J. Stover and M. S. Field, no conflicts of interest.
Abbreviations used: AICAR Tfase, aminoimidazolecarboxamide ribonucleotide transformylase; DHF, dihydrofolate; DHFR, dihydrofolate reductase; FDH, formyltetrahydrofolate dehydrogenase; GAR Tfase, glycinamide ribonucleotide transformylase; MTHFD1, methylenetetrahydrofolate dehydrogenase; MTHFR, methylenetetrahydrofolate reductase MTHFS, methenyltetrahydrofolate synthetase; SHMT, serine hydroxymethyltransferase; THF, tetrahydrofolate; TYMS, thymidylate synthase.
Literature Cited
- 1.Srere PA. Enzyme concentrations in tissues. Science. 1967;158:936–7 [DOI] [PubMed] [Google Scholar]
- 2.Srere PA. Channeling: the pathway that cannot be beaten. J Theor Biol. 1991;152:23. [DOI] [PubMed] [Google Scholar]
- 3.Ovadi J, Saks V. On the origin of intracellular compartmentation and organized metabolic systems. Mol Cell Biochem. 2004;256–257:5–12 [DOI] [PubMed] [Google Scholar]
- 4.Shatalin K, Lebreton S, Rault-Leonardon M, Velot C, Srere PA. Electrostatic channeling of oxaloacetate in a fusion protein of porcine citrate synthase and porcine mitochondrial malate dehydrogenase. Biochemistry. 1999;38:881–9 [DOI] [PubMed] [Google Scholar]
- 5.Velot C, Mixon MB, Teige M, Srere PA. Model of a quinary structure between Krebs TCA cycle enzymes: a model for the metabolon. Biochemistry. 1997;36:14271–6 [DOI] [PubMed] [Google Scholar]
- 6.Qiu A, Jansen M, Sakaris A, Min SH, Chattopadhyay S, Tsai E, Sandoval C, Zhao R, Akabas MH, et al. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 2006;127:917–28 [DOI] [PubMed] [Google Scholar]
- 7.Zhao R, Matherly LH, Goldman ID. Membrane transporters and folate homeostasis: intestinal absorption and transport into systemic compartments and tissues. Expert Rev Mol Med. 2009;11:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low PS, Kularatne SA. Folate-targeted therapeutic and imaging agents for cancer. Curr Opin Chem Biol. 2009;13:256–62 [DOI] [PubMed] [Google Scholar]
- 9.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44 [DOI] [PubMed] [Google Scholar]
- 10.Suh JR, Herbig AK, Stover PJ. New perspectives on folate catabolism. Annu Rev Nutr. 2001;21:255–82 [DOI] [PubMed] [Google Scholar]
- 11.Zakrzewski SF. Evidence for the chemical interaction between 2-mercaptoethanol and tetrahydrofolate. J Biol Chem. 1966;241:2957–61 [PubMed] [Google Scholar]
- 12.Stites TE, Bailey LB, Scott KC, Toth JP, Fisher WP, Gregory JF III. Kinetic modeling of folate metabolism through use of chronic administration of deuterium-labeled folic acid in men. Am J Clin Nutr. 1997;65:53–60 [DOI] [PubMed] [Google Scholar]
- 13.Kim DW, Huang T, Schirch D, Schirch V. Properties of tetrahydropteroylpentaglutamate bound to 10-formyltetrahydrofolate dehydrogenase. Biochemistry. 1996;35:15772–83 [DOI] [PubMed] [Google Scholar]
- 14.Clifford AJ, Heid MK, Muller HG, Bills ND. Tissue distribution and prediction of total body folate of rats. J Nutr. 1990;120:1633–9 [DOI] [PubMed] [Google Scholar]
- 15.Horne DW, Patterson D, Cook RJ. Effect of nitrous oxide inactivation of vitamin B12-dependent methionine synthetase on the subcellular distribution of folate coenzymes in rat liver. Arch Biochem Biophys. 1989;270:729–33 [DOI] [PubMed] [Google Scholar]
- 16.Schirch V, Strong WB. Interaction of folylpolyglutamates with enzymes in one-carbon metabolism. Arch Biochem Biophys. 1989;269:371–80 [DOI] [PubMed] [Google Scholar]
- 17.Pawelek PD, Allaire M, Cygler M, MacKenzie RE. Channeling efficiency in the bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase domain: the effects of site-directed mutagenesis of NADP binding residues. Biochim Biophys Acta. 2000;1479:59–68 [DOI] [PubMed] [Google Scholar]
- 18.Strong WB, Schirch V. In vitro conversion of formate to serine: effect of tetrahydropteroylpolyglutamates and serine hydroxymethyltransferase on the rate of 10-formyltetrahydrofolate synthetase. Biochemistry. 1989;28:9430–9 [DOI] [PubMed] [Google Scholar]
- 19.Shin YS, Chan C, Vidal AJ, Brody T, Stokstad EL. Subcellular localization of gamma-glutamyl carboxypeptidase and of folates. Biochim Biophys Acta. 1976;444:794–801 [DOI] [PubMed] [Google Scholar]
- 20.Lin BF, Huang RF, Shane B. Regulation of folate and one-carbon metabolism in mammalian cells. III. Role of mitochondrial folylpoly-gamma-glutamate synthetase. J Biol Chem. 1993;268:21674–9 [PubMed] [Google Scholar]
- 21.Panetta JC, Wall A, Pui CH, Relling MV, Evans WE. Methotrexate intracellular disposition in acute lymphoblastic leukemia: a mathematical model of gamma-glutamyl hydrolase activity. Clin Cancer Res. 2002;8:2423–9 [PubMed] [Google Scholar]
- 22.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–90 [DOI] [PubMed] [Google Scholar]
- 23.Winter-Vann AM, Kamen BA, Bergo MO, Young SG, Melnyk S, James SJ, Casey PJ. Targeting Ras signaling through inhibition of carboxyl methylation: an unexpected property of methotrexate. Proc Natl Acad Sci USA. 2003;100:6529–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stead LM, Jacobs RL, Brosnan ME, Brosnan JT. Methylation demand and homocysteine metabolism. Adv Enzyme Regul. 2004;44:321–33 [DOI] [PubMed] [Google Scholar]
- 25.Appling DR. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB J. 1991;5:2645–51 [DOI] [PubMed] [Google Scholar]
- 26.Barlowe CK, Appling DR. In vitro evidence for the involvement of mitochondrial folate metabolism in the supply of cytoplasmic one-carbon units. Biofactors. 1988;1:171–6 [PubMed] [Google Scholar]
- 27.Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem. 2002;277:38381–9 [DOI] [PubMed] [Google Scholar]
- 28.MacFarlane AJ, Liu X, Perry CA, Flodby P, Allen RH, Stabler SP, Stover PJ. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem. 2008;283:25846–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy EA, Titus SA, Taylor SM, Jackson-Cook C, Moran RG. A mutation inactivating the mitochondrial inner membrane folate transporter creates a glycine requirement for survival of Chinese hamster cells. J Biol Chem. 2004;279:33829–36 [DOI] [PubMed] [Google Scholar]
- 30.McBurney MW, Whitmore GF. Isolation and biochemical characterization of folate deficient mutants of Chinese hamster cells. Cell. 1974;2:173–82 [DOI] [PubMed] [Google Scholar]
- 31.Taylor RT, Hanna ML. Folate-dependent enzymes in cultured Chinese hamster ovary cells: impaired mitochondrial serine hydroxymethyltransferase activity in two additional glycine–auxotroph complementation classes. Arch Biochem Biophys. 1982;217:609–23 [DOI] [PubMed] [Google Scholar]
- 32.Pike ST, Rajendra R, Artzt K, Appling DR. Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem. 2010;285:4612–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Pietro E, Sirois J, Tremblay ML, MacKenzie RE. Mitochondrial NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is essential for embryonic development. Mol Cell Biol. 2002;22:4158–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Pietro E, Wang XL, MacKenzie RE. The expression of mitochondrial methylenetetrahydrofolate dehydrogenase-cyclohydrolase supports a role in rapid cell growth. Biochim Biophys Acta. 2004;1674:78–84 [DOI] [PubMed] [Google Scholar]
- 35.Bolusani S, Young BA, Cole NA, Tibbetts AS, Momb J, Bryant JD, Solmonson A, Appling DR. Mammalian MTHFD2L encodes a mitochondrial methylenetetrahydrofolate dehydrogenase isozyme expressed in adult tissues. J Biol Chem. 2011;286:5166–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen KE, MacKenzie RE. Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. Bioessays. 2006;28:595–605 [DOI] [PubMed] [Google Scholar]
- 37.Tibbetts AS, Appling DR. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81 [DOI] [PubMed] [Google Scholar]
- 38.Nijhout HF, Reed MC, Budu P, Ulrich CM. A mathematical model of the folate cycle: new insights into folate homeostasis. J Biol Chem. 2004;279:55008–16 [DOI] [PubMed] [Google Scholar]
- 39.Nijhout HF, Reed MC, Ulrich CM. Mathematical models of folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:45–82 [DOI] [PubMed] [Google Scholar]
- 40.Green JM, MacKenzie RE, Matthews RG. Substrate flux through methylenetetrahydrofolate dehydrogenase: predicted effects of the concentration of methylenetetrahydrofolate on its partitioning into pathways leading to nucleotide biosynthesis or methionine regeneration. Biochemistry. 1988;27:8014–22 [DOI] [PubMed] [Google Scholar]
- 41.Kang SS, Zhou J, Wong PW, Kowalisyn J, Strokosch G. Intermediate homocysteinemia: a thermolabile variant of methylenetetrahydrofolate reductase. Am J Hum Genet. 1988;43:414–21 [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlivan EP, Davis SR, Shelnutt KP, Henderson GN, Ghandour H, Shane B, Selhub J, Bailey LB, Stacpoole PW. Methylenetetrahydrofolate reductase 677C->T polymorphism and folate status affect one-carbon incorporation into human DNA deoxynucleosides. J Nutr. 2005;135:389–96 [DOI] [PubMed] [Google Scholar]
- 43.Woeller CF, Anderson DD, Szebenyi DM, Stover PJ. Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J Biol Chem. 2007;282:17623–31 [DOI] [PubMed] [Google Scholar]
- 44.Anderson DD, Woeller CF, Stover PJ. Small ubiquitin-like modifier-1 (SUMO-1) modification of thymidylate synthase and dihydrofolate reductase. Clin Chem Lab Med. 2007;45:1760–3 [DOI] [PubMed] [Google Scholar]
- 45.Anderson DD, Stover PJ. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS ONE. 2009;4:e5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prem veer Reddy G, Pardee AB. Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Natl Acad Sci USA. 1980;77:3312–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boorstein RJ, Pardee AB. Coordinate inhibition of DNA synthesis and thymidylate synthase activity following DNA damage and repair. Biochem Biophys Res Commun. 1983;117:30–6 [DOI] [PubMed] [Google Scholar]
- 48.Noguchi H. Prem veer Reddy G, Pardee AB. Rapid incorporation of label from ribonucleoside disphosphates into DNA by a cell-free high molecular weight fraction from animal cell nuclei. Cell. 1983;32:443–51 [DOI] [PubMed] [Google Scholar]
- 49.Pontarin G, Fijolek A, Pizzo P, Ferraro P, Rampazzo C, Pozzan T, Thelander L, Reichard PA, Bianchi V. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc Natl Acad Sci USA. 2008;105:17801–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mathews CK, Slabaugh MB. Eukaryotic DNA metabolism. Are deoxyribonucleotides channeled to replication sites? Exp Cell Res. 1986;162:285–95 [DOI] [PubMed] [Google Scholar]
- 51.Brown SS, Neal GE, Williams DC. Subcellular distribution of some folic acid-linked enzymes in rat liver. Biochem J. 1965;97:C34–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samsonoff WA, Reston J, McKee M, O'Connor B, Galivan J, Maley G, Maley F. Intracellular location of thymidylate synthase and its state of phosphorylation. J Biol Chem. 1997;272:13281–5 [DOI] [PubMed] [Google Scholar]
- 53.Wong NA, Brett L, Stewart M, Leitch A, Longley DB, Dunlop MG, Johnston PG, Lessells AM, Jodrell DI. Nuclear thymidylate synthase expression, p53 expression and 5FU response in colorectal carcinoma. Br J Cancer. 2001;85:1937–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bissoon-Haqqani S, Moyana T, Jonker D, Maroun JA, Birnboim HC. Nuclear expression of thymidylate synthase in colorectal cancer cell lines and clinical samples. J Histochem Cytochem. 2006;54:19–29 [DOI] [PubMed] [Google Scholar]
- 55.Poon PP, Storms RK. Thymidylate synthase is localized to the nuclear periphery in the yeast Saccharomyces cerevisiae. J Biol Chem. 1994;269:8341–7 [PubMed] [Google Scholar]
- 56.Stover P, Schirch V. 5-Formyltetrahydrofolate polyglutamates are slow tight binding inhibitors of serine hydroxymethyltransferase. J Biol Chem. 1991;266:1543–50 [PubMed] [Google Scholar]
- 57.Chen YL, Eriksson S, Chang ZF. Regulation and functional contribution of thymidine kinase 1 in repair of DNA damage. J Biol Chem. 2010;285:27327–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA. 1997;94:3290–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaoka T, Kondo M, Honda S, Iwahana H, Moritani M, Ii S, Yoshimoto K, Itakura M. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis. J Biol Chem. 1997;272:17719–25 [DOI] [PubMed] [Google Scholar]
- 60.Yamaoka T, Yano M, Kondo M, Sasaki H, Hino S, Katashima R, Moritani M, Itakura M. Feedback inhibition of amidophosphoribosyltransferase regulates the rate of cell growth via purine nucleotide, DNA, and protein syntheses. J Biol Chem. 2001;276:21285–91 [DOI] [PubMed] [Google Scholar]
- 61.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–6 [DOI] [PubMed] [Google Scholar]
- 62.An S, Kyoung M, Allen JJ, Shokat KM, Benkovic SJ. Dynamic regulation of a metabolic multi-enzyme complex by protein kinase CK2. J Biol Chem. 2010;285:11093–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Field MS, Anguera MC, Page R, Stover PJ. 5,10-Methenyltetrahydrofolate synthetase activity is increased in tumors and modifies the efficacy of antipurine LY309887. Arch Biochem Biophys. 2009;481:145–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Field MS, Szebenyi DM, Perry CA, Stover PJ. Inhibition of 5,10-methenyltetrahydrofolate synthetase. Arch Biochem Biophys. 2007;458:194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Field MS, Szebenyi DM, Stover PJ. Regulation of de novo purine biosynthesis by methenyltetrahydrofolate synthetase in neuroblastoma. J Biol Chem. 2006;281:4215–21 [DOI] [PubMed] [Google Scholar]