Abstract
The function of non-coding RNA genes largely depends on their secondary structure and the interaction with other molecules. Thus, an accurate prediction of secondary structure and RNA–RNA interaction is essential for the understanding of biological roles and pathways associated with a specific RNA gene. We present web servers to analyze multiple RNA sequences for common RNA structure and for RNA interaction sites. The web servers are based on the recent PET (Probabilistic Evolutionary and Thermodynamic) models PETfold and PETcofold, but add user friendly features ranging from a graphical layer to interactive usage of the predictors. Additionally, the web servers provide direct access to annotated RNA alignments, such as the Rfam 10.0 database and multiple alignments of 16 vertebrate genomes with human. The web servers are freely available at: http://rth.dk/resources/petfold/
INTRODUCTION
RNA folding follows a hierarchical pathway where the primary sequence dictates the formation of the secondary structure, which in turn leads to the functional tertiary structure. Since the RNA secondary structure dominates the free energy of the tertiary conformation, the secondary structure is critical for a large number of RNA mediated processes such as regulation of gene expression, RNA maturation, splicing and translation (1). Additionally, many non-protein-coding RNAs (ncRNAs) and structured mRNA elements exhibit their function through binding to proteins, other RNA molecules or both. Besides post-transcriptional gene regulation through mRNA binding by microRNAs and siRNAs, RNA–RNA interactions occur between many small RNAs in bacteria, e.g. CopA–CopT and FinP–traJ 5′-UTR, small nucleolar RNAs and small nuclear RNAs as well as ribosomal RNAs for their methylation and pseudouridylation, or even between certain long non-coding RNAs (lncRNAs) and microRNAs to regulate their activity or guide RNA editing. Thereby, the accessibility of nucleotides in the RNA sequence, which is constrained by the intra-molecular structure, has a large impact on the possible interaction sites.
Thermodynamic methods, such as RNAfold (2) or Mfold (3), employ a dynamic programming algorithm to find the thermodynamically most stable secondary structure by minimizing the free energy of the folded molecule. However, it is known that due to several reasons, such as interactions with proteins or other RNAs and processing of RNAs, the minimum free energy (MFE) structure is often not the functional structure of the molecule. Instead, the functional structure is a suboptimal structure, often in the vicinity of the MFE structure. Comparative methods use information from multiple homologous sequences for secondary structure prediction by considering compensatory mutations that maintain important structural elements over the course of evolution, e.g. Pfold (4). The integration of both the thermodynamic and evolutionary paradigms into one model was the motivation behind PETfold (5) and we have shown that our combined PET model increases the accuracy of comparative structure prediction compared to previous methods. We further extended the PET model into PETcofold (6,7) for predicting conserved RNA–RNA interactions.
Two web servers for RNA secondary structure prediction from multiple sequence alignments have been developed previously, RNAalifold [Vienna RNA web suite (8)] and Pfold (4). As mentioned in (5), the approach here employs a scheme based on the principles of these two programs. Interactions between two RNA molecules can be predicted with, e.g. IntaRNA [Freiburg RNA Tools (9)]. However, simultaneous RNA cofolding and prediction of conserved interactions from multiple sequence alignments are not supported. Additionally, none of these web servers allows the direct access to such alignments of known ncRNAs and UTR elements as well as human genome blocks.
The web servers presented here offer the prediction of conserved RNA secondary structures (PETfold) and RNA–RNA interactions (PETcofold) on multiple sequence alignments. The web servers are accompanied by a graphical interface that enables optimal exploitation of the methods when working on a specific data set. As an example, we demonstrate PETfold's structure prediction from a multiple alignment of the microRNA lin-4. As an example for RNA-RNA interaction with PETcofold, we demonstrate the web server functionality on the bacterial inhibitory antisense-target RNA complex between the two conserved structured RNA sequences FinP and the 5′-UTR of traJ.
ALGORITHM
The PETfold algorithm is described in depth in (5) and the extended algorithm of PETcofold in (6). In short, the maximum expected accuracy (MEA) approach of PETfold integrates evolutionary structure reliabilities and thermodynamic structure probabilities in one scoring scheme. First, base pairs with high evolutionary reliability score are fixed to take functional conserved structure elements into account and then the weighted sum of reliabilities is calculated. The consensus structure with maximal expected overlap of a multiple alignment is computed by a Nussinov style algorithm using the combined reliabilities. The evolutionary reliability of paired and unpaired bases is calculated by Pfold from a phylogenetic tree, a substitution model and a stochastic context-free (SCFG) grammar stochastically describing the building of RNA structures. The thermodynamic probability of a base to be paired or unpaired is calculated by RNAfold as the partition function in the energetic equilibrium.
The PETfold algorithm is extended in PETcofold by hierarchical and constrained folding to predict conserved interactions between two RNA alignments taking the non-accessibility of highly reliable intra-molecular stems into account. The hierarchical folding consists of two steps: (i) intra-molecular folding of both RNA alignments which is (ii) followed by constrained expected accuracy scoring, which is an extension of PETfold's MEA for constrained folding. This strategy requires the introduction of a partial structure probability for highly reliable stems in each alignment that are predicted in step 1 to be inaccessible during the interaction of two RNA molecules. In step 2, constrained base pairs are handled as single stranded, so that the Nussinov style algorithm for nested structures allows the prediction of loop–loop interactions between stable intra-molecular hairpin-loops without increased time consumption.
WEB SERVER
Usage
The web server offers a separate input page for PETfold and PETcofold. The required input is one multiple sequence alignment for PETfold or two multiple sequence alignments with at least three shared sequence identifiers for PETcofold in FASTA format. Both input pages also offer direct access to Rfam 10.0 (10) seed alignments with or without paralogs and/or human (hg18) 17-way MULTIZ alignments for predicting secondary structures and interactions. MULTIZ alignments are free of paralogs by definition, in contrast, many Rfam seed alignments contain several instances of a family in the same organism. Thus, both servers offer a no paralogs version of each Rfam seed that is mandatory for the input of PETcofold to get the same identifiers in both alignments. The no paralogs alignment has the Rfam identifiers mapped to organism-specific MULTIZ identifiers and only the sequence that has the closest distance to the mean distance in the phylogenetic tree is selected for each group of paralogs. In addition, the user can specify a phylogenetic tree in Newick format as well as one or two consensus secondary structures in dot-bracket format for calculating their score in the PET-model. Several other parameters can be set for each submission. The default values for both the PETfold and PETcofold models are set to the optimal values that we found in the application to previous data sets. Detailed description of the input formats and sample files are provided on the website.
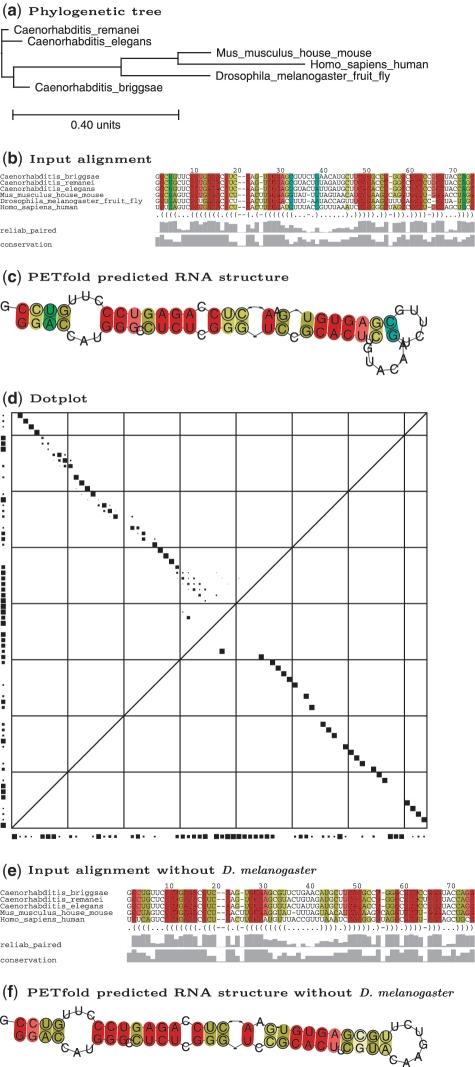
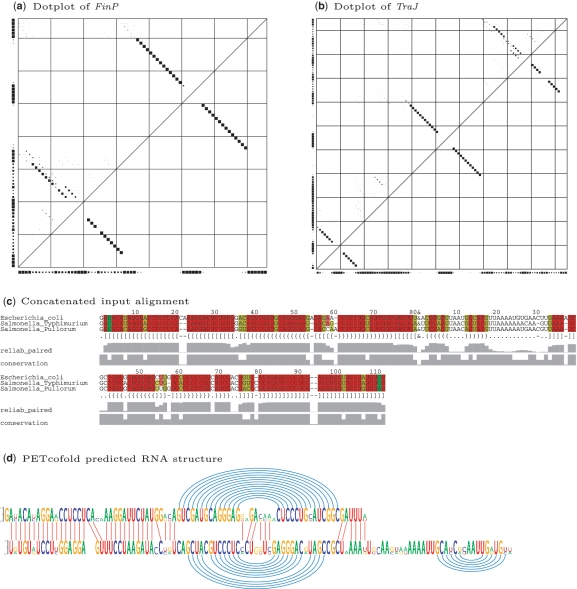
After submission, the user can access his results via a link that contains a secure unique identifier. Optionally, an e-mail notification on the completion of the computation can be selected upon submission. The results of each submission are displayed on an HTML page, which is accessible up to 60 days after completion of the computation. The output page shows either the user-specified phylogenetic tree or the tree calculated by the neighbor joining algorithm. The PETfold or PETcofold plain text output includes the predicted (joint) consensus RNA secondary structure in dot-bracket notation, its score and length normalized score. An example of the PETfold output for a typical microRNA is shown in Figure 1. The alignment shows the conservation of the primary sequence and the reliability scores for base pairs, while the predicted consensus secondary structure is plotted underneath using the same color scheme. The dotplot shows the base pair reliabilities (upper triangle) and the base pairs that take part in the predicted consensus secondary structure (lower triangle). Single stranded reliabilities are shown on the X and Y axis of the plot. The PETcofold output contains a joint multiple alignment including intra- and inter-molecular base pairs in dot-bracket notation (Figure 2a). The predicted joint secondary structure is plotted by connecting base pairs in the intra-molecular structures as arcs and RNA-RNA interactions as straight lines (Figure 2b). The input alignments are denoted by sequence logos (11), which show the sequence conservation for each alignment position. The dotplot is extended by reliabilities for inter-molecular base pairs. All plots can be downloaded as postscript and PDF file.
Figure 1.
The PETfold output for the microRNA lin-4 based on the sequence alignment from Rfam 10.0: (a)–(d) are seed sequences without paralogs and (e)–(f) are seed sequences without paralogs and Drosophila melanogaster. The output consists of a phylogenetic tree (a), the respective input alignment with indication of the sequence conservation and the predicted RNA structure in dot-bracket format and pairing reliabilities (b and e), the predicted RNA structure (c and f), and finally the dotplot with base pair reliabilities in the upper left triangle (size of squares is linear correlated to reliabilities) and MEA-structure in lower right triangle (d). The vertical and horizontal lines stand for base indices dividable by 10. In (b,c,e,f) compensatory mutations supporting the consensus structure are marked by the Vienna RNA coloring schema.
Figure 2.
The PET(co)fold output for the antisense RNA FinP regulation of the 5′-UTR of the major F-plasmid transcriptional activator TraJ based on the sequence alignment without paralogs from Rfam 10.0: (a) the dotplot of FinP with base pair reliabilities in upper left triangle and MEA-structure in lower right triangle produced by PETfold; (b) the dotplot of TraJ produced by PETfold; (c) the concatenated Rfam seed alignments with sequence conservation indicated, the PETcofold predicted structure in dot-bracket format and base paired reliabilities; (d) the PETcofold predicted RNA binding structure. (a) and (b) show the output produced by the PETfold web server and (c) and (d) show the output produced by the PETcofold web server. Intra-molecular pairing is denoted by round brackets in (c) and arcs in (d) and inter-molecular pairing is denoted by curly brackets in (c) and straight lines between both sequences in (d).
Implementation
The web server runs Apache and uses PHP scripts together with the Smarty template engine to process the input and render the HTML pages. Local installations of the Rfam database and the UCSC browser provide access to multiple alignments of ncRNA families and genome-wide MULTIZ alignments, respectively. Structure prediction requests are submitted to a queueing system that distributes jobs on a compute cluster. PETfold and PETcofold are implemented in C and Perl. Phylogenetic trees are drawn by drawphyl and dotplots of structural reliabilities are drawn by drawdot, which both come with the Pfold package. A multiple alignment is drawn by an adapted version of the Vienna RNA suite program coloraln.pl, which includes a scheme for coloring compensatory base pairs as well as a bar plot for both conservation and paired reliabilities for each alignment column. Additionally, a secondary structure of a single RNA molecule is drawn by RNAplot of the Vienna RNA suite. Figures for joint secondary structures including RNA–RNA interactions, as predicted by PETcofold, are drawn by RIplot implemented as part of our web server.
RESULTS AND DISCUSSION
The quality of RNA secondary structure predictions is dependent on the input alignment(s) and suitable parameter settings. For instance, Figure 1b shows that the lin-4 sequence in Drosophila melanogaster consists of 8 bases that are not consistent with the base pair pattern of the homologs in 5 other organisms. The removal of D. melanogaster from the alignment, see Figure 1e and 1f, increases the PET-score of the MEA consensus structure from 42 to 45 and extends the stem inside the larger loop of the Rfam annotated seed alignment structure. In general, the analyses of the phylogenetic tree for clusters or outliers and the base pair conservation in the alignment can help to improve the accuracy of the prediction through the submission of an adapted alignment. The resubmission is supported by a link to the original submission form at the top of the result page. Additionally, the pairing reliabilities in Figure 1b and individual base pair reliabilities in Figure 1d give information about possible alternative structures. Thus, the reliabilities for the lin-4 prediction also show that the base pair between bases 32 and 38 is not very reliable.
The pairing reliability of a base describes how often it is base paired in the ensemble of all RNA structures. The substitution of several adjacent bases with high pairing reliabilities will change the structure dramatically and likely change or destroy the RNA molecules functionality. This information may be relevant for RNA structure probing. Regions of high pairing reliabilities can be found in the alignment plots of the servers such as in Figure 1b and the upper triangle of Figure 1d.
In antisense regulation, stable RNA-RNA complexes require the propagation of initial loop-loop interactions (12). PETcofold addresses this feature through the prediction of inter-molecular kissing hairpins. The 5′-UTR of the major F-plasmid transcriptional activator TraJ and its antisense ncRNA FinP have Rfam entries that can be selected on the web servers. The alignment of the antisense ncRNA FinP is folded by PETfold to have two conserved stems of which the 5′ stem is more unstable as shown by many alternative base pairs in the upper triangle of the dotplot (Figure 2a). TraJ folds in 3 stems of which the 3′ stem is predicted to be the most unstable, also supported by an interior loop as shown in the lower triangle of the dotplot (Figure 2b). The RNA–RNA interaction of TraJ and FinP is initiated by two loop–loop interactions between both hairpin loops of FinP and the two last hairpin loops of TraJ. In previous studies on the duplexing of FinP and traJ (13) as well as the antisense RNA CopA and its target mRNA CopT (12), the destabilization of stems by bulges adjacent to loops had been shown to be necessary for propagation of the inter-molecular base pairing from the initial kissing complex to a more fully paired and therefore more stable RNA–RNA interaction structure. This observation is supported by the PETcofold prediction of a duplex structure that maintains only the stable stem-loops (curly brackets in Figure 2c and blue arcs in Figure 2d) whereas the loop–loop interaction of more unstable stems is extended to longer antisense pairing (squared brackets in Figure 2c and red lines on the left side of Figure 2d). Here, we show that degeneration of stem-loops is driven by low pairing reliabilities and competing structures, which implies interior loops. The features that govern the high efficiency of natural antisense RNA are, thus, related to the secondary structures of interaction partners.
CONCLUSION
We have presented web servers for analysis of aligned RNA sequences with respect to common structures as well as interactions. The web servers provide the user with a graphical output that allows for immediate insight in the result, overview of the reliabilities (confidence) of intra- as well as inter-molecular RNA base pairs, and identification of columns with compensatory changes.
Compensatory base pair changes have been shown to preserve functional RNA structures during evolution, e.g. in the aforementioned microRNA lin-4. Providing RNA multiple alignments where such base changes are preserved is a tedious task that is addressed by the Sankoff algorithm for structural alignment. In practise, the implementations are slow and rely on heuristics and reduced search space, e.g. FOLDALIGN (14,15) and LocaRNA (16). Hence, the PETfold and PETcofold web servers provide access to hand curated alignments such as seed alignments of known RNA families in Rfam and sequence alignments such as MULTIZ alignments, which are still the best resources of phylogenetic information. However, even Rfam seed alignments especially of long RNAs contain many gaps and misalignments that result in wrong structure predictions. The interactivity of the web servers, thus, allows the adaption of alignments for optimal structure predictions.
To comply with the future need for optimized refinement of the multiple alignment, integration with RNA editors such as SARSE (17) may increase the ability for curation. Other possible improvements are the removal of the single non-stacked base pairs inherited from Pfold and hierarchical folding in PETfold as already applied in PETcofold to allow for prediction of intra-molecular pseudoknots. On the graphical side incorporating structure logo (18) features in the output to indicate the degree of compensating base changes would further ease the interpretation of the results.
FUNDING
Danish Strategic and Independent Research Councils, through the committee for strategic growth technologies and research council for Technology and Production; the Danish Center for Scientific Computing. Funding for open access charge: Danish Strategic Research Council.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Bjarne Knudsen and the Vienna RNA community for Pfold, RNAfold and applied graphical tools.
REFERENCES
- 1.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 2.Hofacker I, Fontana W, Stadler P, Bonhoeffer L, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh Chem. 1994;125:167–188. [Google Scholar]
- 3.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol. Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
- 4.Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seemann SE, Gorodkin J, Backofen R. Unifying evolutionary and thermodynamic information for RNA folding of multiple alignments. Nucleic Acids Res. 2008;36:6355–6362. doi: 10.1093/nar/gkn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seemann SE, Richter AS, Gorodkin J, Backofen R. Hierarchical folding of multiple sequence alignments for the prediction of structures and RNA-RNA interactions. Algorithms Mol. Biol. 2010;5:22. doi: 10.1186/1748-7188-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seemann SE, Richter AS, Gesell T, Backofen R, Gorodkin J. PETcofold: predicting conserved interactions and structures of two multiple alignments of RNA sequences. Bioinformatics. 2011;27:211–219. doi: 10.1093/bioinformatics/btq634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber AR, Lorenz R, Bernhart SH, Neubck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373–W377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider TD, Stephens R. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb FA, Westhof E, Ehresmann C, Ehresmann B, Wagner EG, Romby P. Bulged residues promote the progression of a loop-loop interaction to a stable and inhibitory antisense-target RNA complex. Nucleic Acids Res. 2001;29:3145–3153. doi: 10.1093/nar/29.15.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arthur DC, Ghetu AF, Gubbins MJ, Edwards RA, Frost LS, Glover JNM. FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 2003;22:6346–6355. doi: 10.1093/emboj/cdg607. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havgaard JH, Torarinsson E, Gorodkin J. Fast pairwise structural RNA alignments by pruning of the dynamical programming matrix. PLoS Comput. Biol. 2007;3:1896–1908. doi: 10.1371/journal.pcbi.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havgaard JH, Lyngsø RB, Gorodkin J. The FOLDALIGN web server for pairwise structural RNA alignment and mutual motif search. Nucleic Acids Res. 2005;33:W650–W653. doi: 10.1093/nar/gki473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring non-coding RNA families and classes by means of genome-scale structure-based clustering. PLOS Comput. Biol. 2007;3:e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen ES, Lind-Thomsen A, Knudsen B, Kristensen SE, Havgaard JH, Torarinsson E, Larsen N, Zwieb C, Sestoft P, Kjems J, et al. Semiautomated improvement of RNA alignments. RNA. 2007;13:1850–1859. doi: 10.1261/rna.215407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorodkin J, Heyer LJ, Brunak S, Stormo GD. Displaying the information contents of structural RNA alignments: the structure logos. Comput. Appl. Biosci. 1997;13:583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]