Abstract
WebFR3D is the on-line version of ‘Find RNA 3D’ (FR3D), a program for annotating atomic-resolution RNA 3D structure files and searching them efficiently to locate and compare RNA 3D structural motifs. WebFR3D provides on-line access to the central features of FR3D, including geometric and symbolic search modes, without need for installing programs or downloading and maintaining 3D structure data locally. In geometric search mode, WebFR3D finds all motifs similar to a user-specified query structure. In symbolic search mode, WebFR3D finds all sets of nucleotides making user-specified interactions. In both modes, users can specify sequence, sequence–continuity, base pairing, base-stacking and other constraints on nucleotides and their interactions. WebFR3D can be used to locate hairpin, internal or junction loops, list all base pairs or other interactions, or find instances of recurrent RNA 3D motifs (such as sarcin–ricin and kink-turn internal loops or T- and GNRA hairpin loops) in any PDB file or across a whole set of 3D structure files. The output page provides facilities for comparing the instances returned by the search by superposition of the 3D structures and the alignment of their sequences annotated with pairwise interactions. WebFR3D is available at http://rna.bgsu.edu/webfr3d.
INTRODUCTION
Although the number of atomic-resolution RNA 3D structures deposited in the Protein Data Bank [PDB, (1)] and Nucleic Acids Database [NDB, (2)] databases remains small compared to the numbers of protein or DNA structures, it is steadily growing. Significantly, it contains a number of very large and complex supra-molecular assemblies, including, with the addition of the 40 S subunit from Tetrahymena (3), ribosomes from all three domains of life. The complexity of such large 3D structures requires the use of computational tools to extract information for specific chemical and biological analyses.
The hierarchical nature of structured RNA molecules has been noted by a number of workers (4,5). Besides helical elements defining the secondary structure, most RNA molecules contain recurrent structural modules that share the same 3D shape and interaction patterns and serve as anchoring points for tertiary interactions and binding sites for proteins, small molecules or other RNAs. Recurrent 3D motifs in general are small enough to evolve independently in unrelated RNA molecules (6–8). Examples of recurrent RNA motifs include sarcin–ricin, kink-turn and C-internal loops as well as TPsiC- and GNRA hairpin loops. Although usually shown in 2D diagrams as unstructured ‘loops’, such motifs are generally highly structured. To further our understanding of their structures, sequence variations and evolution it is important to be able to identify and catalog these motifs, which necessitates the development of software to enable detailed analysis of atomic-resolution 3D structures.
FR3D (9), a suite of MATLAB programs developed for this purpose, has been available for download since 2008 as source code (http://rna.bgsu.edu/FR3D). As FR3D is under constant development and improvement, and as the RNA structure database is constantly growing, maintaining a current local version of the software and up-to-date collection of annotated structure files is a burden to users. Therefore, we have developed WebFR3D (http://rna.bgsu.edu/webfr3d), which has the same familiar interface as the standalone version, offers extensive help and runs the current version of FR3D, including access to up-to-date annotated sets of RNA 3D structures from the PDB, including a non-redundant set of files.
WebFR3D complements other web servers dedicated to searching RNA 3D structures. First we describe the key features of existing web servers and then we note the complementary features that WebFR3D offers. RNA FRABASE (10) uses primary and/or secondary structure to search in a database that stores pre-computed annotations of all PDB-derived RNA structures and is regularly updated. It includes modified nucleotides and allows one to limit pseudo-rotation parameters, sugar pucker amplitude and torsion angles. It can search for large RNA fragments, but has a limited capability for specifying detailed interaction constraints between nucleotides in the query, as WebFR3D does.
With FASTR3D (11), the user can specify a range of nucleotides from a PDB file as a query. The server uses secondary structure information and backbone torsion angles to look for similar structures in a list of PDB files. Alternatively, it can take primary and/or secondary structures as an input. Unlike FR3D, FASTR3D only allows secondary structure constraints on searches. Moreover, it does not appear that the library of PDB files available for FASTR3D searches is regularly updated.
FRASS (12) is capable of handling large RNA fragments and is designed for global similarity searching. The user can select an entire chain from a PDB file or upload a structure to the server. The searching method is based on Gauss integrals that are used to compare the shapes of backbones of RNA molecules.
ARTS (13) employs a geometric approach enhanced by heuristics to find the largest number of phosphorous atoms and base pairs that can be superimposed in the two input structures. The output is a global superposition of the query structures or discovery of the maximal structural similarities between them. Both FRASS and ARTS are designed for global structural comparison of structures and are of limited use for exhaustively searching for and comparing instances of recurrent 3D motifs.
DIAL (14) applies a dynamic programming approach to align pairs of lists of annotated dihedral angles, representing RNA chain segments. The method can optionally take into account base sequence and base pairing within the input structures.
SARSA (15) uses vector quantization in order to obtain a structural alphabet of RNA backbone conformations. The input structures are represented using this alphabet, and structural alignment problem is carried out by classical sequence alignment.
Both DIAL and SARSA have multiple alignment modes, of which pairwise semi-global modes are most similar to WebFR3D. These modes, although based on different approaches, can detect query RNA 3D motifs in the target structure. However, neither program provides for imposing detailed constraints on the query, and, as with all pairwise methods, neither is suitable for quick high-throughput analysis of RNA 3D motifs.
SARA (16) applies unit-vector root mean square approach to pairwise structural alignment. It can also assign RNA structures to functional classes as defined in the SCOR database (17). SARA is not applicable for aligning structures smaller than 20 nt, which makes it less relevant for RNA 3D motif search and discovery.
Another web service developed by our group is WebR3DAlign (18). While WebFR3D is designed to search for individual recurrent RNA motifs, WebR3DAlign addresses a related problem, to identify all motifs conserved in the 3D structures of two possibly homologous RNA molecules. WebR3DAlign produces a nucleotide-to-nucleotide alignment of two 3D structures from which one may readily identify conserved motifs, while allowing for differences in the global structure of the molecules (for example, domain motions). In WebR3DAlign, the two 3D structures are decomposed into a large number of overlapping 4-nt neighborhoods, which are compared with each other geometrically using the same base-centric approach implemented in WebFR3D. The structural alignment is produced by systematically combining alignments of locally similar neighborhoods.
In summary, several features distinguish WebFR3D (and FR3D) from methods implemented on other web servers:
WebFR3D uses a base-centric geometric approach so that searches return all geometrically similar motifs, regardless of differences in their backbone topologies (9);
WebFR3D can perform purely geometric searches, and thus it does not rely solely on pre-computed structural annotations, which are limited by current understanding of recurrent RNA structures; and
WebFR3D allows placing constraints on the interactions formed by pairs of nucleotides in the query. This gives FR3D the unique capability to conduct purely symbolic searches as well as geometric searches with additional symbolic constraints.
INPUT AND OUTPUT
Description of input
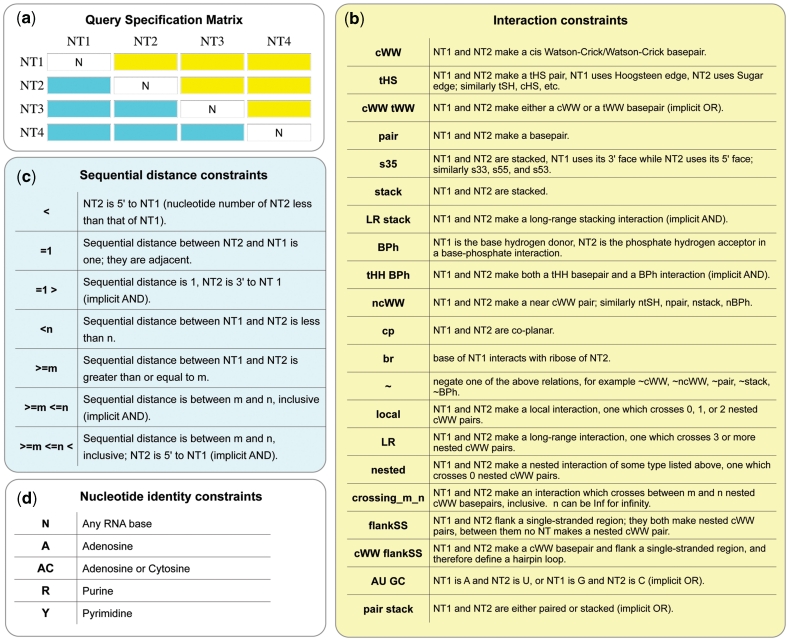
WebFR3D allows users to perform geometric and symbolic searches for structural motifs in RNA-containing 3D structures from the PDB. In the symbolic search mode, the user can specify collections of up to 15 nt and a variety of pairwise base–base and base–backbone interactions to find all RNA fragments that satisfy the constraints (Figure 1). Using symbolic search, it is possible to find all instances of a specific structural motif in a given RNA structure or across a whole set of 3D structure files; for example, all GNRA hairpin loops, sarcin–ricin internal loops, or A-minor interactions and most kissing loop interactions. This mode can also be used more generally to find all hairpin, internal or junction loop motifs in a given set of 3D structures. Or it can be used to simply list all pairwise interactions of a specific type; for example, all base pairs belonging to a particular geometric family, or to check whether a particular sequence has been observed in the 3D database in some structural context.
Figure 1.
Data inputs for defining and restricting symbolic and geometric searches in WebFR3D and FR3D. (a) Query Specification Matrix (QSM) for a 4-nt query. The user can specify constraints between any pairs of nucleotides in the query in the QSM by entering any of the codes shown in (b) in the cells with yellow background. Distance constraints are entered using any of the codes shown in (c) in the cells with cyan background. Nucleotide identity constraints shown in (d) are entered in the cells along the diagonal of the QSM. (b) Codes for interaction constraints. The syntax is illustrated for nucleotides 1 and 2 of the query (‘NT1’ and ‘NT2’). (c) Codes for sequential distance constraints. (d) Some of the available codes for nucleotide identity constraints. Detailed information on the use of constraints is available in the Help section of the WebFR3D website.
In the geometric search mode, the user can select a fragment from any RNA-containing structure deposited in the PDB and search for geometrically similar fragments in other 3D structures. The algorithm implemented for geometric search in WebFR3D is the same as FR3D and guarantees finding all similar fragments within a user-specified geometric discrepancy (9). Geometric search is slower but more robust than symbolic search, as it does not rely on pre-computed structural annotations. The user can focus and speed up geometric search by specifying symbolic constraints. This is especially useful when searching for known RNA 3D motifs with specific patterns of interactions, such as sarcin–ricin loops, kink-turns or C-loops. Specifying backbone connectivity constraints also speeds up searches. However, WebFR3D can be run to identify motifs that contain insertions of arbitrary length or arbitrary topologies.
WebFR3D is updated weekly with new X-ray, NMR and cryo-EM RNA structures as they become available in PDB. Users can choose one or more individual PDB files to search. Alternatively, the user can select one of the pre-compiled non-redundant lists of X-ray structures, grouped according to minimum resolution (from 1.5 to 4.0 Å). These lists are determined by an automated implementation of the procedure outlined in (19), and are also regularly updated. The current non-redundant lists can be accessed at http://rna.bgsu.edu/nrlist.
User input is extensively error-checked prior to submission of a search to ensure, for example, that all nucleotides in query motifs for geometric searches actually exist. In geometric search, the user can preview the 3D structure of the query fragment to ensure that the correct RNA fragment was selected before the search is submitted. WebFR3D also provides for entry of an email address to receive notification when the results of a search become available.
To assist the user in preparing a query, WebFR3D is equipped with a contextual help system, which the user can consult to view options for relevant search parameters. A tutorial is accessible from the main page, as well as examples of geometric and symbolic searches. Users are encouraged to contact the WebFR3D team with questions and suggestions using the built-in contact form.
Experience with WebFR3D shows that it is generally more efficient to begin with more restricted queries and then to gradually relax the search parameters in subsequent searches, based on analysis of the output. Thus, when searching geometrically for similar hairpin loops, an effective way to reduce the search space significantly is to set the distance between adjacent nucleotides to a number larger than any expected insertion between adjacent nucleotides, for example, specifying ‘≤5' to allow insertion of up to 4 nt. This constraint can be adjusted or removed in subsequent searches depending on the results.
Description of output
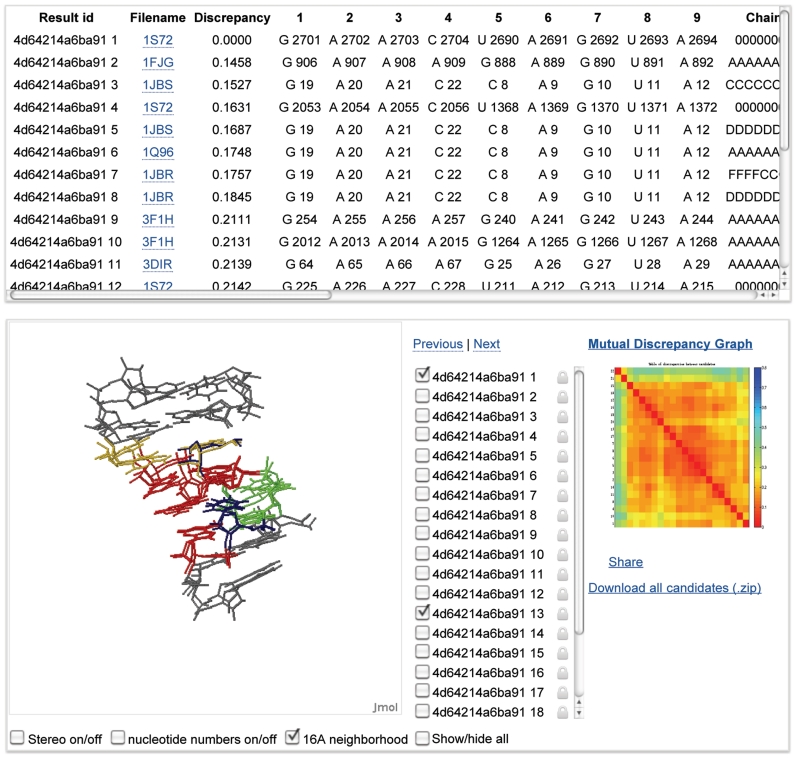
WebFR3D presents the user with an annotated list of the fragments that satisfy the search criteria specified in the query. The fragments are also visualized interactively using a Jmol applet (http://www.jmol.org), which allows one to compare fragments by superposition and to explore the structural context in which they occur by displaying the neighboring nucleotides (Figure 2). All fragments satisfying the query can be downloaded in PDB format and viewed locally.
Figure 2.
Output of a WebFR3D search for a recurrent RNA 3D motif showing the annotated instances (top), Jmol superpositions and neighboring nucleotides (bottom left), and the mutual discrepancy heat map (bottom right).
The mutual geometric discrepancy between all fragments is calculated and displayed as a heat map (Figure 2, bottom right). Low geometric discrepancy (red) corresponds to similar structures, while high geometric discrepancy (blue) indicates differing structures.
The results are stored on the server indefinitely with stable URLs, which makes it easy for collaborators to share search results or to provide interactive supplementary material for publications.
METHOD
The computations are carried out by FR3D, a suite of programs written in MATLAB (9). FR3D has the capability to perform rapid searches for RNA structural fragments matching all pairwise constraints given in the query. For geometric searches, FR3D finds all instances that match the query motif within a user-specified geometric discrepancy, calculated as described in (9). The upper limit on the geometric discrepancy is effectively a pairwise constraint on the distance between two nucleobases. In symbolic mode, FR3D searches pre-computed annotations of nucleotide interactions to find all structural fragments matching the user-specified criteria. Mixed searches involve geometric search using a query motif as well as symbolic search criteria. In geometric or mixed searches, FR3D ranks candidate motifs according to the geometric discrepancy from the query motif and only returns those below the user-specified cutoff discrepancy. Motifs matching all the search criteria are also aligned and compared to each other by geometric discrepancy to cluster them into geometrically similar groups. The clusters are displayed using a heat map.
FR3D annotates base pair interactions in RNA 3D structures according to the Leontis–Westhof classification (20), stacking consistent with the RNA Ontology (21), base–phosphate interactions (22) and backbone conformations (21,23). FR3D identifies nested, local and long-range interactions by comparison to the secondary structure defined by the Watson–Crick pairs following the IR heuristic in (24).
IMPLEMENTATION
The server is hosted by the RNA structural bioinformatics laboratory at BGSU. The user interface is implemented in HTML and CSS. Validation is performed using Ajax and JavaScript, which must be enabled in the user's browser for the website to work properly. The server-side implementation involves Perl and PHP scripting and a MySQL database. The main computations are performed in MATLAB.
The server is capable of processing multiple requests simultaneously. The time required for searching is dependent on the size and the nature of the query (smaller and more restricted queries run more quickly) and on the number of PDB files searched. Well-designed searches are usually completed within 3–10 min. Note that after 20 min the execution is aborted and the user is notified. It is recommended to use standalone FR3D installations to perform intense computations that take more time.
WebFR3D shares the same performance characteristics as its core program, FR3D, with a small performance hit caused by the additional processing required for web output. The operation count of FR3D and WebFR3D is of order O(n2+[(m-1)/2]), where n is the number of nucleotides in the file(s) being searched and m is the number of nucleotides in the query motif (9). This applies to the worst-case scenario, when the search is purely geometric with no symbolic constraints; symbolic constraints reduce search time, sometimes dramatically. For benchmark examples of FR3D performance the reader is referred to the original publication (9). WebFR3D’s real-world performance and execution time depend strongly on the number of imposed constraints and the servers’ workload.
Site usage is monitored using the Google Analytics tracking system. When the demand for the service increases, WebFR3D will be migrated to a more powerful computational facility.
FUNDING
National Institutes of Health (grant numbers 1R01GM085328-01A1 to C.L.Z. and N.B.L. and 2R15GM055898-05 to N.B.L.). Funding for open access charge: NIH grant (1R01GM085328-01A1). Meetings supported by National Science Foundation Grant No. 0443508 (RNA Ontology Consortium) accelerated the development of WebFR3D.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Blake Sweeney and Ryan Rahrig for helpful suggestions while developing the server, and Michael Sarver and Jesse Stombaugh for developing and testing the original versions of FR3D.
REFERENCES
- 1.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40 S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 4.Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl Acad. Sci. USA. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Leontis NB, Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 7.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasalean L, Stombaugh J, Zirbel CL, Leontis NB. In: Non-Protein Coding RNAs. Walter NG, Woodson SA, Batey RT, editors. Vol. 13. Berlin Heidelberg: Springer; 2009. pp. 1–26. [Google Scholar]
- 9.Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J. Math. Biol. 2008;56:215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popenda M, Szachniuk M, Blazewicz M, Wasik S, Burke EK, Blazewicz J, Adamiak RW. RNA FRABASE 2.0: an advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures. BMC Bioinformatics. 2010;11:231. doi: 10.1186/1471-2105-11-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai C-E, Tsai M-Y, Liu Y-C, Wang C-W, Chen K-T, Lu CL. FASTR3D: a fast and accurate search tool for similar RNA 3D structures. Nucleic Acids Res. 2009;37:W287–W295. doi: 10.1093/nar/gkp330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirillova S, Tosatto SC, Carugo O. FRASS: the web-server for RNA structural comparison. BMC Bioinformatics. 2010;11:327. doi: 10.1186/1471-2105-11-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dror O, Nussinov R, Wolfson HJ. The ARTS web server for aligning RNA tertiary structures. Nucleic Acids Res. 2006;34:W412–W415. doi: 10.1093/nar/gkl312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferre F, Ponty Y, Lorenz WA, Clote P. DIAL: a web server for the pairwise alignment of two RNA three-dimensional structures using nucleotide, dihedral angle and base-pairing similarities. Nucleic Acids Res. 2007;35:W659–W668. doi: 10.1093/nar/gkm334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YF, Huang YL, Lu CL. SARSA: a web tool for structural alignment of RNA using a structural alphabet. Nucleic Acids Res. 2008;36:W19–W24. doi: 10.1093/nar/gkn327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capriotti E, Marti-Renom MA. SARA: a server for function annotation of RNA structures. Nucleic Acids Res. 2009;37:W260–W265. doi: 10.1093/nar/gkp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura M, Hendrix DK, Klosterman PS, Schimmelman NR, Brenner SE, Holbrook SR. SCOR: Structural Classification of RNA, version 2.0. Nucleic Acids Res. 2004;32:D182–D184. doi: 10.1093/nar/gkh080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahrig RR, Leontis NB, Zirbel CL. R3D Align: global pairwise alignment of RNA 3D structures using local superpositions. Bioinformatics. 2010;26:2689–2697. doi: 10.1093/bioinformatics/btq506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stombaugh J, Zirbel C, Westhof E, Leontis N. Frequency and isostericity of RNA base pairs. Nucleic Acids Res. 2009;37:2294–2312. doi: 10.1093/nar/gkp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoehndorf R, Batchelor C, Bittner T, Dumontier M, Eilbeck K, Knight R, Mungall CJ, Richardson JS, Stombaugh J, Westhof E, et al. The RNA Ontology (RNAO): an ontology for integrating RNA sequence and structure data. Appl. Ontol. 2011;6:53–89. [Google Scholar]
- 22.Zirbel CL, Sponer JE, Sponer J, Stombaugh J, Leontis NB. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res. 2009;37:4898–4918. doi: 10.1093/nar/gkp468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, et al. RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smit S, Rother K, Heringa J, Knight R. From knotted to nested RNA structures: a variety of computational methods for pseudoknot removal. RNA. 2008;14:410–416. doi: 10.1261/rna.881308. [DOI] [PMC free article] [PubMed] [Google Scholar]