Abstract
MemPype is a Python-based pipeline including previously published methods for the prediction of signal peptides (SPEP), glycophosphatidylinositol (GPI) anchors (PredGPI), all-alpha membrane topology (ENSEMBLE), and a recent method (MemLoci) that specifically discriminates the localization of eukaryotic membrane proteins in: ‘cell membrane’, ‘internal membranes’, ‘organelle membranes’. MemLoci scores with accuracy of 70% and generalized correlation coefficient (GCC) of 0.50 on a rigorous homology-unbiased validation set and overpasses other predictors for subcellular localization. The annotation process is based both on inheritance through homology and computational methods. Each submitted protein first retrieves, when available, up to 25 similar proteins (with sequence identity ≥50% and alignment coverage ≥50% on both sequences). This helps the identification of membrane-associated proteins and detailed localization tags. Each protein is also filtered for the presence of a GPI anchor [0.8% false positive rate (FPR)]. A positive score of GPI anchor prediction labels the sequence as exposed to ‘Cell surface’. Concomitantly the sequence is analysed for the presence of a signal peptide and classified with MemLoci into one of three discriminated classes. Finally the sequence is filtered for predicting its putative all-alpha protein membrane topology (FPR <1%). The web server is available at: http://mu2py.biocomp.unibo.it/mempype.
INTRODUCTION
In Eukaryotes, most protein functional features are constrained by the different cell compartments and their enclosing membranes (1–3). Functional features of biological membranes strictly depend on proteins that specifically interact with them. Membrane proteins can be classified into two major classes: integral membrane proteins, which span the lipid bilayer [transmembrane (TM) proteins (TPs)] or covalently bind a lipid molecule, and peripheral membrane proteins, which physically interact with the membrane surfaces. About 30% of eukaryotic proteins in SwissProt are annotated with the keyword ‘membrane’ (48 963 sequences out of 166 219), and 75% of them are also annotated as ‘transmembrane’ (37 659 sequences). In most cases, the experimental determination of the structure and function of membrane proteins is presently hampered by technical problems and their function is often annotated on the basis of sequence similarity. Our annotation procedure takes advantage of both inheritance of annotation (annotation transfer) after homology search and annotation by predicting features with different machine learning approaches. To this purpose MemPype integrates methods that are specifically suited to predict the presence of signal peptides, lipid anchors, membrane protein localization and topology of all-alpha membrane proteins, thus providing an integrated computational resource for annotation of eukaryotic membrane proteins. However, the main novelty in MemPype is the integration of MemLoci, a method that allows a reliable classification of both eukaryotic integral and peripheral membrane proteins into three classes: cell membrane (CM), organelle membranes (OMs) and internal membranes (IMs) (4). This is a key step for functional annotation of membrane proteins in relation to their membrane type (5,6). We propose MemPype to support annotation of membrane proteomes of eukaryotic organisms with the unique feature of also identifying proteins present on the cell surface. These chains are likely candidates to be characterized as biomarkers and/or targets for new drugs.
MemPype WORKFLOW
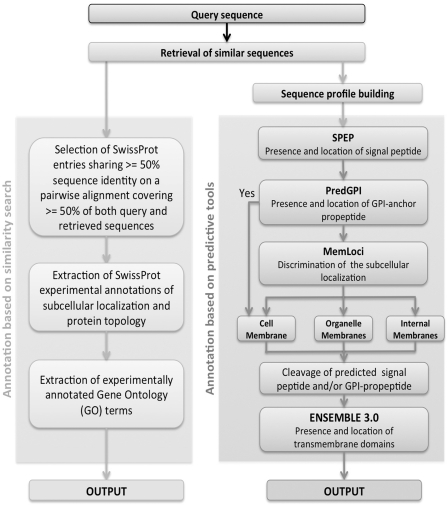
MemPype includes two flows of annotation (Figure 1). The first collects information directly from SwissProt in terms of keywords and Gene Ontology (GO) terms associated with proteins sharing high similarity with the target sequence (≥50% sequence identity with an alignment coverage ≥50% on both sequences, see below). The second parallel flow of annotation includes machine learning-based methods that score at the state of the art for the specific problem at hand. Each sequence is filtered for the presence of: (i) signal peptides with SPEP (7); (ii) presence and location of glycophosphatidylinositol (GPI)-anchoring domains with PredGPI (8); then (iii) the subcellular localization of both integral and peripheral membrane proteins is predicted with MemLoci, a recent predictor based on support vector machine (SVM); and finally (iv) the location and topology of all-alpha integral membrane proteins is predicted with ENSEMBLE 3.0 (9). The only input is the residue sequence of the target protein. The first step of the pipeline is a BLAST search against SwissProt that produces alignments of the target sequence with an E-value ≤10−3 (leftmost path in Figure 1). Homologous sequences are used both for performing annotation transfer by sequence similarity and for compiling the sequence profiles that are used as input to most of the predictive methods included in the pipeline (rightmost path in Figure 1). Both flow outputs are given as a result of MemPype running (Figure 2). The results of the first search gives at the most 25 aligned sequences and their features as derived from SwissProt. This information can or cannot be present depending on the target sequence. The second output is always present and gives computed features whose reliability is statistically computed according to the different predictors and can be inspected in relation to the results of the SwissProt search when available. The platform integrates predictors that have been previously described and validated on their specific task. Presently a set of proteins with experimentally validated features to be used in cross-validation for the joint combination of all the predictors is not available. Prediction performances are therefore calculated independently for each method with never seen before proteins carrying along the experimentally validated property to be predicted.
Figure 1.
Workflow of the MemPype annotation pipeline. MemPype performs annotation with homology search and prediction tools. See text for further details.
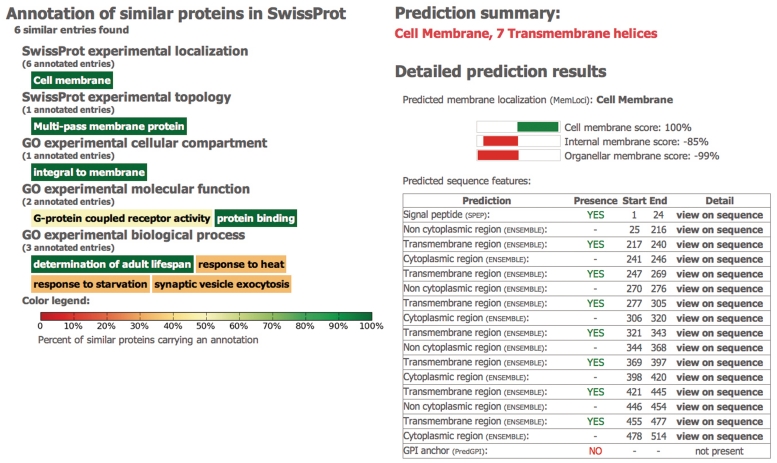
Figure 2.
MemPype output results. Two outputs are returned: (i) a list of at the most 25 proteins sharing sequence identity ≥50% on an alignment covering ≥50% of both sequence lengths (when available). Both keywords and GO terms can be transferred on the basis of sequence similarity to the query sequence. (ii) A list of all the predicted features including signal peptide [with SPEP (7)], GPI-anchor [with PredGPI (8)], all-alpha TM topology [with ENSEMBLE3.0 (9)] and prediction of subcellular localization [with MemLoci (4)]. See text for further details.
ANNOTATION THROUGH INHERITANCE
Transfer of annotation on the basis of sequence similarity is a widely adopted procedure that relies on the assumption that similar sequences share similar structural and functional features (10). The threshold value of sequence similarity necessary for ensuring a reliable inference of function depends on the specific task. It is well known that the overall protein structure is conserved for proteins sharing some ≥30% identical residues, while the conservation of molecular function requires higher identity thresholds [≥50% (11)]. In relation to subcellular localization, sequence identity ≥30% ensures a reliable annotation transfer within non-membrane proteins (12). However, to our knowledge, the same threshold has not yet been determined for membrane proteins. To this aim, we collected from SwissProt 24 640 membrane proteins endowed with experimental annotation of subcellular localization [the set is described in (4)]. Twelve localization classes are considered. Upon an extensive pairwise alignment procedure, we determined that the subcellular localization is conserved in 99.7% cases, when two proteins share ≥50% sequence identity with coverage ≥50% on both sequences (data not shown). The MemPype annotation transfer procedure considers therefore only the set of annotated SwissProt sequences fulfilling these constraints with respect to the target proteins. When many annotated sequences with identity ≥50% and coverage ≥50% are retrieved, only the most similar 25 are taken into account. If existing, the annotations reported in the ‘KEYWORD’ field of the retrieved sequences and referring to structural and localization features are collected, as well as the GO annotations coming from experimental evidences. All the annotation terms are then represented as a tag cloud, where each tag is coloured with a scale representing the frequency of each keyword in the set (Figure 2). By pointing over each tag, the detailed statistics of each annotation appears. The set of entries promoting a specific annotation can then be retrieved by clicking on the corresponding tag. In some cases, the annotation transfer procedure allows a very specific and detailed annotation such as ‘Endoplasmic reticulum-Golgi intermediate compartment membrane.’ Moreover, the system can be useful for annotating proteins endowed with multiple localizations. It is not always possible to find annotated proteins fulfilling the constraints of sequence identity necessary for a reliable transfer of annotation based on homology search. A complementary approach is therefore the adoption of predictive methods that run in the same platform and whose results can be either compared/confirmed with those obtained with the homology search or provides the unique annotation resource.
PREDICTION OF SIGNAL PEPTIDE AND GPI ANCHOR
The first step of the prediction pipeline is to determine the sequence of the mature protein, where N-terminal signal peptides and/or the GPI-anchoring propeptides, when present, are cleaved. To this aim, SPEP in its version for eukaryotic sequences (7) and PredGPI (8) are applied. Both methods analyse the residue sequence and efficiently determine the presence of peptides as well as the position of the cleavage sites. SPEP is a neural network (NN)-based system, trained on 2300 eukaryotic proteins endowed with experimental annotation (13). Two NNs scan the 65-residue long N-terminal segment of the query sequence, scoring the probability of each residue to be part of a signal peptide and to be the cleavage site, respectively. The allowed signal peptide length ranges between 11 and 59 residues. A signal peptide is predicted if the sum of the outputs of the NNs are greater than a threshold that was selected in order to optimize the performance. By this, when performing the discrimination task on the training data set with a cross-validation procedure, SPEP scores with a Matthews correlation coefficient (CC) as high as 0.91 and overall accuracy (Acc) equal to 95% (7). Here a validation set consisting of 1287 eukaryotic proteins has been extracted from (14) with the exclusion of sequences present in the SPEP training set. The results of the blind validation are reported in Table 1 and show a performance consistent with the scores obtained in cross-validation (CC = 0.87 and Acc = 93%). PredGPI is trained on a data set comprising 340 and 10 630 GPI- and non-GPI-anchored proteins, respectively (8). It includes a SVM, whose discrimination threshold is selected in order to limit the false positive rate (FPR) to 0.5% on the training set. By this, the cross-validation performances are CC = 0.78 and Acc = 99% (8). When a protein is predicted as GPI anchored, the cleavage site is predicted with a hidden Markov model (HMM) that casts the features of the cleaved propeptide and its surrounding regions. Here we collect a validation set consisting of 19 GPI-anchored proteins (with unknown cleavage site) released after training PredGPI, and 391 non-GPI-anchored proteins released after Jan 2011. On this blind set PredGPI scores with CC = 0.87 and Acc = 99.2%, with FPR of the GPI-anchored class as low as 0.8% (Table 1). MemPype outputs list, when present, cleaved peptides highlighted along the sequence. Sequence and sequence profile of the mature protein are then obtained by deleting the sequence segments corresponding to the cleaved peptides. When a sequence contains a GPI-anchor domain, its subcellular localization is labelled ‘cell membrane’ (15). The low FPR of PredGPI ensures that the rate of wrong localization annotation due to misprediction of GPI anchor is about 1%. Irrespective of this labelling, the sequence is predicted by the complete pipeline and results of MemLoci and the possible presence of TM helices are reported (see next sections). To further assess the error rate that could arise from the combination of PredGPI and MemMoci, PredGPI was also scored on a blind validation subset of MemLoci comprising 68 proteins in OM and IM with the exclusion of CM proteins. Only one protein is wrongly predicted as GPI anchored and thus reported as ‘cell membrane’, confirming the low FPR of PredGPI.
Table 1.
Performance of the different predictors included in MemPype on never seen before validation sets
| Method | Blind validation set | Sen, % | Sp, % | FPR, % | Acc, % | CC |
|---|---|---|---|---|---|---|
| SPEP | 543 proteins with SP | 89 | 95 | 3 | 93 | 0.87 |
| 744 proteins without SP | 97 | 91 | 11 | |||
| PredGPIa | 19 GPI-anchored proteins | 89 | 85 | 0.8 | 99 | 0.87 |
| 391 non-GPI-anchored proteins | 99 | 99 | 11 | |||
| ENSEMBLE3.0a | 15 TM proteins | 100 | 83 | 0.4 | 99 | 0.91 |
| 208 non-TM proteins | 99 | 100 | 0 | |||
| MemLocia | 32 CM proteins | 56 | 75 | 9 | 70 | 0.50b |
| 18 OM proteins | 50 | 56 | 9 | |||
| 50 IM proteinsc | 86 | 72 | 34 |
aThe validation set collects never seen before chains by the method and deposited after January 2010. Predictions are scored with the following indexes: Sen: sensitivity = (no. of correctly predicted proteins in the class)/(total no. of proteins in the class); Sp: specificity = (no. of correctly predicted proteins in the class)/(total no. of proteins predicted in the class); FPR = (no. of mispredicted proteins in the class)/(total no. of proteins in the complementary class); Acc = (no. of correctly predicted proteins)/(total no. of proteins); Matthews CC is adopted for binary classifications, while GCC (b) is computed for multiclass classifications (22).
cIMs comprising all the endomembrane system except the cell membrane. All the validation sets are available at the MemPype website in the ‘Info’ page.
PREDICTION OF SUBCELLULAR LOCALIZATION
Prediction of subcellular localization of eukaryotic membrane proteins is performed with MemLoci [4], a SVM-based method able to discriminate the localization of membrane proteins within three classes: CM, OMs and IMs. The OM class comprises proteins located at mitochondrial or plastidial membranes: the IM class comprises all the remaining intracellular membranes (the endoplasmic reticulum, the nuclear membranes, the Golgi apparatus, the vesicles, the vacuoles, the lysosomes, the peroxisome, the microsomes and the endosome). MemLoci is the first tool specifically suited to predict the subcellular localization of both integral and peripheral membrane proteins. Other available predictors of subcellular localization explicitly exclude membrane proteins from their training sets (16,17), group all the membrane proteins into a single class referred as ‘membrane’ or ‘cell membrane’ (18,19), or focus on specific membrane types and organisms (20,21). MemLoci scores with generalized CC (GCC) (22) in the range of 0.50 when tested on both the 10 634 sequences included in the training set and the 100 sequences of an independent validation set (Table 1). For each sequence, MemPype lists the localizations predicted with MemLoci and three values scoring their likelihood. The highest value indicates the most likely prediction.
TOPOLOGY PREDICTION AND DISCRIMINATION AND OF ALL-ALPHA TPs
The mature sequence (after signal peptide and GPI-anchor propetide cleavage) is predicted for the presence and topology of all-alpha TM domains with ENSEMBLE3.0, an updated version of ENSEMBLE (9) and based on an ensemble prediction of different machine learning tools that analyse the information contained in sequence profiles, including the capability of discriminating between all-alpha membrane and globular protein. ENSEMBLE 3.0 is trained on a non-redundant data set of 138 all-alpha membrane proteins (including only three eukaryotic chains), whose structure is known with atomic resolution and was deposited in the Protein Data Bank (PDB) before January 2010. Performing a rigorous cross-validation, ENSEMBLE3.0 is able to correctly locate the TM segments of 126 proteins (91%) and to predict the correct orientation with respect to the membrane plane of 119 proteins (86%) of the training/testing set, respectively. Here we test ENSEMBLE 3.0 on a validation set of 15 independent membrane proteins sharing low identity (≤25%) with the training set and whose structures have been deposited after January 2010. This set includes only three proteins from eukaryotes, and two of these are endowed with one validated and one putative signal peptide, respectively. When the sequences of all 15 mature proteins are predicted, ENSEMBLE3.0 correctly computes the topology of all of them. Alternatively, when the full-length sequence of the 15 proteins is submitted to ENSEMBLE 3.0, the topology of only 13 proteins is correctly predicted (87%), with the exclusion of the two eukaryotic proteins endowed with signal peptide. These proteins are correctly predicted when SPEP is combined with ENSEMBLE3.0. In order to test whether ENSEMBLE3.0 is capable of discriminating membrane from globular proteins, we trained a filter on a data set also including 1611 globular structural domains, relative to proteins sharing <25% sequence similarity with the training set and released before January 2010 [extracted from PDB with PISCES (23)]. On a validation set comprising 208 never seen before globular domains (in proteins released after January 2010 and with sequence identity ≤25% to the training set) and the 15 TM proteins, FPR was 0 and 0.4%, respectively (Table 1). When the total set of eukaryotic full-length globular and membrane proteins (67 and 3, respectively) were jointly predicted by SPEP and ENSEMBLE, FPR was 0 and 2%, respectively. For TPs, MemPype lists the membrane spanning segments and their topological organization (cytoplasmic, non-cytoplasmic; Figure 2). When the sequence does not contain predicted membrane-spanning segments or GPI-anchored domains, a warning message is visualized indicating that MemLoci prediction should be taken with caution and possibly validated by merging features derived from the homology search.
WEB SERVER
The MemPype web server requires protein sequences in FASTA format as input. Each sequence must at least be 50-residue long. Upon request submission the server displays the prediction result page that is periodically updated until the completion of the prediction procedure. This page can be bookmarked and accessed later. Moreover, a unique identifier marks each prediction request as a future reference to retrieve prediction results. For each sequence the current queue state is reported, and upon completion the prediction results are shown. These are stored in a local database and will remain available for at least 1 month. The web server can be accessed either from anonymous or registered users. Registration is free of charge. Registered users can submit up to five sequences per request and up to 30 different requests per hour, while, to enforce a fair use policy, anonymous users are allowed for only 1 sequence per request and 10 requests per hour. For facilitating the retrieval of the results the web server provides a ‘Recent Jobs’ page, where the predictions of anonymous users are publicly available, while registered users can retrieve their own jobs in the private ‘My Jobs’ page. All the software used to build MemPype (except for BLAST+) is written in Python language. The web server runs on a web2py engine, and the annotated sequences are stored in SQLite database adopting the BioSQL schema. Parsing of SwissProt annotation data is performed with the BioPython uniprot-xml parser. HMMs and SVMs needed for all the prediction steps were implemented in Python as well.
FUNDING
MIUR-FIRB (Fondo per gli Investimenti della Ricerca di Base) 2003/LIBI-International Laboratory for Bioinformatics (to R.C., in part). Funding for open access charge: Fondo Ordinario per le Università (FFO) 2010 (to R.C. and P.L.M.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
C.S. and V.I. are PhD students supported by Ministero Italiano della Università e Ricerca (MIUR) and CIRC, respectively.
REFERENCES
- 1.Sachs JN, Engelman DM. Introduction to the membrane protein reviews: the interplay of structure, dynamics, and environment in membrane protein function. Annu. Rev. Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 2.White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 3.Almén MS, Nordström KJV, Friedriksson R, Schiöt HB. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierleoni A, Martelli PL, Casadio R. MemLoci: predicting subcellular localization of membrane proteins in Eukaryotes. Bioinformatics. 2011;27:1224–1230. doi: 10.1093/bioinformatics/btr108. [DOI] [PubMed] [Google Scholar]
- 5.Imai K, Nakai K. Prediction of subcellular locations of proteins: where to proceed? Proteomics. 2010;10:3970–3983. doi: 10.1002/pmic.201000274. [DOI] [PubMed] [Google Scholar]
- 6.Casadio R, Martelli PL, Pierleoni A. The prediction of protein subcellular localization from sequence: a shortcut to functional genome annotation. Brief. Funct. Genomics Proteomics. 2008;7:63–73. doi: 10.1093/bfgp/eln003. [DOI] [PubMed] [Google Scholar]
- 7.Fariselli P, Finocchiaro G, Casadio R. SPEPLip: the detection of signal peptide and lipoprotein cleavage sites. Bioinformatics. 2003;19:2498–2499. doi: 10.1093/bioinformatics/btg360. [DOI] [PubMed] [Google Scholar]
- 8.Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martelli PL, Fariselli P, Casadio R. An ENSEMBLE machine learning approach for the prediction of all-alpha membrane proteins. Bioinformatics. 2003;19:i205–i211. doi: 10.1093/bioinformatics/btg1027. [DOI] [PubMed] [Google Scholar]
- 10.Loewenstein Y, Raimondo D, Redfern OC, Watson J, Frishman D, Linial M, Orengo C, Thornton J, Tramontano A. Protein function annotation by homology-based inference. Genome Biol. 2009;10:207. doi: 10.1186/gb-2009-10-2-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rost B. Enzyme function less conserved than anticipated. J. Mol. Biol. 2002;318:595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- 12.Nair R, Rost B. Sequence conserved for subcellular localization. Prot. Sci. 2002;11:2836–2847. doi: 10.1110/ps.0207402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menne KM, Hermjakob H, Apweiler R. A comparison of signal sequence prediction methods using a test set of signal peptides. Bioinformatics. 2000;16:741–742. doi: 10.1093/bioinformatics/16.8.741. [DOI] [PubMed] [Google Scholar]
- 14.Nugent T, Jones DT. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee S, Mayor S. The GPI-anchor and protein sorting. Cell. Mol. Life. Sci. 2001;58:1969–1987. doi: 10.1007/PL00000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair R, Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005;348:85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 17.Pierleoni A, Martelli PL, Fariselli P, Casadio R. BaCelLo: a balanced subcellular localization predictor. Bioinformatics. 2006;22:e408–e416. doi: 10.1093/bioinformatics/btl222. [DOI] [PubMed] [Google Scholar]
- 18.Briesemeister S, Rahnenführer J, Kohlbacher O. Going from where to why–interpretable prediction of protein subcellular localization. Bioinformatics. 2010;26:1232–1238. doi: 10.1093/bioinformatics/btq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurila K, Vihinen M. PROlocalizer: integrated web service for protein subcellular localization prediction. Amino Acids. 2011;40:975–980. doi: 10.1007/s00726-010-0724-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Dunbrack RL., Jr PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Res. 2005;33:W94–W98. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]