Abstract
The BioExtract Server (bioextract.org) is an open, web-based system designed to aid researchers in the analysis of genomic data by providing a platform for the creation of bioinformatic workflows. Scientific workflows are created within the system by recording tasks performed by the user. These tasks may include querying multiple, distributed data sources, saving query results as searchable data extracts, and executing local and web-accessible analytic tools. The series of recorded tasks can then be saved as a reproducible, sharable workflow available for subsequent execution with the original or modified inputs and parameter settings. Integrated data resources include interfaces to the National Center for Biotechnology Information (NCBI) nucleotide and protein databases, the European Molecular Biology Laboratory (EMBL-Bank) non-redundant nucleotide database, the Universal Protein Resource (UniProt), and the UniProt Reference Clusters (UniRef) database. The system offers access to numerous preinstalled, curated analytic tools and also provides researchers with the option of selecting computational tools from a large list of web services including the European Molecular Biology Open Software Suite (EMBOSS), BioMoby, and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The system further allows users to integrate local command line tools residing on their own computers through a client-side Java applet.
INTRODUCTION
Scientific workflows attempt to automate repetitive computation and analysis by chaining together related processes. Automating repetitive time-consuming tasks allows scientists to keep pace with ever-growing volumes of data (1). Furthermore, workflows can aid in the reproducibility of scientific computations by providing a formal declaration of an analysis. Reproducibility is central to the scientific method, and detailed workflow provenance information ensures an analysis can be reproduced and extended (1). It has been suggested that in addition to references, scientific publications should include documentation of computational methods that allows readers to easily reproduce their results, increasing reader engagement with scientific publications (2).
Biology is a prominent domain for applications of workflows and workflow management systems. Researchers need to access exponentially growing amounts of DNA and protein sequence data along with a large number of bioinformatic analytic tools (3). The data is distributed in different formats and in heterogeneous data structures and information systems. Analytic tools are made available to scientists in a variety of ways including as downloadable tools for local installation, web pages and web services. By enabling researchers to work effectively through multiple interconnected tools and handling multiple data formats and large data quantities, workflows with embedded data pipelines and analysis scripts are a powerful alternative to using standalone, off-grid applications (4).
The BioExtract Server (5) is a web-based system that offers researchers a flexible environment for analyzing genomic data. The BioExtract Server provides: (i) a flexible querying and retrieval interface to National Center for Biotechnology Information (NCBI) nucleotide and protein databases, European Molecular Biology Laboratory (EMBL-Bank) non-redundant nucleotide databases, and the European Bioinformatics Institute's (EBI) Universal Protein Resource (UniProt) and UniProt Reference Clusters (UniRef) databases; (ii) the ability to filter search results and use them as input into analytic tools; (iii) the facility to save search results as data extracts that are automatically integrated into the system as searchable data resources; (iv) access to analytic tools including a large list of curated web services, as well as generic access to SOAP-based web services and command line tools residing on the user's local workstation; (v) the ability to save a series of BioExtract Server tasks (e.g. query a data source, save a data extract and execute an analytic tool) as a workflow; and (vi) the opportunity for researchers to share their data extracts, analytic tools and workflows with collaborators.
QUERYING DATA SOURCES AND MANAGING DATA EXTRACTS
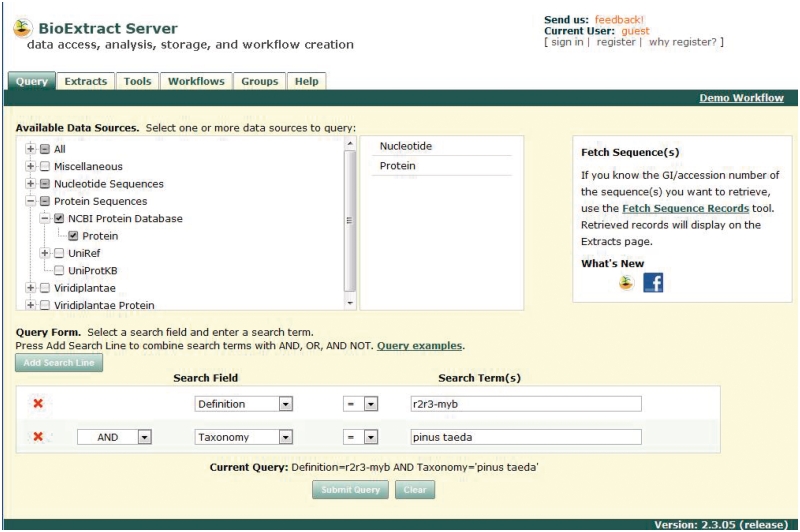
Queries are constructed by first checking the box for one or more data sources and composing a query via a web form. Figure 1 presents an example of querying NCBI Nucleotide and Protein databases for the R2R3-MYB genes in Pinus taeda (Loblolly pine). Queries executed against data sources residing at the BioExtract Server respond quickly. For example, a query of Viridiplantae returns in a matter of seconds. Response times for queries against data sources residing at other sites (e.g. NCBI) are consistent with response times for queries made directly at those sites.
Figure 1.
BioExtract Server queries are executed through the ‘Query’ tab of the system by checking the box for the desired data resource(s), constructing the query using the query form, and submitting the query.
The execution of the query results in the creation of a data extract that is displayed on the ‘Extracts’ tab of the system. Individual records may be viewed locally or through an external link to the original data source. In addition, data extracts may be filtered, exported and used as input into analytic tools. While most of the BioExtract Server's functionality is available to all users, the ability to save a data extract is available only to users who have registered with the system. Users who are signed into the system as registered users (Figure 1, top right) also have the option of saving a data extract simply by clicking the ‘Save Extract’ button on the ‘Extracts’ tab and entering the name and description of the extract. Once saved, the researcher's privately owned data extracts are integrated into the BioExtract Server platform and are listed with other available data resources on the ‘Query’ tab for the registered user only.
Data extracts may also be created within the system by providing a list of sequence record identifiers (ids) such as accession numbers. On the ‘Query’ tab, researchers are able to fetch sequence records by entering or uploading a list of ids and specifying from what data source the records should be retrieved.
All data extracts created within the system are privately owned by the researcher and are only made available to others by explicitly sharing them with a group. This is accomplished by: (i) navigating to the ‘Groups’ tab; (ii) creating a group (under additional actions); (iii) clicking on the new group; (iv) selecting the ‘Extracts’ tab for the new group; and finally (v) clicking the ‘Add Elements’ button to select the extract to share.
EXECUTING, ADDING AND MODIFYING ANALYTIC TOOLS
A number of well-established and unique bioinformatic analytic tools are made available through the BioExtract Server, with the majority integrated as curated web services. Researchers access analytic tools through the list of available tools on the ‘Tools’ tab. The source of the analytic tool's input(s) may be: (i) the current data extract (i.e. the records listed on ‘Extracts’ tab); (ii) the output from a previously executed tool; or (iii) private data provided by the researcher (uploaded or entered in a text box). Analytic tool parameters may be selected or modified before execution and resulting output files may be viewed, downloaded and used as input into subsequently executed analytic tools.
The BioExtract Server offers researchers the ability to add additional analytic tools to their workspace. Users may select such tools from a large list of web services, including EMBOSS SoapLab, BioMoby and KEGG, with the integration of BioMart (www.biomart.org) currently in development. The system also offers generic support for other SOAP-based web services and lets users integrate local command line tools residing on their own workstations through the use of a client-side Java applet. Analytic tools specifically added by researchers may be annotated through the ‘Customize Your Tools’ functionality, which allows users to provide detailed descriptions of the tools as well as a help link to additional information.
BIOEXTRACT SERVER WORKFLOWS
As researchers perform tasks (e.g. execute a query, save a data extract or execute an analytic tool) within the BioExtract Server, their actions are recorded and saved in the background. The series of recorded tasks may be saved as a workflow through the ‘Workflow’ tab after the user provides a name and description of the workflow. Workflows are represented within the BioExtract Server as directed acyclic graphs with the graph nodes representing specific tasks and the arcs representing the data dependencies. Reuse of workflows is realized through the easily modified workflow inputs, queries and analytic tool parameter settings. Copying a workflow for future editing is accomplished by executing the workflow and providing a name and description for the copy.
Once a task within a workflow has been executed, specific task information may be accessed by clicking on the executed node within the workflow graph. Additionally, a provenance report related to an executed workflow provides detailed information such as a general description of the workflow, analytic tool descriptions, parameter settings, input/output data, query information and saved data extract descriptions. Provenance reports may be downloaded as PDF files.
BioExtract Server workflows may be shared with the research community in a variety of ways. First, researchers can export a workflow as an XML file and share it with collaborators. When a workflow XML file is imported into the BioExtract Server by a registered user, a new instance of that workflow is created and privately owned by the user. A second method of sharing a workflow is through myExperiment (myexperiment.org). MyExperiment is a virtual research environment designed to provide a mechanism for collaboration and sharing workflows. This was achieved by adopting a social web approach that is tailored to the particular needs of the scientist (6). BioExtract Server workflows imported into myExperiment may be additionally annotated through their system and executed directly through their platform. Third, along with data extracts and analytic tools, workflows can be shared with collaborative groups through the BioExtract Server ‘Groups’ tab.
BIOEXTRACT SERVER WORKFLOW EXAMPLE
To explain how a simple workflow is created within the BioExtract Server, consider the task of carrying out a multiple sequence alignment analysis using a known protein sequence record as input. To complete this analysis, a researcher would:
Sign into the BioExtract Server by clicking the ‘Sign in’ link in the upper right corner of the screen.
Execute the NCBI blastp tool (7) located under the ‘Similarity Search Tools’ node in the Available Tools tree on the ‘Tools’ tab. The execution of blastp results in the creation of a data extract that can be viewed on the ‘Extracts’ tab.
Execute the Xmknr analytic tool, a simple shell script that employs Vmatch (8) (www.vmatch.de) to remove duplicate sequences, found under the ‘Edit Tools’ node. The input into this tool is specified as ‘Use records on Extract Page formatted as FASTA’. Its execution results in the modification of the data extract
Execute the ClustalW (9) tool to create the multiple sequence alignment with the input specified as ‘Use records on Extract Page formatted as FASTA’.
Navigate to the ‘Workflow’ tab and click the ‘Create and Import Workflows’ node within the ‘Workflows’ tree. Enter a name and description for the workflow. Through the description, researchers are able to annotate the workflow with information that will assist others in understanding decisions made during its creation.
The multiple sequence alignment could serve as input for subsequent steps; for example, phylogenetic or motif analyses.
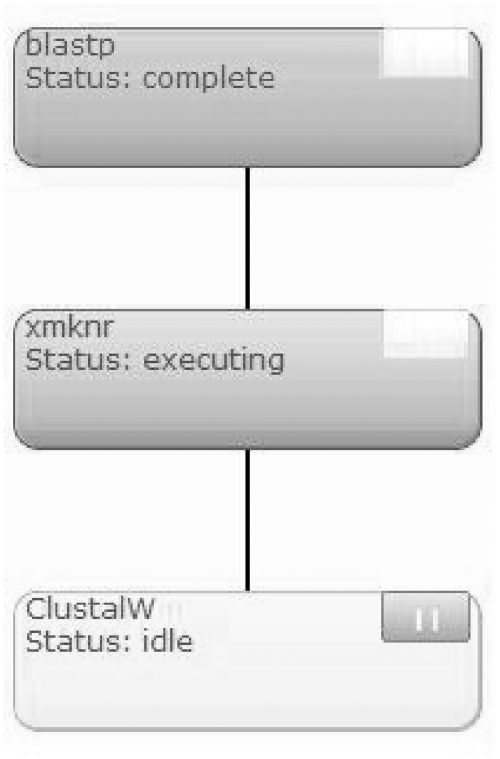
Figure 2 shows the resulting workflow graph. More information related to the creation of workflows (including this video demonstration) can be found through the follow link: http://bioservices.usd.edu/bioextract/videos/blastn-clustalw-example.html.
Figure 2.
Example of a simple BioExtract Server workflow. The top node denotes a tool that has completed execution, the middle node represents an executing tool, and bottom node represents an idle tool waiting to execute.
To further illustrate the advantages of the BioExtract Server platform, we discuss the mRNA Markup workflow. This workflow represents typical steps undertaken in the primary analysis and comprehensive annotation of a set of transcript sequences. Such sets are currently abundantly generated from full-length cDNA sequencing projects or assembly of EST sequences, and with increasing read lengths of novel sequencing technologies, there will be similar assemblies of RNA-Seq data from many species and sampling conditions. Using NCBI BLAST (7), MuSeqBox (10) and Linux shell scripting, the workflow partitions the input transcript set successively according to matching targets. Specifically, mRNA Markup identifies potential vector or bacterial contaminants (sequencing artifacts), potential chimeras (assembly artifacts), likely full-length protein-coding mRNAs, as well as matches from a comprehensive protein database and a protein domain database. What remains after these identifications include presumably novel transcripts for further analysis with other programs. While conceptually simple, implementation and maintenance of the programs in individual labs would be hugely time- and cost-ineffective compared to a publicly available workflow system. For illustration, the workflow is implemented with a sample configuration consisting of a transcript set of Arabidopsis EST assemblies derived from PlantGDB (www.plantgdb.org) and target databases consisting of UniVec (www.ncbi.nlm.nih.gov/VecScreen/UniVec.html) for vector contaminants, an Escherichia coli genome for determining bacterial contamination, the Arabidopsis reference protein set TAIR10 (www.arabidopsis.org), a comprehensive protein subject database containing Viridiplantae entries from UniRef90 (www.uniprot.org/help/uniref) and the Conserved Domain Database (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Users can easily change any of the inputs. The final step of the workflow produces a summary report detailing how many sequences were matched during each step as well as how many potentially novel sequences remain. The matched and unmatched sequences themselves are presented as well. The help URL bioservices.usd.edu/mrnamarkup.html provides more information regarding this specific workflow. The workflow is presented graphically at bioextract.org/scripts/. When the user clicks the mRNA Markup label within the list of workflows, a workflow graph will be displayed. The user can click any node in the workflow graph to acquire more information regarding the analytic step it represents as well as the output it produces.
CONCLUSION
There exist many biological data and analytic tool integration systems that offer powerful and comprehensive workflow creation functionality. Taverna (11), Kepler (12), Triana (13), UGene (14) and Trident (15) are some examples of workflow management systems that are primarily stand-alone applications requiring installation of client software. Through a ‘drag-and-drop’ user interface, these systems allow researchers to develop complex workflows from distributed and local resources. The Taverna Workbench, part of the myGrid project (16), is arguably the most well-known bioinformatic workflow development system. It is an application that gives users the ability to easily integrate the growing number of molecular biology tools and databases available on the web, especially web services.
GenePattern (17), Mobyle (18) and Galaxy (19) are web-based workflow management systems. GenePattern is a genomic analysis platform that provides access to tools for gene expression analysis, proteomics and common data-processing tasks. A web-based interface provides easy access to these tools and allows the creation of multi-stepanalysis pipelines. Mobyle provides a flexible web environment for defining and running bioinformatics analysis by embedding management features allowing users to combine tools using a hierarchical typing system. Galaxy is an open source workflow system with which researchers are able to create complex workflows that can be shared, cloned, annotated and edited. A researcher's activities are recorded and may be saved as a workflow. The primary objective of the Galaxy system is to provide support for accessible, reproducible and transparent science (19).
The BioExtract Server is entirely web-accessible, and while allowing completely flexible customization including integration of locally accessed scripts and tools, the interface and functionality of the site are specifically designed for the novice and implementation detail-uninterested user whose primary need is ease of use and quality results. These design criteria seek to explicitly circumvent the gap between workflow developers and the user community, which from our experience is still substantial.
FUTURE DIRECTION
Future enhancements to the BioExtract Server will focus on improving workflow reproducibility, flexibility and provenance information. Functionality is currently being developed that will provide better feedback regarding a tool's compatibility with the user-selected input file format. In order for workflows to be reproducible, it is imperative that the required analytic tools be reliable. This underscores the need for curation of analytic tools to ensure they are both available and well defined (4). One approach will be to integrate the BioExtract Server analytic tool system with BioCatalogue. BioCatalogue is a comprehensive, curated registry of biological web services that aims to improve process reliability and validation through individual, automatic and community curation (3).
Recognizing that the application of web services within a computational experiment can compromise its reproducibility (19), version information will be incorporated into all of the BioExtract Server's publicly accessible analytic tools. Such information will be included in workflow provenance reports.
Communicating the intent of a workflow to the research community is an important aspect of a computational experiment. Providing researchers with the capacity to annotate and search workflow provenance information, advances the ability to understand the decisions made during the experimental design and execution. The BioExtract Server provenance information will be expanded to allow for additional user annotations and ad hoc querying of provenance data.
FUNDING
National Science Foundation (DBI-0606909 to V.B and C.M.L.) Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Joe Reynoldson and Brent Anderson for their help in the development of the BioExtract Server.
REFERENCES
- 1.Gil Y, Deelman E, Ellisman M, Fahringer T, Fox G, Gannon D, Goble C, Livny M, Moreau L, Myers J. Examining the challenges of scientific workflows. IEEE Comput. 2007;40:24–32. [Google Scholar]
- 2.Mesirov JP. Computer science. Accessible reproducible research. Science. 2010;327:415–416. doi: 10.1126/science.1179653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhagat J, Tanoh F, Nzuobontane E, Laurent T, Orlowski J, Roos M, Wolstencroft K, Aleksejevs S, Stevens R, Pettifer S, et al. BioCatalogue: a universal catalogue of web services for the life sciences. Nucleic Acids Res. 2010;38:W689–W694. doi: 10.1093/nar/gkq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goble CRD. Curating scientific web services and workflow. EDUCAUSE Rev. 2008;43:10–11. [Google Scholar]
- 5.Lushbough C, Bergman MK, Lawrence CJ, Jennewein D, Brendel V. BioExtract server–an integrated workflow-enabling system to access and analyze heterogeneous, distributed biomolecular data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2010;7:12–24. doi: 10.1109/TCBB.2008.98. [DOI] [PubMed] [Google Scholar]
- 6.Goble C, Bhagat J, Aleksejevs S, Cruickshank D, Michaelides D, Newman D, Borkum M, Bechhofer S, Roos M, Li P. myExperiment: a repository and social network for the sharing of bioinformatics workflows. Nucleic Acids Res. 2010;38:W677–W682. doi: 10.1093/nar/gkq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abouelhoda MI, Kurtz S, Ohlebusch E. Replacing suffix trees with enhanced suffix arrays. J. Dis. Algor. 2004;2:53–86. [Google Scholar]
- 9.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing L, Brendel V. Multi-query sequence BLAST output examination with MuSeqBox. Bioinformatics. 2001;17:744–745. doi: 10.1093/bioinformatics/17.8.744. [DOI] [PubMed] [Google Scholar]
- 11.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ludäscher B, Altintas I, Berkley C, Higgins D, Jaeger E, Jones M, Lee EA, Tao J, Zhao Y. Scientific workflow management and the Kepler system. Concurr. Comput. Pract. Exp. 2006;18:1039–1065. [Google Scholar]
- 13.Harrison A, Taylor I, Wang I, Shields M. WS-RF workflow in Triana. Int. J. High Perform. Comput. Appl. 2008;22:268–283. [Google Scholar]
- 14.Fursov MY, Oshchepkov DY, Novikova OS. UGENE: interactive computational schemes for genome analysis. Proc. Fifth Moscow Int. Congress Biotechnol. 2009;3:14–15. [Google Scholar]
- 15.Barga R, Jackson J, Araujo N, Guo D, Gautam N, Simmhan Y. IEEE Fourth International Conference. 2008. The Trident Scientific Workflow Workbench eScience, 2008; pp. 317–318. [Google Scholar]
- 16.Oinn T, Li P, Kell DB, Goble C, Goderis A, Greenwood M, Hull D, Stevens R, Daniele T, Zhao J. Taverna/myGrid: aligning a workflow system with the life sciences community. In: Taylor I, Deelman E, Gannon D, Shields M, editors. Workflows in e-Science. New York: Springer; 2006. pp. 300–319. [Google Scholar]
- 17.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 18.Neron B, Menager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C. Mobyle: a new full web bioinformatics framework. Bioinformatics. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]