Abstract
R.E.D. Server is a unique, open web service, designed to derive non-polarizable RESP and ESP charges and to build force field libraries for new molecules/molecular fragments. It provides to computational biologists the means to derive rigorously molecular electrostatic potential-based charges embedded in force field libraries that are ready to be used in force field development, charge validation and molecular dynamics simulations. R.E.D. Server interfaces quantum mechanics programs, the RESP program and the latest version of the R.E.D. tools. A two step approach has been developed. The first one consists of preparing P2N file(s) to rigorously define key elements such as atom names, topology and chemical equivalencing needed when building a force field library. Then, P2N files are used to derive RESP or ESP charges embedded in force field libraries in the Tripos mol2 format. In complex cases an entire set of force field libraries or force field topology database is generated. Other features developed in R.E.D. Server include help services, a demonstration, tutorials, frequently asked questions, Jmol-based tools useful to construct PDB input files and parse R.E.D. Server outputs as well as a graphical queuing system allowing any user to check the status of R.E.D. Server jobs.
INTRODUCTION
Force field-based molecular dynamics (MD) simulations are today the theoretical methods of choice to study large biomolecular systems (glycoconjugates, proteins, nucleic acids and complexes). The functional form of a force field contains both bonded terms related to atoms that are connected by covalent bonds, and non-bonded terms describing the long-range electrostatic and van der Waals interactions (1,2). Most additive force fields use a ‘non-polarizable fixed-charge’ model by which a single value for the atomic charge is assigned to each atom independently of the electrostatic environment. Force field parameter sets are derived and validated by using both experimental data and high-level quantum mechanical (QM) calculations. The parameters required to represent the potential functions are available in empirical force fields [AMBER (3,4), CHARMM (5–7), GLYCAM (8) and OPLS (9) among many others] except for the atomic charges required for calculating the electrostatic energy. Each force field provides its own charge set for the standard residues in a specific format. However, for any non-standard residue or molecule charge values need to be derived and validated, and a force field library has to be built anew, which are limiting steps for computational biologists involved in structural studies.
Numerous methods have been developed for deriving effective non-polarizable atomic charge values for the additive force field model (10–20). Difficulties in such an approach arise from the facts that (i) atomic charges are fictitious and are not observables and that (ii) no real criterion has been established to rigorously define the ‘quality’ of atomic charges. It has been proposed that charges should be independent of the computational method, basis set, molecular orientation or conformation, or should be able to reproduce experimental multipole moments, or should be transferable and conform to atom electronegativities. Consequently, many approaches have been developed for atomic charge derivation. Unfortunately, no charge model has proved to be the best in all respects (21,22). Among the different approaches developed, QM computed Molecular Electrostatic Potential (MEP)-based charges, or ElectroStatic Potential (ESP) and Restrained ESP (RESP) derived charges are widely used (18,20,21,23–25). These types of charges are recognized as optimally handling inter-molecular properties, which are essential in condensed phase MD simulations. Historically, RESP charges are strongly connected to the AMBER Cornell et al. force field and its successive adaptations (3,26). The GLYCAM force field uses also a RESP charge model defined specifically for carbohydrates (8,27). The CHARMM force fields are not related to ESP or RESP charges (5–7), but CHARMM developers have indiscriminately used MEP-based charges in MD simulations (28,29). A RESP charge OPLS-like model has also been reported (30), making RESP and ESP charge derivation an ensemble of approaches highly popular and extensively used in the entire computational chemistry community.
Here, we report on R.E.D. Server, a unique web service designed to derive non-polarizable RESP and ESP charges and to build force field libraries for new molecules/molecular fragments in the context of an additive force field model. It provides the resources i.e. the software and a cluster of computers required for charge derivation and force field library building to any computational biologist, who wishes to use MEP-based atomic charges in MD simulations. R.E.D. Server is freely available at http://q4md-forcefieldtools.org/REDS/, and is open to all users without any mandatory registration or login requirement.
METHODOLOGY
ESP and RESP atomic charges are fitted to reproduce MEP computed via QM. The strategy describing the procedure has been already reported (18,20,21,23–25). We also summarized many aspects related to RESP and ESP charge derivation in the previously published articles about the R.E.DD.B. database and the R.E.D. tools (31,32). Here, we will focus on the latest features developed in the Ante_R.E.D. 2.0 and R.E.D. IV programs interfaced by R.E.D. Server. Although a modular approach has been followed in the Ante_R.E.D. 2.0 and R.E.D. IV programs allowing the development of large numbers of user-defined options, the actual version of R.E.D. Server only enables pre-defined settings used in published charge derivation approaches (8,26,33).
New features developed in Ante_R.E.D. 2.0
Ante_R.E.D. 2.0 is an entirely new code developed independently of the previously published Ante_R.E.D. 1.x suite of programs. It converts input structures in the PDB file format (http://www.wwpdb.org/documentation/format32/v3.2.html) into the P2N file format, which practically corresponds to the PDB file format with two columns of atom names (http://q4md-forcefieldtools.org/Tutorial/Tutorial-1.php, see also the Supplementary Data for examples of P2N files taken from the case study reported below). Features required for the rigorous definition of a force field library for a new molecule or molecular fragment are calculated, checked and corrected when errors are detected. Particular attention has been paid to the development of new algorithms enabling the automatic correction of improper PDB input files. Considering that two atoms belonging to a given residue cannot share the same name in a force field library, atom names are checked and corrected when duplicates are found. Following a similar principle and considering that a force field library usually contains a single residue, residue names are verified and renamed when needed. The molecular topology is calculated based on inter-atomic distances. When an open valency is detected due to an inaccurate input geometry, the correct molecular topology can be reconstructed using an iterative approach. Following the principle that two chemically equivalent atoms should bear the same charge values in MD simulations a tree of atom connectivities is constructed to define chemical equivalences required for RESP and ESP charge derivation. Finally, the atom order of the input molecule can also be modified by the user allowing the intuitive definition of atom orders with the identification of methylene and methyl groups. This facilitates atom equivalencing necessary in RESP charge fitting (23,27).
New features developed in R.E.D. IV
The R.E.D. program is a tool for developing highly reproducible RESP and ESP charges and building the force field libraries for new molecules and molecular fragments in the Tripos mol2 file format (http://tripos.com/data/support/mol2.pdf). Although atomic charges and force field libraries can be obtained separately, these two concepts are strongly connected in R.E.D. Charges are derived for whole molecules and/or molecular fragments using intra-, inter-molecular charge constraint(s) as well as inter-molecular charge equivalencing. A force field library is then built for the whole molecule and molecular fragment designed during the fitting step. Chemical elements up to Bromine are automatically handled. A modular approach has been followed allowing the straightforward development of large numbers of charge fitting protocols. R.E.D. IV is a direct expansion of the R.E.D. III.x suite of programs. R.E.D. IV extends many features of R.E.D. III.x, and handles complex charge derivation. It enables to use a large set of orientations, conformations and molecules in RESP and ESP charge derivation, and it makes it possible to build force field library practically for any type of molecular fragments. The latest developments include (i) the reinforcement of charge reproducibility by differentiating translation and rotation in MEP computation, (ii) the building of a complex set of force field library or Force Field Topology DataBases (FFTopDB) for any type of bio-organic and bio-inorganic molecules, (iii) the generation of all-atom force field libraries or united-carbon force field libraries and (iv) the development of a statistic module allowing charge value comparisons (this represents a first attempt toward validating charge values).
DEVELOPMENT
R.E.D. Server is developed using the C-Shell, PERL, XHTML, PHP, MySQL, Javascript and Java languages. The corresponding database is implemented using MySQL (version 14.12, distribution 5.0.32) and consists of six interconnected tables, while web-based forms and queries to the database are written in PHP (version 4.4.4). The web site is hosted on a Linux server located at the Université de Picardie - Jules Verne in Amiens, and is available at the q4md-forcefieldtools domain. Moreover, since the web site uses the Jmol program (http://jmol.sourceforge.net/) to visualize atomic charge values and molecular structures, the J2SE Runtime Environment (http://www.java.com/en/download/manual.jsp) has to be installed on the user front end machine. R.E.D. Server is not browser specific, and any popular web browser under any operating system can be used to interact with R.E.D. Server. The first version of R.E.D. Server was released in April 2009, and was visited by more than 250 users. Using the experience gained with this first version and the numerous tests carried out, R.E.D. Server 2.0 has been released in November 2010.
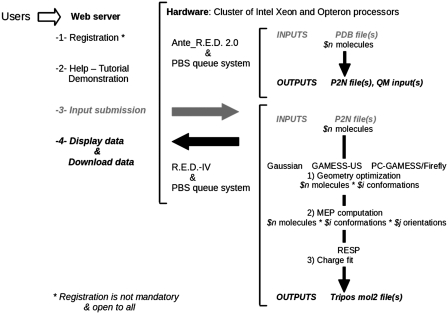
R.E.D. Server provides the software and hardware required for RESP and ESP atomic charge derivation and force field library building for new molecules and/or new molecular fragments. It interfaces the latest version of the Gaussian (34), GAMESS-US (35) and Firefly (36) QM programs, a standalone version of the RESP program (23, http://q4md-forcefieldtools.org/RED/resp/), and the latest version of the Ante_R.E.D. 2.0 and R.E.D. IV programs developed at q4md-forcefieldtools.org. A cluster of quadri-core Intel Xeon® (processor E5430) and AMD dual-Core Opteron™ (processors 8212 and 8220) with 16 GB RAM per node is the machine on which R.E.D. Server is installed and where calculations are performed. Ante_R.E.D. and R.E.D. jobs are executed using one and eight processor cores, respectively. The description of R.E.D. Server is reported in Figure 1. Numerous services related to charge derivation and force field library building are also provided. They are reported below.
Figure 1.
Description of R.E.D. Server and services provided by R.E.D. Server.
INTERACTING WITH THE SERVER
Conditions of usage
R.E.D. Server is open to all users and can be anonymously used through a public account. Registration to use R.E.D. Server is not mandatory. However, R.E.D. Server provides the opportunity for a user to create a private space of work or account. In agreement with the developers of the Gaussian (34), GAMESS-US (35) and Firefly (36) programs, the direct use of R.E.D. Server without any registration step is limited to GAMESS-US and Firefly, while the execution of the Gaussian program is restricted to academic users after registration. The private account obtained after registration is temporary, and one retains it only when one keeps using it. Thus, after three months of inactivity a private account is automatically deleted, and a user has to re-register to obtain a new login.
Input submission
A R.E.D. Server user uploads inputs to R.E.D. Server from the ‘Submit’ service available at the R.E.D. Server home page. When interfacing the Ante_R.E.D. 2.0 program, a user has to upload the PDB input file(s) to be converted into the P2N file format. R.E.D. Server can execute Ante_R.E.D. 2.0 in two different modes. In the automatic mode, which should be fully effective when a proper input geometry and a correct PDB input file format are provided, the default setups are used. In the non-automatic mode a user can modify the tasks carried out by the program, and adjust the molecular topology and chemical equivalencing, atom naming, residue definition and atom ordering. When the R.E.D. IV program is used, two different inputs can be uploaded and submitted to R.E.D. Server depending on the mode in which R.E.D. IV is executed. In Mode 1, the program automatically performs the geometry optimization step and the MEP computation—charge fitting step. In this case, a user only needs to provide a P2N file for each molecule. In Mode 2, which is the default, only the MEP computation—charge fitting step is performed, and P2N file(s) with the corresponding geometry optimization output(s) computed using a QM program have to be uploaded. To complete the system, a Jmol-based builder through various Java applets has been developed allowing any user to create PDB input files, to correct improper input geometries and to add hydrogen atoms to a structure when required.
Status of a R.E.D. Server job and queuing system
When using R.E.D. Server a project is identified by a name chosen by the user and by a code, which is automatically incremented. The TORQUE resource manager (version 2.5.4, http://www.clusterresources.com/products/torque-resource-manager.php) and Maui scheduler (version 3.3, http://www.clusterresources.com/products/maui/docs/pbsintegration.shtml) are used to handle jobs on the R.E.D. Server cluster through a queuing system. To each job submitted to R.E.D. Server a queue job number is assigned ensuring an efficient tracking system for R.E.D. Server jobs. A maximum of three simultaneous jobs per account (private and public logins) is allowed on R.E.D. Server. Other jobs are automatically sent to the R.E.D. Server queuing system. These simple rules ensure that every R.E.D. Server user gets an equal access to the computational resources provided.
When a job is directly submitted to R.E.D. Server, without any private account, a user has the opportunity to provide an email address at the beginning of the input submission procedure. In this case, R.E.D. Server is able to send information about the status (initiation and termination) of a job to its owner. If a wrong email address is provided or if a user refuses to provide an email address, the job can still be submitted to R.E.D. Server. In this case, as the submission procedure cannot guess when a R.E.D. Server job starts or ends and cannot contact the user, the status of the job cannot be provided the user. Consequently, a graphical interface of the Qstat command is provided allowing any user to find out when a R.E.D. Server job terminates. When the email is provided during the job submission or during the registration for a private account, R.E.D. Server sends three emails to the user. The first one is sent when the inputs have been successfully uploaded to R.E.D. Server, while the second and third ones are sent when the job effectively starts and ends, respectively.
Parsing and downloading R.E.D. Server outputs
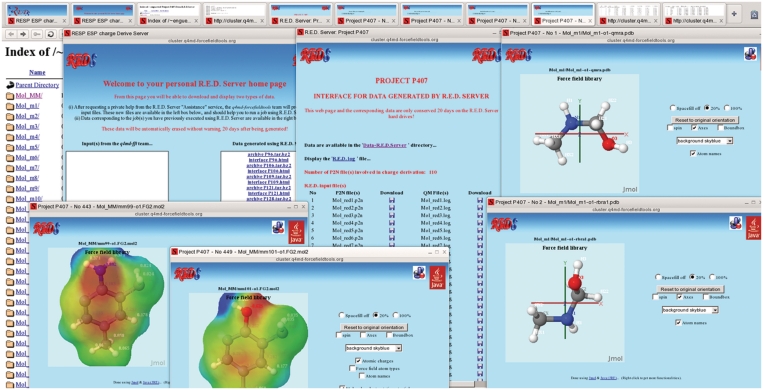
For each project a Jmol-based graphical interface is generated in the XHTML format allowing any user to parse and display R.E.D. Server outputs. This graphical interface provides a modular environment with Java applets useful for displaying PDB, P2N and Tripos mol2 files, and uses the browser-integrated file manager for searching the project file system and the operating system file editors for viewing plain text files. A screen snapshot of such a graphical interface corresponding to the case study related to calix[n]arenes described below is presented in Figure 2.
Figure 2.
A screen snapshot of the graphical interface generated for the case study related to calix[n]arenes presented in this work.
For each project computed by R.E.D. Server a specific internet address for downloading results is also generated. The corresponding web link is provided to the user at the end of the input submission procedure. This web address is also sent by email to the user by R.E.D. Server if the user email is known. Considering that the public account can be extensively used and that tremendous amounts of data can be computed, R.E.D. Server projects generated using this account are kept only for five days on the R.E.D. Server hard drives. When a researcher uses a private account, the projects generated can be downloaded via the ‘Download’ service available at the R.E.D. Server home page, and the corresponding data are kept for twenty days.
Documentation, help and demonstration
Documentation includes detailed frequently asked questions and extensive tutorials covering RESP and ESP charge derivation and force field library building for potentially any type of biomolecules and biomolecular fragments (http://q4md-forcefieldtools.org/Tutorial/Tutorial-3.php). A demonstration service has been also developed allowing any user to quickly execute examples using the Ante_R.E.D. 2.0 and R.E.D. IV programs. Help is provided to registered users via a ‘private assistance’ service, while general questions can be posted to the q4md-fft mailing list. All these services are directly available at the R.E.D. Server home page.
Miscellaneous and q4md-forcefieldtools.org
Miscellaneous features related to R.E.D. Server include the distribution of the R.E.D. Server source code under the GNU General Public License 3.0, as well as the full integration of R.E.D. Server within the q4md-forcefieldtools.org Internet site. This allows the direct submission to the R.E.DD.B. database for the projects generated using R.E.D. Server, developing common services related to charge derivation and force field library building such as tutorials and the q4md-fft mailing list. In addition, a standalone distribution of the R.E.D. III.x tools is also available (32).
CASE STUDY
Calixarenes are macrocycles, which feature a hydrophobic cavity. They are widely used for their ability to complex small molecules with potential biological applications. An extensive variety of structurally related molecules derived from calixarenes has been synthesized. This includes calix[n]arenes with functionalized ligands (37). Since topological fragments required for building calix[n]arenes with functionalized ligands are not available in the Cornell et al. force field (26) [and in its successive modifications as the additive Amber99SB force field (3)] we developed a new FFTopDB compatible with a selection of organic functions. The calix[n]arenes studied in this work include oligomers of 2,4-dimethylaniline, 2,4-dimethylphenol, 2,4-dimethylthiophenol (with the corresponding anions), substituted or not at the C4 position by different alkyl groups (methyl, propyl, isopropyl, butyl, tertiobutyl, 1,1,2,2-tetramethylpropyl and allyl) and via the heteroatom at the C1 position by various functionalized aliphatic ligands (with alcohol, ketone, ether, amine, amide, thiol, thioether, ester and carboxylic acid organic functions).
Charge derivation involving multiple orientations, multiple conformations and multiple molecules (i.e. 310 structures) and force field library building for calix[n]arene-based systems have been automatically carried out using the R.E.D. IV program (version June 2010) available in R.E.D. Server. Theory levels involved in QM calculations were selected to be in agreement with these used in RESP charge derivation for the Cornell et al. force field. The different building blocks used here were optimized using the HF/6-31G** theory level (38) and the version 7.1.G of the Firefly program (36). The lowest energy minimum for each aromatic building block was included in charge derivation. For the different alkyl molecules and functionalized ligands, one or two energy minima were selected after conformational search. A lowest minimum was considered only if no canonical intra-molecular hydrogen bond [donor (D)–acceptor (A) distance lower than 3.20 Å and the D-H…A angle between 120–180° (39)] was observed in a structure. MEP computation involved the Connolly surface algorithm (40) and the HF/6-31G* theory level (38) implemented in the Firefly program. For each aromatic moiety four molecular orientations and for each alkyl molecule and ligand, two molecular orientations based on the Rigid-Body Reorientation Algorithm (RBRA) implemented in R.E.D. were involved in MEP computation assuring the reproducibility of charge values (information about the atoms involved in the RBRA procedure is available in the PDB files of the ‘F-87’ R.E.DD.B. project; see the ‘REMARK REORIENT’ keyword). The molecular fragments required for MD simulations were constructed by setting intra-molecular charge constraints within the aromatic moieties and inter-molecular charge constraints between the aromatic moieties and the alkyl groups or ligands during the charge fitting step. RESP charge fitting was carried out using the standalone version of the RESP program available at q4md-forcefieldtools.org and following a two-stage fitting procedure with a hyperbolic restraint function, using weighting factor of 0.0005 and 0.001 for the two stages (23), respectively. A Relative Root Mean Square (RRMS) value of 0.067 between the MEP calculated by QM and that generated using the derived charge values was obtained for the last charge fitting step. A RRMS value of 0.068 was also obtained in the absence of intra- and inter-molecular charge constraints. The relative small RRMS values as well as the small difference of RRMS between the charge fitting steps carried out without and with intra- and inter-molecular charge constraints demonstrate the accuracy of the fitting step, and the weak effect of the constraints used. By directly defining new intra-molecular charge constraints set to the averaged 0.1494, 0.1768 and 0.1163 values obtained in this project for the methyl groups of the NH-, O- and S-families of ligands, respectively, a potentially infinite number of new fragments (R-NH, R-O and R-S) can be added to the present list, and could constitute addons to this present project.
The corresponding data have been submitted to the R.E.DD.B. database (31), and are available for download under the ‘F-87’ R.E.DD.B. code. The computational data, Cartesian coordinates of the optimized geometries, the force field libraries and the corresponding LEaP script allowing the construction of input files for MD simulations for the Amber suite of programs (41) are accessible at the corresponding web link.
Charge derivation and FFTopDB building for the calix[n]arenes studied in this work have been performed using two 2.66 GHz quadri-core Intel Xeon® processors. The geometry optimization step (77 and 33 molecules represented by a single and two conformations, respectively), the MEP computation step (sets of two or four molecular orientations per conformation representing a total of 310 MEPs) as well as the charge fitting, FFTopDB building (113 Tripos mol2 files generated) and statistical analyses were performed in two steps. Geometry optimization was first carried out in a standalone approach, and the corresponding QM outputs and P2N files were involved in a single R.E.D. Server/R.E.D. IV run. The geometry optimization step used a tight gradient convergence tolerance and required a total of one and an half days (cumulative wall clock time), while MEP computation and charge fitting, FFTopDB building and statistical analyses took around one hour and one minute, respectively. These calculation times does not take into account the conformational search and frequency computation performed before and after the geometry optimization step.
CONCLUSION AND PERSPECTIVES
We have presented R.E.D. Server, a web service devoted to RESP and ESP charge derivation and force field library building for new biomolecules and new biomolecular fragments. R.E.D. Server provides to computational biologists the means to derive rigorously QM MEP-based charges embedded in force field libraries that are ready to be used in force field development, charge validation and/or MD simulations. New functionalities are under active development. They include RESP and ESP charge derivation for all the elements of the periodic table up to Lawrencium with a version 2.2 of the RESP program, the implementation of user-defined options (related to QM theory levels and basis sets, atomic radii required in MEP computation and hyperbolic restraints used during charge fitting), the handling of lone-pairs and extra-points for the oxygen, sulfur and nitrogen atoms (42), the extension of the statistics module for charge validation, and the interface of the i-resp program to derive charges for non-additive polarizable force fields (43).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The computer cluster on which R.E.D. Server has been developed was funded by the ‘Contrat Plan Etat Région 2007–2011’ and the Conseil Régional de Picardie. Funding for the open access charge: the ‘Centre National pour la Recherche Scientifique’ and the “Bonus Qualité Recherche” of the Université de Picardie – Jules Verne.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
F.-Y. Dupradeau is grateful to Alineos SA (Linux solutions provider for high-performance numerical computation, Bourron-Marlotte, France) for supporting R.E.D. Server. P. C. acknowledges support from National Institute of Health, grant GM079383.
REFERENCES
- 1.Cheatham TE, III, Young MA. Molecular dynamics simulation of nucleic acids: successes, limitations and promise. Biopolymers. 2001;56:232–256. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 2.Ponder JW, Case DA. Force fields for protein simulations. Adv. Prot. Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 3.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct. Funct. Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general AMBER force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 5.MacKerell AD, Jr, Banavali N, Foloppe N. Development and current status of the CHARMM force field for nucleic acids. Biopolymers. 2001;56:257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 6.Mackerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 7.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Lachele Foley B, Woods RJ. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 10.Löwdin PO. On the non-orthogonality problem connected with the use of atomic wave functions in the theory of molecules and crystals. J. Chem. Phys. 1950;18:365–375. [Google Scholar]
- 11.Mulliken RS. Electronic population analysis on LCAO-MO molecular wave functions. I. J. Chem. Phys. 1955;23:1833–1840. [Google Scholar]
- 12.Hagler AT, Huler E, Lifson S. Energy functions for peptides and proteins. I. Derivation of a consistent force field including the hydrogen bond from amide crystals. J. Am. Chem. Soc. 1974;96:5319–5327. doi: 10.1021/ja00824a004. [DOI] [PubMed] [Google Scholar]
- 13.Bader RWF, Nguyen-Dang TT. Quantum theory of atoms in molecules-Dalton revisited. Adv. Quantum Chem. 1981;14:63–124. [Google Scholar]
- 14.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 15.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 16.Momany FA. Determination of partial atomic charges from ab initio molecular electrostatic potentials. Application to formamide, methanol, and formic acid. J. Phys. Chem. 1978;82:592–601. [Google Scholar]
- 17.Cox SR, Williams DE. Representation of the molecular electrostatic potential by a net atomic charge model. J. Comput. Chem. 1981;2:304–323. [Google Scholar]
- 18.Singh UC, Kollman PA. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 1984;5:129–145. [Google Scholar]
- 19.Chirlian LE, Francl MM. Atomic charges derived from electrostatic potentials: a detailed study. J. Comput. Chem. 1987;8:894–905. [Google Scholar]
- 20.Breneman CM, Wiberg KB. Determining atom-centered monopoles from molecular electrostatic potentials. The need for high sampling density in formamide conformational analysis. J. Comput. Chem. 1990;11:361–373. [Google Scholar]
- 21.Williams DE. Net atomic charge and multipole models for the Ab initio molecular electric potential. In: Lipkowitz KB, Boyd DB, editors. Reviews in Computational Chemistry. Vol. 2. New York: VCH Publishers; 1991. pp. 219–271. [Google Scholar]
- 22.Bachrach SM. Population analysis and electron densities from quantum mechanics. In: Lipkowitz KB, Boyd DB, editors. Reviews in Computational Chemistry. Vol. 5. New York: VCH Publishers; 1994. pp. 171–227. [Google Scholar]
- 23.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for determining atom-centered charges: the RESP Model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 24.Cornell WD, Cieplak P, Bayly CI, Kollman PA. Application of RESP charges to calculate conformational energies, hydrogen bond energies and free energies of solvation. J. Am. Chem. Soc. 1993;115:9620–9631. [Google Scholar]
- 25.Cieplak P, Cornell WD, Bayly CI, Kollman PA. Application of the multimolecule and multiconformational RESP methodology to biopolymers: charge derivation for DNA, RNA and proteins. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- 26.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 27.Woods RJ, Chappelle R. Restrained electrostatic potential atomic partial charges for condensed-phase simulations of carbohydrates. J. Mol. Struct.: THEOCHEM. 2000;527:149–156. doi: 10.1016/S0166-1280(00)00487-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayaan E, Moser A, MacKerell AD, Jr, York DM. CHARMM force field parameters for simulation of reactive intermediates in native and thio-substituted ribozymes. J. Comput. Chem. 2007;28:495–507. doi: 10.1002/jcc.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight JL, Brooks CL., III Validating CHARMM parameters and exploring charge distribution models in structure-based drug design. J. Chem. Theory Comput. 2009;5:1680–1691. doi: 10.1021/ct900079t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henchman RH, Essex JW. Generation of OPLS-like charges from molecular electrostatic potential using restraints. J. Comput. Chem. 1999;20:483–498. [Google Scholar]
- 31.Dupradeau F-Y, Cezard C, Lelong R, Stanislawiak E, Pecher J, Delepine JC, Cieplak P. R.E.DD.B.: a database for RESP and ESP atomic charges, and force field libraries. Nucleic Acids Res. 2008;36:D360–D367. doi: 10.1093/nar/gkm887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupradeau F-Y, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010;12:7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Jr, Weiner P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- 34. Gaussian 09, Revision B.1, Frisch,M.J., Trucks,G.W., Schlegel,H.B., Scuseria,G.E., Robb,M.A., Cheeseman,J.R., Scalmani,G., Barone,V. et al. (2009) Gaussian, Inc., Wallingford CT.
- 35.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- 36.Nemukhin AV, Grigorenko BL, Granovsky AA. Molecular modeling with the PC GAMESS. Moscow Univ. Chem. Bull. 2004;45:75–102. [Google Scholar]
- 37.Gutsche CD. Calixarenes. Cambridge: Royal Society of Chemistry; 1989. [Google Scholar]
- 38.Jensen F. Introduction to Computational Chemistry. Chichester: John Wiley & Sons; 1999. [Google Scholar]
- 39.Jeffrey GA. An Introduction to Hydrogen Bonding. New York, Oxford: Oxford University Press; 1997. [Google Scholar]
- 40.Connolly ML. Analytical molecular surface calculation. J. Appl. Cryst. 1983;16:548–558. [Google Scholar]
- 41.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods R. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixon RW, Kollman PA. Advancing beyond the atom-centered model in additive and nonadditive molecular mechanics. J. Comput. Chem. 1997;18:1632–1646. [Google Scholar]
- 43.Cieplak P, Dupradeau F-Y, Duan Y, Wang J. Polarization effects in molecular mechanical force fields. J. Phys.: Condens. Matter. 2009;21:333102. doi: 10.1088/0953-8984/21/33/333102. [DOI] [PMC free article] [PubMed] [Google Scholar]