Abstract
microRNAs (miRNAs) are small endogenous RNA molecules that are implicated in many biological processes through post-transcriptional regulation of gene expression. The DIANA-microT Web server provides a user-friendly interface for comprehensive computational analysis of miRNA targets in human and mouse. The server has now been extended to support predictions for two widely studied species: Drosophila melanogaster and Caenorhabditis elegans. In the updated version, the Web server enables the association of miRNAs to diseases through bibliographic analysis and provides insights for the potential involvement of miRNAs in biological processes. The nomenclature used to describe mature miRNAs along different miRBase versions has been extensively analyzed, and the naming history of each miRNA has been extracted. This enables the identification of miRNA publications regardless of possible nomenclature changes. User interaction has been further refined allowing users to save results that they wish to analyze further. A connection to the UCSC genome browser is now provided, enabling users to easily preview predicted binding sites in comparison to a wide array of genomic tracks, such as single nucleotide polymorphisms. The Web server is publicly accessible in www.microrna.gr/microT-v4.
INTRODUCTION
microRNAs are small endogenous RNA molecules that affect many biological processes by regulating gene expression in a post-transcriptional way. They are ∼21–22 nt in length whose primary role is to regulate gene expression through translational repression and/or mRNA degradation (1). The first miRNA molecules and miRNA targets were identified in 1993 via classical genetic techniques in Caenorhabditis elegans (2). Since then, there has been a dramatic increase in the number of miRNAs registered in miRBase (3). In parallel, the development of the first computational target prediction programs (4–7) led to the experimental identification of dozens of miRNA targets (8), and emphasized the need to provide miRNA target predictions in an efficient way to assist biologists in experimental design.
The previous version of the DIANA-microT Web server (9) presented extensive information for predicted miRNA target gene interactions in a user-friendly interface. It offers links to nomenclature, sequence and protein databases, information for experimentally verified targets through TarBase (8) and targets predicted by PicTar (6) or TargetScan (10). Also, users are facilitated by being able to search for targeted genes using different names or functional features.
Here, we describe an extensive update of this Web server with several important improvements: (i) an advanced bibliographic analysis which correlates miRNAs to diseases, (ii) support for two additional species, (iii) a graphical display with all relevant functional information from the UCSC genome browser, (iv) tracking of changes in miRNA nomenclature and (v) user personalized sessions allowing personal query history and bookmarks.
METHODS AND RESULTS
Relation of miRNAs to functional features, diseases and medical descriptors
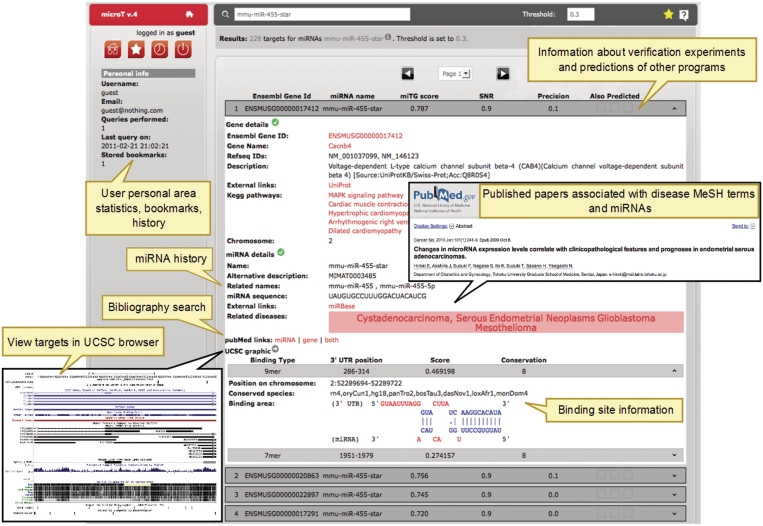
DIANA microT Web server provides functional analysis of miRNAs that reaches beyond a simple listing of miRNA targets through integration of knowledge extracted from bibliography and known biological pathways. In the previous version of the Web server, bibliographic integration considered automated searches in PubMed providing publications related to a miRNA, each target gene or combination of the two. Now, an additional feature noted as ‘Related diseases’ has been added that directly associates a miRNA to publications connected to one or several diseases. This feature is based on information included in the title or the abstract of publications found in PubMed. All abstracts associated with a miRNA are retrieved from PubMed, based on the presence of the name of the miRNA or a member of its family, as defined by miRBase, in the title or abstract of the publication. The retrieved publications are associated with Medical Subject Headings (MeSH), the National Library of Medicine's controlled vocabulary thesaurus, through their metadata. All disease associated MeSH terms for a miRNA are counted and visualized through a tag cloud (Figure 1), where MeSH terms appear in a size proportional to the number of publications reporting this miRNA-disease association. The MeSH terms of the tag cloud also serve as hyperlinks to the relevant publications. For example, in Figure 2 miR-455-star (miR-455*) has been associated with a publication indicating that lower expression of this miRNA correlates with poor overall survival in endometrial serous adenocarcinoma (11).
Figure 1.
An example of a tag cloud for hsa-miR-1 showing relevant disease associated MeSH terms.
Figure 2.
Example of a DIANA-microT Web server results page. Balloons indicate and explain important features of the results page. ‘Related diseases’ tag cloud contains links to PubMed and specifies all papers which associate the particular disease with the corresponding miRNA. The field ‘PubMed links’ provides automated bibliography searches based only on the name of miRNAs, protein coding genes or the combination of both. The ‘UCSC graphic’ link presents the predicted binding sites in a UCSC genome browser window along with tracks such as SNPs and repeat elements. The left side of the page is devoted to the administration of the user personal space and reports their latest searches and bookmarks.
The feature that allowed the user to filter the targets of a miRNA based on KEGG pathways (12) is now integrated in the initial input query form. The user has now the option to enter a miRNA together with names of pathways of interest and provided immediately with only the miRNA target genes found in the pathways of interest. For example, a query for hsa-miR-221 would return 113 predicted targets at a score threshold of 0.5 while the combined search of the miRNA with the term ‘MAPK-signaling-pathway’ would filter the results to 21 relevant targets.
In addition, the positions of the binding sites on the mRNA of the target gene are graphically presented through the UCSC genome browser. This automatic upload can be used to provide information in comparison to other tracks of interest such as single nucleotide polymorphisms (SNPs), repeat elements and alternative 3′-UTR splice forms.
Extraction of miRNA history from miRBase
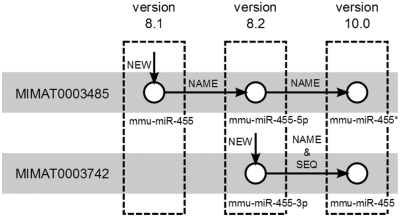
The relation of miRNAs to function and diseases described above is based on miRNA nomenclature. Since miRNA biology is still a field in flux, it can occur that a miRNA may change name between two successive versions of miRBase. Due to such changes, researchers may lose track of a miRNA full history and related literature searches will remain incomplete. To address this issue, an extended analysis on 13 versions of miRBase (versions 7.1. to 14) is performed, and the nomenclature history of each miRNA is extracted. The analysis uses version 13 of miRBase as the reference database. This version includes 1884 mature miRNAs for the four species supported in the Web server. Each miRNA is assigned a unique identification number denoted as ‘MIMAT id’ and one associated miRNA specific name. Among versions, changes are found in 77 MIMAT ids (38 in human, 37 in mouse and 2 in Drosophila) and 151 miRNA names (76 in human, 71 in mouse and 4 in Drosophila). This indicates that name changes are more frequent than changes in MIMAT ids. To keep track of these changes, a history index is integrated in the Web server. This index information is made available to the user through a specific feature called ‘miRNA history’ which is also used for miRNA related bibliography searches. For example, mmu-miR-455 first appeared in miRBase v8.1. Its name was later changed to mmu-miR-455-5 p in version 8.2 and later appeared as mmu-miR-455* in version 10.0 (Figure 3). This information can be retrieved from the miRNA history, but is also used to extend the association of mmu-mir-455* with endometrial serous adenocarcinoma in the publication where it is referred to as miR-455-5 p (Figure 2).
Figure 3.
Graphic presentation of the changes involved in the history of miRNA mmu-miR-455. Initially, MIMAT0003485 was presented in version 8.1 as mmu-miR-455 but its name changed consecutively to mmu-miR-455-5 p in version 8.2 and mmu-miR-455-star (mmu-miR-455*) in version 10.0. Similarly, MIMAT0003742 was first presented in version 8.2 as mmu-miR-455-3 p, while in version 10.0, its sequence changed and it was renamed to mmu-miR-455.
Target prediction and supported miRNAs
The first version of DIANA-microT Web server was designed to support the functional analysis of human and mouse miRNAs. Now the server has been updated with predictions for two additional species and newer versions of miRBase (miRBase 13) and Ensembl (Ensembl 54) (13). In total predictions for 723 new miRNAs have been added, out of which 147 correspond to Drosophila melanogaster, 154 to C. elegans and the rest being new Homo sapiens and Mus musculus miRNAs. This results in an approximately doubled number of predicted targets in comparison to the previous version, totaling to more than six million predicted target genes. The Web server can support different prediction algorithms and currently provides the targets of an updated version of the previously used algorithm (14). While microT-v3.0 is based on features separating real and mock (shuffled) miRNAs the current version, microT-v4.0, uses high throughput experimental data for the same purpose (15).
Personalized user sessions
To allow users to take full advantage of the Web server's functions, several functional improvements have been implemented (Figure 2). The most important is an integrated personal user space in which users can easily save important searches and results that they wish to keep for future analysis. In particular, the system keeps the most recent user searches, giving the opportunity to repeat searches. A bookmarking mechanism provides the opportunity to save interesting results along with user comments. The personal space provides usage statistics regarding the most recent searches, thus, enabling them to keep track of their latest findings. It is noted that researchers may use any feature of the Web server irrespectively of the personal user space feature. Finally, special attention has been given to the Web Server documentation introducing hovering help notes for important fields.
CONCLUSION
In recent years it has become apparent that Web applications are an essential tool for researchers to decipher complex biological processes. As one of these, the DIANA microT Web server has been updated to integrate different additional resources in order to offer insight for putative miRNA involvement in biological processes, and to allow researchers to supplement their knowledge with already published scientific material. Performing an extended analysis on the versioning pattern of miRBase the history of each miRNA has been extracted, offering a unique feature in computational miRNA analysis. We believe that the current Web server update provides important tools for biological analysis and improves interaction with the complicated interconnections regarding miRNA functionality.
FUNDING
The Hellenic Republic, General Secretariat for Research and Technology (09SYN-13-1055). Funding for open access charge: 09SYN-13-1055.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We thank the anonymous reviewers for their helpful comments.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Current Protocols in Bioinformatics. 2010;Chapter 12:11–10. doi: 10.1002/0471250953.bi1209s29. [DOI] [PubMed] [Google Scholar]
- 4.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 6.Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Hiroki E, Akahira J, Suzuki F, Nagase S, Ito K, Suzuki T, Sasano H, Yaegashi N. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101:241–249. doi: 10.1111/j.1349-7006.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]