Abstract
Identifying new indications for existing drugs (drug repositioning) is an efficient way of maximizing their potential. Adverse drug reaction (ADR) is one of the leading causes of death among hospitalized patients. As both new indications and ADRs are caused by unexpected chemical–protein interactions on off-targets, it is reasonable to predict these interactions by mining the chemical–protein interactome (CPI). Making such predictions has recently been facilitated by a web server named DRAR-CPI. This server has a representative collection of drug molecules and targetable human proteins built up from our work in drug repositioning and ADR. When a user submits a molecule, the server will give the positive or negative association scores between the user’s molecule and our library drugs based on their interaction profiles towards the targets. Users can thus predict the indications or ADRs of their molecule based on the association scores towards our library drugs. We have matched our predictions of drug–drug associations with those predicted via gene-expression profiles, achieving a matching rate as high as 74%. We have also successfully predicted the connections between anti-psychotics and anti-infectives, indicating the underlying relevance of anti-psychotics in the potential treatment of infections, vice versa. This server is freely available at http://cpi.bio-x.cn/drar/.
INTRODUCTION
More than 90% of drug candidates fail during development (1), which makes pharmaceutical R&D extremely expensive and time consuming. Identifying novel indications for existing drugs, or drug repositioning, can enhance drug safety, maximize the potential of the drugs and lower R&D costs (2,3). Many drugs such as sildenafil citrate (Viagra®) and raloxifene hydrochloride (Evista®) have already been repositioned for other indications after reports of side effects in clinical trials (4). Adverse drug reaction (ADR) has always been a world-wide concern as one of the leading causes of death among hospitalized patients (5,6). Since both new indications and ADRs are caused by unexpected chemical–protein interactions (7–14), which may be indirect or complex at the mechanism level, it is reasonable to try to predict these interactions based on mining the chemical–protein interactome (CPI).
Understanding drug–drug associations can not only benefit the discovery of novel indications and therapies (15) but also prevent serious negative outcomes (16). The use of a large database of transcriptional responses to identify connections between small molecules which share the same mechanisms and processes within diseases (17,18) can reveal unexpected similarities between drugs so as to indicate potential repositioning uses (19–23) or unexpected adverse reactions (24). Though high throughput technology such as microarray has the potential to generate large quantities of data for analyzing drug–drug associations, this methodology, although robust, can also be costly (25,26) while reliability and quality measures still need to be improved (27,28).
Here we introduce the DRAR-CPI server, for predicting Drug Repositioning potential and ADR via CPI (29). This server has a comprehensive collection of the 385 structural models of targetable human proteins and 254 active forms of small molecules with known descriptions, indications and ADRs. When a user submits a molecule, docking programs can be applied to calculate the binding energy between the uploaded molecule and the targets. The server will give the positive or negative association scores between the user’s molecule and our library drugs based on their interaction profiles across 385 human proteins and will also suggest candidate off-targets that tend to interact with it. Since our library drugs have a comprehensive annotation of their indications and ADRs, users can predict potential indications or ADRs based on the association scores of their molecule across our library molecules. We have matched our in silico predictions of drug–drug associations with those predicted via gene-expression profiles, achieving a matching rate as high as 74% while significantly reducing time and cost. Information on drug–drug associations can lead to new indications for existing drugs, such as the application of anti-psychotics in the potential treatment of bacterial infections, vice versa. The server is freely available at http://cpi.bio-x.cn/drar/.
METHODS
Preparation of the target set and the library drugs
Using the criteria (29,30) and preparation method (31) described in our previous research, we achieved 385 pocket models of 353 proteins with known functions derived from UniProt. We then chose 254 active forms of 166 small molecules from DrugBank (24) with known descriptions, indications and ADR as our library drugs based on the collection criteria of the background drugs in our previous work (30). As all the proteins are human proteins from third-party targetable protein databases, and all drug molecules are from our previous study, we did not add any subjectively selected protein or drug based on our interest in drug repositioning, so as to make it a representative set of the background distribution for both proteins and drug molecules. We will continue to update the targets and library drugs in DRAR-CPI, and users can subscribe to our updates through RSS feeds.
Preparation of the library interactome
We prepared an in silico hybridization using the DOCK program (32), generating a library interactome of 254 library ligands towards 385 protein pockets in the form of a docking score matrix of 254 × 385 elements. Docking scores ≥ 0 were treated as missing values according to our previous scoring process pipeline (31). The two-directional Z-transformation (2DIZ) was applied to process the original docking-score matrix so that the docking scores were normalized to the direction of drugs as Z-scores and then to the direction of targets as Z′-scores to increase accuracy (31).
Evaluation of the drug–drug associations
When one drug is uploaded, it is ‘hybridized’ with all targets using the DOCK program (32). The docking scores of all the library drugs plus the uploaded drug towards all the targets are transformed into a matrix of Z′-scores containing 255 × 385 elements for the calculation of the enrichment score. We developed an algorithm based on connectivity analytics (17) to calculate an association score Si and a P-value between the uploaded drug and each library drug i. For one uploaded drug, after 2DIZ (31), we treat the targets towards the uploaded drug with a Z′-score <−1 as the favorable targets and those with Z′-score >1 as unfavorable targets. Both the favorable and unfavorable targets construct the query signature at this stage. For each library drug, say drug i, we compute an enrichment score for the set of favorable or unfavorable targets in the signature, 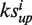 and
and 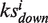 , respectively. To calculate
, respectively. To calculate  , we set n as the total number of all the targets and t as the number of the favorable targets. We sort the favorable targets by Z′-scores towards the library drug i in ascending order and get their positions (1 … t) as list T. Then we sort all the targets in the same way and get their positions (1 … n) as list N. For each favorable target, we get its position in list T as j and its corresponding position in list N as v, and calculate the following values:
, we set n as the total number of all the targets and t as the number of the favorable targets. We sort the favorable targets by Z′-scores towards the library drug i in ascending order and get their positions (1 … t) as list T. Then we sort all the targets in the same way and get their positions (1 … n) as list N. For each favorable target, we get its position in list T as j and its corresponding position in list N as v, and calculate the following values:
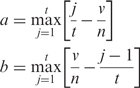 |
Set 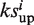 = a, if a > b or
= a, if a > b or 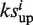 = −b if b > a. Then calculate
= −b if b > a. Then calculate 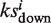 for the unfavorable targets in the same way. Set the association score Si = 0 if
for the unfavorable targets in the same way. Set the association score Si = 0 if 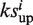 and
and 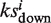 have the same algebraic sign. Otherwise, set si =
have the same algebraic sign. Otherwise, set si = 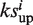 −
− 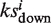 . Scan across all the library drugs and get the maximum and minimum of si as
. Scan across all the library drugs and get the maximum and minimum of si as 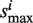 and
and 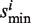 , respectively. Set the association score Si = si/
, respectively. Set the association score Si = si/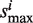 when si > 0 or Si = −si/
when si > 0 or Si = −si/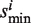 when si < 0. The association score Si is calculated from
when si < 0. The association score Si is calculated from 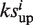 and
and 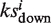 and the P-value is calculated using the Kolmogorov–Smirnov statistic.
and the P-value is calculated using the Kolmogorov–Smirnov statistic.
INPUT AND OUTPUT
Users need to upload a drug molecule in mol2 format with charges and hydrogens added. When the user submits a drug molecule, our server checks the format suitability and calculates the interaction profile of this drug towards all the targets in the database using DOCK6 (32). The parameters of DOCK6 used in back end are listed in Supplementary Table S1, inherited from our previous experience of constructing CPI (29–31). Users can view the real-time progress online, and the page showing the current docking status of the uploaded drug will also be provided for bookmarking. It takes between 6 and 20 h to finish a one-molecule task and an email will be sent on completion. The outputs comprise the two following major elements:
Library drugs which share similar (or opposite) interaction profile with the user's molecule, ranked by the similarity (or disparity) with known indications and ADR information, suggesting the underlying new indication and ADR of the user’s molecule.
The candidate off-targets that tend to interact with the user's molecule. The server will visualize the drug–protein interactions, with amino acid residues around 6 Å of the molecule highlighted.
RESULTS
Prediction of the drug–drug associations
Drug–drug associations based on gene-expression profiles can be used in drug repositioning (19). To test whether the CPI-based docking score profiles corresponded with drug–drug associations, we input the drugs used by Lamb et al. (17) in our server to generate query signatures and compared our result with their reports as the gold standard to test the associations among the drugs. Of the 87 associations in our server, we found that 64 associations (74%) matched with the correlations indicated in their article (Supplementary Table S2).
The data sets used by us are completely independent as we inherited our method from the algorithm in Connectivity Map (cMap) for connectivity analytics without training it with any experimental data. To evaluate our method, the 74% matching rate towards true positive of the gene-expression profiles measured the sensitivity; however, the specificity can not be accurately scaled since it is difficult to define which two drugs are totally not associated with each other.
Case study 1: predicting drug–drug associations for rosigliazone
Rosiglitazone is an anti-diabetic drug of the thiazolidinedione class. Under the stimulation of insulin, it binds to the peroxisome proliferator-activated receptors (PPARs) in fat cells to make the cells more responsive (33). To find the potential indications and ADRs for rosiglitazone, we uploaded an active form of the drug and checked the results (Table 1). We found that
The drug sharing the closest similarity (association score of 1 and P-value 0.0270) to rosiglitazone is fulvestrant, a known anti-estrogenic drug (34), which is used in the treatment of hormone receptor positive metastatic breast cancer in post-menopausal women (35). As rosiglitazone’s binding target PPARγ is significantly related to human primary and metastatic breast adenocarcinomas (36), our server suggested a new indication for rosiglitazone in the treatment of breast cancer.
The seventh nearest drug to rosiglitazone is pravastatin (association score −0.909 and P-value 0.0590), which is used in the treatment of hypercholesterolemia to reduce the risk of myocardial infarction. Since rosiglitazone was associated with a significant increase in the risk of myocardial infarction (37), this opposite association suggested the potential ADR of rosiglitazone for causing myocardial infarction.
After clicking on the ‘CPI’ button, we see that using Z′-scores our server ranked PPARγ to the top.
Table 1.
Associations of library drugs towards rosiglitazone
| Rank | Library drug | Indication | ADR | Association score | P-value |
|---|---|---|---|---|---|
| 1 | Fulvestrant | For the treatment of hormone receptor positive metastatic breast cancer in post-menopausal women with disease progression following antiestrogen therapy. | N/A | 1 | 0.0270 |
| 2 | Geldanamycin | N/A | N/A | −1 | 0.0742 |
| 3 | Rosiglitazone | For the treatment of Type II diabetes mellitus | LongQT | 0.977 | 0.0000 |
| 4 | Risperidone 4 | For the treatment of schizophrenia in adults and in adolescents, ages 13 to 17, and for the short-term treatment of manic or mixed episodes of bipolar I disorder in children and adolescents ages 10 to 17. | Rhabdomyolysis | −0.939 | 0.1215 |
| 5 | 17-allylamino-17-demethoxygeldanamycin | N/A | N/A | −0.934 | 0.1066 |
| 6 | Galantamine 2 | For the treatment of mild to moderate dementia of the Alzheimer’s type. | N/A | −0.931 | 0.0122 |
| 7 | Pravastatin 2 | For the treatment of hypercholesterolemia to reduce the risk of myocardial infarction. | Rhabdomyolysis | −0.909 | 0.0590 |
Seven drugs are ranked by association scores at the top of the list.
This prediction provides clues for further studies on rosiglitazone’s new therapeutic applications to breast cancer as well as a warning of rosiglitazone’s ADR in relation to myocardial infarction. If these data are confirmed by specific experiments, the manufacturer would have a more comprehensive guide as to which indication is appropriate and whether to redesign or modify the drug to weaken unexpected bindings on off-targets. With this information, the drug development process can be made quicker and less costly and unexpected lawsuits can be avoided.
Network analysis of drug associations
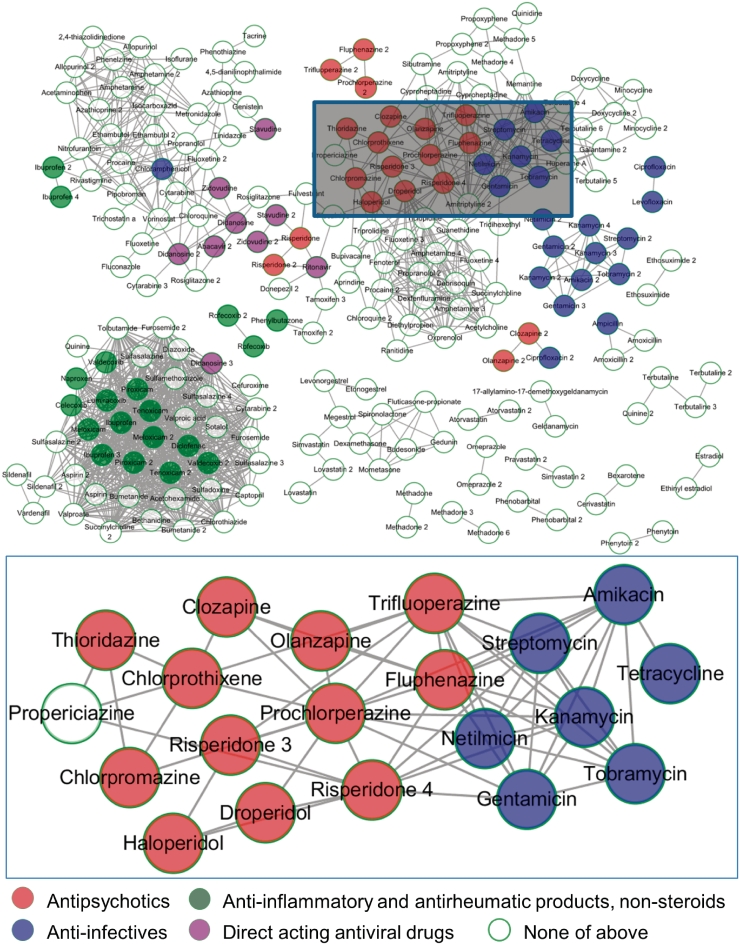
Based on the fact that the drug–drug associations presented by docking results matched the correlations indicated by gene-expression profiles in as many as 74% of cases, we selected the pair-wise associations among all 254 molecules in our library and applied thresholds for association scores of ≥0.6 and P-value ≤0.05. We then visualized the remaining associations in a network layout using a force-directed method based on association scores (Figure 1).
Figure 1.
Drug association network. The drugs are clustered using Cytoscape (47) and employing a force-directed method based on association scores. Partial nodes are coloured according to ATC codes. Five phenothiazine anti-psychotics (chlorpromazine, fluphenazine, prochlorperazine, thioridazine and trifluoperazine) and six non-phenothiazine anti-psychotics (chlorprothixene, clozapine, droperidol, haloperidol, olanzapine and risperidone) are retrieved by our server (shown in red circles, ATC code N05A). Seven anti-infectives are nearby (shown in blue circles, ATC code S01A), while six of them are aminoglycosides (gentamicin, streptomycin, netilmicin, amikacin, kanamycin and tobramycin, ATC code J01G). Background nodes and edges are hidden in the bottom image. The associations revealed potential novel applications for the anti-psychotics and anti-infectives.
Case study 2: the potential for repositioning anti-psychotics as anti-infectives
Five phenothiazine (chlorpromazine, fluphenazine, prochlorperazine, thioridazine and trifluoperazine) and two non-phenothiazine (haloperidol and clozapine) anti-psychotics showed positive connections in terms of gene expression profile (17). From our network, we found all seven drugs tightly clustered (Figure 1, shown in red). In addition, based on the edges at which they connected to other nearby drugs, we also found four other anti-psychotics (chlorprothixene, droperidol, olanzapine and risperidone) holding the same anatomical therapeutic chemical (ATC) code N05A (38–44) (Figure 1, shown in red). Among the 11 typical anti-psychotics (structures are shown in Supplementary Figure S1), 6 anti-psychotics, including chlorprothixene, clozapine, droperidol, haloperidol, olanzapine and risperidone are non-phenothiazines, which have distinct structures. Furthermore, propericiazine at the left of this cluster is used as adjunctive medication in some psychotic patients (http://www.drugbank.ca/drugs/DB01608), which is also highly related to anti-psychotic treatment. By using the drug–drug associations predicted by our server, we successfully recalled 11 anti-psychotics and one medication, indicating that new drug molecules falling within this cluster might have an effect in the treatment of psychology disorders.
The cluster of the anti-infectives (ATC code S01A, Figure 1, shown in blue) is close to the anti-psychotics, while six out of the seven anti-infectives are aminoglycosides (gentamicin, streptomycin, netilmicin, amikacin, kanamycin and tobramycin, ATC code J01G). Anti-psychotic agent prochlorperazine is reported as offering powerful antimicrobial activity against 157 strains of bacteria in vitro (45), and prochlorperazine and chlorpromazine can typically reduce by >1000-fold the minimum inhibitory concentration (MIC) for aminoglycosides in their synergistic interactions against Burkholderia pseudomallei, the causative agent of melioidosis (46), suggesting a potential novel therapeutic treatment for drug-resistance in bacterial infections. Using the drug–drug associations predicted by our server, we found a novel application of anti-psychotics for anti-infective treatment, vice versa, implying potential connections between two fields of the drugs on both applications and mechanisms.
DISCUSSION
Both chemical–protein interactions and gene expression changes reflect how drug/chemicals perturb biosystems. Gene expression change is a downstream event; however, the chemical–protein interactions are the primary step when drugs enter biosystems. In this study, without mining the microarray data, we demonstrated the power of CPI to represent the perturbation towards the biosystems and how it would be used in measuring drug effect in terms of indication and drug adverse effect. On the other hand, in silico discovery of associations among the interaction profiles of small molecules can be efficient and cheap, and can achieve a high rate of accuracy by matching predictions to gene-expression profiles. Furthermore, the putative targets could also be prioritized for unexpected interactions, and sent for further wet-lab validation.
Knowledge of the existing is important to find clues for the new (48). Drug repositioning combined with the prevention of ADR is a major preoccupation for the manufacturers. Discovering drug–drug associations based on CPI is a novel method which can be simultaneously applied in the prediction of both repositioning and ADRs. With the in silico predictions of potential associations among drugs, researchers may not only find helpful clues for exploring potential mechanisms, but could also save significant time and cost in safely repositioning existing drugs for new indications, or predicting potential ADRs. Uncovering associations among molecules as a means of understanding intricate biological systems is consistent with the current trend of ‘-omics’ analyses.
This server is to serve as a complementary methodology for the analysis of gene-expression profiles in drug repositioning. By applying this method into CPI in combination with our previous algorithm 2DIZ, we evaluated the drug–drug associations based on their interaction profiles to indicate potential therapies and ADRs. The advantage of our method is the low cost of harvesting high-dimensional data in silico instead of in vitro while the outcome can still be predictive. We found the predictions of our server indicated some information that fail to be revealed in cMap. Here is an example.
Estradiol is a sex hormone which can be used in the therapy of hormone replacement while minocycline is bacteriostatic antibiotic for the treatment of infections by microorganisms. From a query signature generated by estradiol, minocycline showed no connectivity (connectivity score = 0) in Result S2 of cMap (17). However, after querying our server by estradiol, both two forms of minocycline achieved positive association scores (0.310 and 0.373, respectively). Since the two drugs showed antioxidative abilities of lipid peroxidation inhibition and DPPH radical scavenging (49), contributed to hormone-modulated anabolic responses in fibroblasts after adjunctive periodontal treatment (50) and could be used in the treatment for prevention of ovariectomy reduced bone mineral density (51), our server successfully predicted the positive association between them for potential indications.
This server is independent and different from that in our previous project, SePreSA (31), in terms of both method and purpose, since the current project is concerned with drug repositioning by searching for similar (or opposite) drugs using associations, while the earlier work focused on populations susceptible to Serious Adverse Drug Reaction (SADR) by searching for potential patient-specific targets using polymorphisms within the binding pockets.
CONCLUSIONS
The main function of the DRAR-CPI server is to evaluate associations between the user’s uploaded drug and the library drugs based on their docking profiles towards the putative targets so as to provide suggestions on potential indications and ADRs of the user’s drug. The accuracy of the drug–drug associations is evaluated by recalling drug–drug associations based on gene expression profiles.
Researchers can not only identify the putative targets towards their drugs of interest, but also view a suggested prioritization of potential indications and ADRs with a given confidence value. An extensive range of decisions can be made from the pool of information thus improving the efficiency of R&D.
Unexpected associations can be revealed thereby advancing the understanding of the underlying mechanisms of different kinds of interactions and potentially indicating novel treatments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The National Natural Science Foundation of China (30900841) and the 973 Program (2010CB529600).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We appreciate the support and help extended to us by Brian Fitzgerald of FDA/CDRH/OSEL, Philippe Sanseau and Pankaj Agarwal of Quantitative Sciences, GlaxoSmithKline. We thank Mr Yunguo Gong for useful suggestions to our article. We thank all the developers of the DOCK program, Cytoscape and Jmol applet. L.Y. conceived and designed the experiments, H.L., J.C. and L.Y. performed the experiments; H.L., L.Y., K.W. and L.S. analysed the data; M.M., H.Z. and L.H. contributed reagents/materials/analysis tools and H.L., L.Y. and L.H. wrote the article.
REFERENCES
- 1.DiMasi JA, Hansen RW, Grabowski HG, Lasagna L. Cost of innovation in the pharmaceutical industry. J. Health Econ. 1991;10:107–142. doi: 10.1016/0167-6296(91)90001-4. [DOI] [PubMed] [Google Scholar]
- 2.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 3.Tobinick EL. The value of drug repositioning in the current pharmaceutical market. Drug News Perspect. 2009;22:119. doi: 10.1358/dnp.2009.22.2.1303818. [DOI] [PubMed] [Google Scholar]
- 4.Tartaglia LA. Complementary new approaches enable repositioning of failed drug candidates. Expert Opin. Investig. Drugs. 2006;15:1295–1298. doi: 10.1517/13543784.15.11.1295. [DOI] [PubMed] [Google Scholar]
- 5.Bandekar MS, Anwikar SR, Kshirsagar NA. Quality check of spontaneous adverse drug reaction reporting forms of different countries. Pharmacoepidemiol. Drug Safety. 2010;19:1181–1185. doi: 10.1002/pds.2004. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Xu L, He L. A CitationRank algorithm inheriting Google technology designed to highlight genes responsible for serious adverse drug reaction. Bioinformatics (Oxford, England) 2009;25:2244–2250. doi: 10.1093/bioinformatics/btp369. [DOI] [PubMed] [Google Scholar]
- 7.Rognan D. Structure-based approaches to target fishing and ligand profiling. Mol. Inform. 2010;29:176–187. doi: 10.1002/minf.200900081. [DOI] [PubMed] [Google Scholar]
- 8.Berger SI, Iyengar R. Role of systems pharmacology in understanding drug adverse events. Wiley interdisciplinary reviews. Sys. Biol. Med. 2011;3:129–135. doi: 10.1002/wsbm.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Franchi E, Schalon C, Messa M, Onofri F, Benfenati F, Rognan D, Romesberg F. Binding of protein kinase inhibitors to synapsin I inferred from pair-wise binding site similarity measurements. PLoS ONE. 2010;5:348–354. doi: 10.1371/journal.pone.0012214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li YY, An J, Jones SJ. A large-scale computational approach to drug repositioning. Genome Inform. 2006;17:239–247. [PubMed] [Google Scholar]
- 12.Lee S, Park K, Kim D. Building a drug-target network and its applications. Expert Opin. Drug Discovery. 2009;4:1177–1189. doi: 10.1517/17460440903322234. [DOI] [PubMed] [Google Scholar]
- 13.Ma XH, Shi Z, Tan C, Jiang Y, Go ML, Low BC, Chen YZ. In-silico approaches to multi-target drug discovery: computer aided multi-target drug design, multi-target virtual screening. Pharmaceut Res. 2010;27:739–749. doi: 10.1007/s11095-010-0065-2. [DOI] [PubMed] [Google Scholar]
- 14.Keiser MJ, Irwin JJ, Shoichet BK. The chemical basis of pharmacology. Biochemistry. 2010;49:10267–10276. doi: 10.1021/bi101540g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia J, Zhu F, Ma X, Cao ZW, Li YX, Chen YZ. Mechanisms of drug combinations: interaction and network perspectives. Nat. Rev. Drug Discovery. 2009;8:111–128. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 16.Yap KYL, Chan A, Chui WK, Chen YZ. Cancer informatics for the clinician: an interaction database for chemotherapy regimens and antiepileptic drugs. Seizure. 2010;19:59–67. doi: 10.1016/j.seizure.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, Murino L, Tagliaferri R, Brunetti-Pierri N, Isacchi A, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl Acad. Sci. USA. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang AP, Butte AJ. Systematic evaluation of drugCDisease relationships to identify leads for novel drug uses. Clin. Pharmacol. Ther. 2009;86:507–510. doi: 10.1038/clpt.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suthram S, Dudley JT, Chiang AP, Chen R, Hastie TJ, Butte AJ. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput. Biol. 2010;6:e1000662. doi: 10.1371/journal.pcbi.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu G, Agarwal P. Human disease-drug network based on genomic expression profiles. PLoS ONE. 2009;4:e6536. doi: 10.1371/journal.pone.0006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang M, Smith S, Thorpe A, Barratt MJ, Karim F. Evaluation of phenoxybenzamine in the CFA model of pain following gene expression studies and connectivity mapping. Mol. Pain. 2010;6:56. doi: 10.1186/1744-8069-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang ZQ, Han LY, Lin HH, Cui J, Jia J, Low BC, Li BW, Chen YZ. Derivation of stable microarray cancer-differentiating signatures using consensus scoring of multiple random sampling and gene-ranking consistency evaluation. Cancer Res. 2007;67:9996. doi: 10.1158/0008-5472.CAN-07-1601. [DOI] [PubMed] [Google Scholar]
- 26.Sbisa E, Catalano D, Grillo G, Licciulli F, Turi A, Liuni S, Pesole G, De Grassi A, Caratozzolo MF, D'Erchia AM, et al. p53FamTaG: a database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. BMC Bioinformatics. 2007;8(Suppl. 1):S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat. Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Chen J, He L. Harvesting candidate genes responsible for serious adverse drug reactions from a chemical-protein interactome. PLoS Comput. Biol. 2009;5:e1000441. doi: 10.1371/journal.pcbi.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Wang K, Chen J, Jegga AG, Luo H, Shi L, Wan C, Guo X, Qin S, He G, et al. Exploring off-targets and off-systems for adverse drug reactions via chemical-protein interactome—clozapine-induced agranulocytosis as a case study. PLoS Comput. Biol. 2011;7:e1002016. doi: 10.1371/journal.pcbi.1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Luo H, Chen J, Xing Q, He L. SePreSA: a server for the prediction of populations susceptible to serious adverse drug reactions implementing the methodology of a chemical-protein interactome. Nucleic Acids Res. 2009;37:W406–W412. doi: 10.1093/nar/gkp312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J. Comput.-Aided Mol. Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 33.Young PW, Buckle DR, Cantello BC, Chapman H, Clapham JC, Coyle PJ, Haigh D, Hindley RM, Holder JC, Kallender H, et al. Identification of high-affinity binding sites for the insulin sensitizer rosiglitazone (BRL-49653) in rodent and human adipocytes using a radioiodinated ligand for peroxisomal proliferator-activated receptor gamma. J. Phar. Expr. Therap. 1998;284:751–759. [PubMed] [Google Scholar]
- 34.Wakeling AE, Dukes M, Bowler J. A potent specific pure antiestrogen with clinical potential. Cancer Res. 1991;51:3867. [PubMed] [Google Scholar]
- 35.Howell A, Robertson JFR, Abram P, Lichinitser MR, Elledge R, Bajetta E, Watanabe T, Morris C, Webster A, Dimery I. Comparison of fulvestrant versus tamoxifen for the treatment of advanced breast cancer in postmenopausal women previously untreated with endocrine therapy: a multinational, double-blind, randomized trial. J. Clin. Oncol. 2004;22:1605. doi: 10.1200/JCO.2004.02.112. [DOI] [PubMed] [Google Scholar]
- 36.Mueller E, Sarraf P, Tontonoz P, Evans RM, Martin KJ, Zhang M, Fletcher C, Singer S, Spiegelman BM. Terminal differentiation of human breast cancer through PPAR [gamma] Mol. Cell. 1998;1:465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 37.Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. New Engl. J. Med. 2007;356:2457. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 38.Davis JM. Dose equivalence of the antipsychotic drugs. J. Psychiat. Res. 1974;11:65. doi: 10.1016/0022-3956(74)90071-5. [DOI] [PubMed] [Google Scholar]
- 39.Altschuler E, Kast R. The atypical antipsychotic agents ziprasidone [correction of zisprasidone], risperdone and olanzapine as treatment for and prophylaxis against progressive multifocal leukoencephalopathy. Med. Hypotheses. 2005;65:585. doi: 10.1016/j.mehy.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 40.Yamanouchi Y, Iwata N, Suzuki T, Kitajima T, Ikeda M, Ozaki N. Effect of DRD2, 5-HT2A, and COMT genes on antipsychotic response to risperidone. Pharmacogenomics J. 2003;3:356–361. doi: 10.1038/sj.tpj.6500211. [DOI] [PubMed] [Google Scholar]
- 41.Schatzman RC, Wise BC, Kuo J. Phospholipid-sensitive calcium-dependent protein kinase: inhibition by antipsychotic drugs. Biochem. Biophys. Res. Commun. 1981;98:669–676. doi: 10.1016/0006-291x(81)91166-9. [DOI] [PubMed] [Google Scholar]
- 42.Frahnert C, Rao ML, Grasm der K. Analysis of eighteen antidepressants, four atypical antipsychotics and active metabolites in serum by liquid chromatography: a simple tool for therapeutic drug monitoring. J. Chromatogr. B. 2003;794:35–47. doi: 10.1016/s1570-0232(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 43.McAllister-Williams RH, Ferrier IN. Rapid tranquillisation: time for a reappraisal of options for parenteral therapy. Br. J. Psychiatry. 2002;180:485. doi: 10.1192/bjp.180.6.485. [DOI] [PubMed] [Google Scholar]
- 44.Rijcken CAW, Monster T, Brouwers JRBJ, de Jong-van den Berg L. Chlorpromazine equivalents versus defined daily doses: how to compare antipsychotic drug doses? J. Clin. Psychopharmacol. 2003;23:657. doi: 10.1097/01.jcp.0000096247.29231.3a. [DOI] [PubMed] [Google Scholar]
- 45.Rani Basu L, Mazumdar K, Kumar Dutta N, Karak P, Dastidar SG. Antibacterial property of the antipsychotic agent prochlorperazine, and its synergism with methdilazine. Microbiol. Res. 2005;160:95–100. doi: 10.1016/j.micres.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Chan YY, Ong YM, Chua KL. Synergistic interaction between phenothiazines and antimicrobial agents against Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2007;51:623. doi: 10.1128/AAC.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protocol. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng C, Han L, Yap C, Ji Z, Cao Z, Chen Y. Therapeutic targets: progress of their exploration and investigation of their characteristics. Pharmacol. Rev. 2006;58:259. doi: 10.1124/pr.58.2.4. [DOI] [PubMed] [Google Scholar]
- 49.Kraus RL, Pasieczny R, Lariosa-Willingham K, Turner MS, Jiang A, Trauger JW. Antioxidant properties of minocycline: neuroprotection in an oxidative stress assay and direct radical-scavenging activity. J. Neurochem. 2005;94:819–827. doi: 10.1111/j.1471-4159.2005.03219.x. [DOI] [PubMed] [Google Scholar]
- 50.Tilakaratne A, Soory M. The modulation of androgen metabolism by estradiol, minocycline, and indomethacin in a cell culture model. J. Periodontol. 2002;73:585–590. doi: 10.1902/jop.2002.73.6.585. [DOI] [PubMed] [Google Scholar]
- 51.Williams S, Wakisaka A, Zeng Q, Barnes J, Martin G, Wechter W, Liang C. Minocycline prevents the decrease in bone mineral density and trabecular bone in ovariectomized aged rats. Bone. 1996;19:637–644. doi: 10.1016/s8756-3282(96)00302-x. [DOI] [PubMed] [Google Scholar]