Abstract
In this article, we present CoPub 5.0, a publicly available text mining system, which uses Medline abstracts to calculate robust statistics for keyword co-occurrences. CoPub was initially developed for the analysis of microarray data, but we broadened the scope by implementing new technology and new thesauri. In CoPub 5.0, we integrated existing CoPub technology with new features, and provided a new advanced interface, which can be used to answer a variety of biological questions. CoPub 5.0 allows searching for keywords of interest and its relations to curated thesauri and provides highlighting and sorting mechanisms, using its statistics, to retrieve the most important abstracts in which the terms co-occur. It also provides a way to search for indirect relations between genes, drugs, pathways and diseases, following an ABC principle, in which A and C have no direct connection but are connected via shared B intermediates. With CoPub 5.0, it is possible to create, annotate and analyze networks using the layout and highlight options of Cytoscape web, allowing for literature based systems biology. Finally, operations of the CoPub 5.0 Web service enable to implement the CoPub technology in bioinformatics workflows. CoPub 5.0 can be accessed through the CoPub portal http://www.copub.org.
INTRODUCTION
Medline abstracts are a very useful source of biomedical information covering topics such as biology, biochemistry, molecular evolution, medicine, pharmacy and health care. This knowledge is useful to better understand the complexity of living organisms and can, for instance, be used to study groups of genes or metabolites in their biological context. In the 2008, Web Service issue of NAR, we presented CoPub as a publicly available text mining system. This system uses Medline abstracts to calculate robust statistics for keyword co-occurrences, to be used for the biological interpretation of microarray data (1,2). Since then, CoPub has been intensively used in the analysis of several microarray experiments and toxicogenomics studies (3–8). However, literature data can be applied far beyond questions related to microarray studies. Therefore, we broadened the scope of CoPub by implementing new technology and adding new thesauri to the database. We developed a new technology called CoPub Discovery, which can be used to mine the literature for new relationships following a simple ABC-principle, in which keyword A and C have no direct relationship, but are connected via shared B-intermediates (9). This technology can, for instance, be used to study mechanisms behind diseases, connect new genes to pathways or to find novel applications for existing drugs.
To reflect all these developments, we created CoPub 5.0, which has a complete new user interface and in which we integrated all CoPub technologies. CoPub 5.0 enables the use of CoPub functionality in a very dynamic interactive manner by easily switching between multiple analysis modes and is very suitable to answer a variety of biological questions. It is also accessible using operations of the CoPub 5.0 Web Service (SOAP or JSON), which makes it possible to embed the CoPub functionality into bioinformatics workflows. CoPub 5.0 and the CoPub 5.0 Web Service can be accessed at the CoPub portal http://www.copub.org.
METHODS
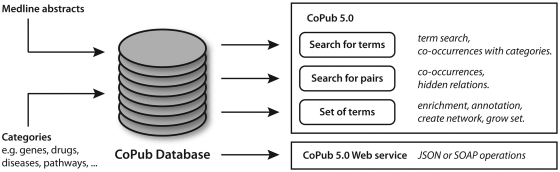
CoPub 5.0 has three analysis modes. A ‘term search’ mode that retrieves abstracts and keyword relations for a single term, a ‘pair search’ mode that analyzes known or new relations between a pair of terms and a set of terms mode that deals with the relation between multiple terms (Figure 1).
Figure 1.
Schematic representation of CoPub. The CoPub database holds co-occurrence information between categories in Medline Abstracts. The CoPub functionality can be used via three modes using the web interface or via the CoPub web services either via SOAP or JSON.
‘Term search’ mode
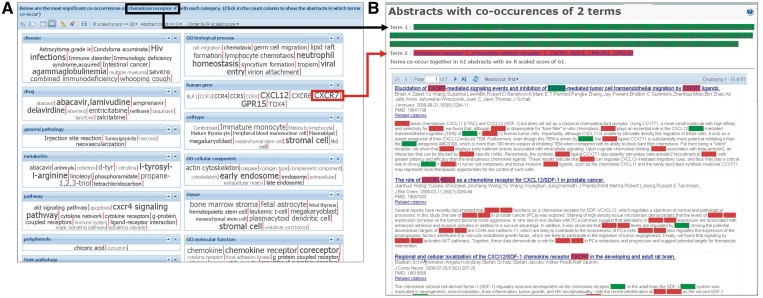
The ‘term search’ mode provides a way to search for keywords and subsequently showing their relations with other categories in the CoPub database. This mode provides a table and cloud view which can be used to answer questions such as ‘to which diseases is this gene related?’ or ‘in which biological processes is my metabolite involved?’ For instance, the cloud view in which strongly connecting terms [i.e. high R-scaled score (1)] are displayed with a larger font, can be used to immediately show the most important relations of the term with keywords from one or more categories in the database (Figure 2A). The evidence for these relations lies in the Medline abstracts in which both terms occur. CoPub retrieves these abstracts, highlights both terms in them and ranks the abstracts which has the most term occurrences as first (Figure 2B). In the example, in Figure 2, it is shown that CXCR4 is strongly connected to its ligand CXCL12 and to CXCR7, with which it forms a heterodimer, and it mediates HIV infections.
Figure 2.
An example of the term search view for the human chemokine receptor 4. In the cloud view, it is immediately clear, by the large font of the terms, that CXCR4 is strongly connected to its ligand CXCL12 and CXCR7, with which it forms a heterodimer (A). Also, CXCR4 is strongly connected to ‘HIV infections’ (category: disease), which is mediated by CXCR4 and to ‘stromal’ cell, to which CXCR4 is linked because of its stromal derived ligand CXCL12. In B an example is shown of the underlying abstracts for the co-occurrences.
Besides co-occurrences, it is also possible to search for new hidden relations between the term and selected categories via shared intermediates using the ‘open discovery’ mode (see ‘Hidden Relations’ section). From the ‘term search’ mode, it is possible to add a term to the current set and switch to the ‘set of terms’ mode.
‘Pair search’ mode
The ‘pair search’ mode can be used to search for specific relations between existing keywords in the CoPub database, e.g. to search for a relation between a gene and a drug. A wizard will guide the user in its search for relations between terms. CoPub will first search for co-occurrences and if no co-occurrence is found, the user can search for hidden relations using the ‘closed discovery’ mode (see ‘Hidden Relations’ section). The pair search mode can be useful to search the literature for more evidence which supports relations found in experiments, for instance, between a drug and a pathway or between a gene and a pathway or which supports hypothesis.
‘Set of terms’ mode
Biological research often involves a better understanding of the complexity of living organisms, for instance, to better understand the development of a disease or to gain more insight into complex signaling pathways (10,11). This requires a systems approach in which groups of genes or metabolites are studied in relation to a disease, drugs or pathways. In CoPub 5.0, we provide such a systems approach via the ‘set of terms mode’. In this mode, a set of keywords can be uploaded either by copy–pasting them or by uploading a text file. Terms can belong to multiple categories (e.g. insulin belongs to the category human gene and to the category drug), which can be further specified to only the desired categories using the ‘Members of category’ option. An uploaded set of terms can be analyzed in a number of ways.
Set enrichment analysis
To see with which categories the set has significant relations, an enrichment analysis can be performed. In this analysis, the relation of a given term from a category with the set is tested using the Fisher exact test against a background set. The calculated P-values are corrected using the Benjamini–Hochberg multiple testing correction method. In case the set consists of multiple categories, only one type of category can be chosen to be used as a background set. For each enriched term, the number of contributors of the set is shown and the contributors can be accessed by clicking on this number. All statistical tests are done using the R Statistics package (http://www.r-project.org).
Set annotation
A set of terms can be annotated by searching for co-occurrences between the set and categories in the database. The cloud view immediately shows for each term in the set, the most significant associated terms (in larger font) per category. Categories from the database can be added or removed from this view. All co-occurring annotation can be downloaded from this view using the download button.
Network
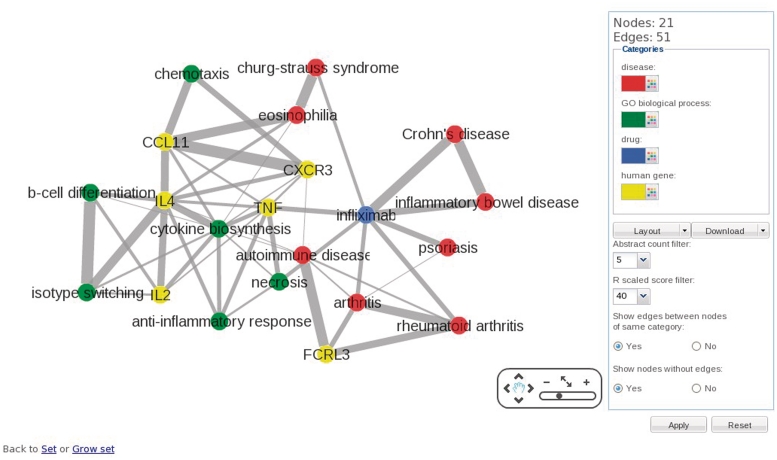
To analyze the relations between terms in the set, a literature network of the set can be created. Subsequently, the network will be visualized using the Cytoscape web plugin (http://cytoscapeweb.cytoscape.org/). Strongly connected terms have thick edges (high R-scaled score), which immediately shows important relations (Figure 3). For large networks (>500 nodes), the network can be downloaded and visualized in a standalone Cytoscape environment.
Figure 3.
Network of a group of mixed terms using the Cytoscape plugin. In the network the gene IL4 has strong connections to the genes IL2 and CCL11 and is also strongly connected to the biological process ‘isotype switching’ and ‘cytokine biosynthesis’. This is indicated by the thick edges between these nodes. Clicking on an edge will show the abstracts in which both terms occur allowing for more detailed analysis of the biological context in which the terms are related.
Add additional terms
At any time an uploaded set can be extended with additional terms. These additional terms can be provided by the user (via ‘add additional terms’), by searching for co-occurrences between the set and categories in the database (via ‘Grow set with co-occurrences’) or by adding a specific term via the ‘term search’ mode, from which it can be added to the set using the ‘Add term to set’ button.
Hidden relations
From the ‘term search’ mode and the ‘pair search’ mode in the website, it is possible to search for hidden relations using the CoPub Discovery technology (9). CoPub Discovery uses an ‘open discovery’ and ‘closed discovery’ process to search for new hidden relations. Both processes follow an ABC principle in which, in case of ‘open discovery’, the user provides a term A (e.g. disease) and searches the literature for hidden relations with a category (C) via intermediates (B) and in case of ‘closed discovery’, the user tests the hypothesis that, for instance, a gene (A) is related to a disease (C) and searches the literature for shared intermediates (B) which support this hypothesis. This technology can be useful to find different roles of genes in new pathways or to get more insight into mechanisms behind diseases.
CoPub Web Service
The operations from the CoPub Web Service allows to embed CoPub functionality into work flows and to use it in an automatic fashion. For this, we provide to use these operations either via SOAP or via JSON. The description of these operations can be found in the help files of the CoPub 5.0 website and an example script, showing how operations can be used, is accessible via the CoPub portal http://www.copub.org.
DISCUSSION AND CONCLUSION
CoPub 5.0 can be used to answer a wide variety of biological questions and bridges the gap between indexed searching of PubMed and dedicated manually curated pathway databases such as Wikipathways (12), Ingenuity Pathway Analysis (http://www.ingenuity.com) and Metacore (GeneGo) (http://www.genego.com/metacore.php).
There are a number of tools that provide part of the technology offered by CoPub. For example, Chilibot (13) is an NLP based tool that retrieves abstracts for user defined pairs of terms, but has no curated dictionaries, meaning that only relations between user defined terms are found, thus limiting the possibility to discover new relations. FACTA (14) offers curated dictionaries but does not provide indirect relation searching or network possibilities. Arrowsmith (15) is a tool for the discovery of hidden relations but does not contain curated ontologies nor does it provide networking possibilities or term mode options. Furthermore, options for analyzing enrichment in terms lists are not provided by these tools, limiting their use for the analysis of approximately omics sets. The advantage of CoPub is that it integrates the approaches offered by the above methods and combines this with advanced graphical output, web service ability and multiple options for analyzing lists of terms and creating networks. The statistical frame work of CoPub 5.0, together with the cloud view functionality is very suitable for the analysis of large ‘omics’ data sets. First, by running a broad scan using enrichment to get a general overview of the data and subsequently by zooming in on relevant pathways, focusing on strong connections (by means of R-scaled score) in the data. Together with the hidden relations technology, this can be used to generate new hypotheses. Future steps could include a better interface to Gene Set Enrichment Analysis (GSEA) software (16) and to incorporate Natural Language Processing (NLP) to be able to even better filter on biological relevant information.
FUNDING
Grants received from the Netherlands Bioinformatics Centre (NBIC) under the BioAssist program and from Merck Sharp & Dohme (MSD). Funding for open access charge: NBIC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank SARA Computing and Networking Services (Amsterdam, The Netherlands) for maintaining and hosting our CoPub database and web server.
REFERENCES
- 1.Alako BT, Veldhoven A, van Baal S, Jelier R, Verhoeven S, Rullmann T, Polman J, Jenster G. CoPub Mapper: mining MEDLINE based on search term co-publication. BMC Bioinformatics. 2005;6:51. doi: 10.1186/1471-2105-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frijters R, Heupers B, van Beek P, Bouwhuis M, van Schaik R, de Vlieg J, Polman J, Alkema W. CoPub: a literature-based keyword enrichment tool for microarray data analysis. Nucleic Acids Res. 2008;36:W406–W410. doi: 10.1093/nar/gkn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frijters R, Verhoeven S, Alkema W, van Schaik R, Polman J. Literature-based compound profiling: application to toxicogenomics. Pharmacogenomics. 2007;8:1521–1534. doi: 10.2217/14622416.8.11.1521. [DOI] [PubMed] [Google Scholar]
- 4.Mitterhuemer S, Petzl W, Krebs S, Mehne D, Klanner A, Wolf E, Zerbe H, Blum H. Escherichia coli infection induces distinct local and systemic transcriptome responses in the mammary gland. BMC Genomics. 11:138. doi: 10.1186/1471-2164-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu T, Krebs S, Bauersachs S, Blum H, Wolf E, Miyamoto A. Actions and interactions of progesterone and estrogen on transcriptome profiles of the bovine endometrium. Physiol. Genomics. 42A:290–300. doi: 10.1152/physiolgenomics.00107.2010. [DOI] [PubMed] [Google Scholar]
- 6.Friberg PA, Larsson DG, Billig H. Transcriptional effects of progesterone receptor antagonist in rat granulosa cells. Mol. Cell. Endocrinol. 315:121–130. doi: 10.1016/j.mce.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 7.Merkl M, Ulbrich SE, Otzdorff C, Herbach N, Wanke R, Wolf E, Handler J, Bauersachs S. Microarray analysis of equine endometrium at days 8 and 12 of pregnancy. Biol. Reprod. 83:874–886. doi: 10.1095/biolreprod.110.085233. [DOI] [PubMed] [Google Scholar]
- 8.Frijters R, Fleuren W, Toonen EJ, Tuckermann JP, Reichardt HM, van der Maaden H, van Elsas A, van Lierop MJ, Dokter W, de Vlieg J, et al. Prednisolone-induced differential gene expression in mouse liver carrying wild type or a dimerization-defective glucocorticoid receptor. BMC Genomics. 11:359. doi: 10.1186/1471-2164-11-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frijters R, van Vugt M, Smeets R, van Schaik R, de Vlieg J, Alkema W. Literature mining for the discovery of hidden connections between drugs, genes and diseases. PLoS Comput. Biol. 6:e1000943. doi: 10.1371/journal.pcbi.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ideker T, Lauffenburger D. Building with a scaffold: emerging strategies for high- to low-level cellular modeling. Trends Biotechnol. 2003;21:255–262. doi: 10.1016/S0167-7799(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 11.Sharan R, Ideker T. Modeling cellular machinery through biological network comparison. Nat. Biotechnol. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 12.Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC bioinformatics. 2004;5:147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuruoka Y, Tsujii J, Ananiadou S. FACTA: a text search engine for finding associated biomedical concepts. Bioinformatics. 2008;24:2559–2560. doi: 10.1093/bioinformatics/btn469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smalheiser NR, Torvik VI, Zhou W. Arrowsmith two-node search interface: a tutorial on finding meaningful links between two disparate sets of articles in MEDLINE. Comput. Meth. Prog. Bio. 2009;94:190–197. doi: 10.1016/j.cmpb.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]