Abstract
iPath2.0 is a web-based tool (http://pathways.embl.de) for the visualization and analysis of cellular pathways. Its primary map summarizes the metabolism in biological systems as annotated to date. Nodes in the map correspond to various chemical compounds and edges represent series of enzymatic reactions. In two other maps, iPath2.0 provides an overview of secondary metabolite biosynthesis and a hand-picked selection of important regulatory pathways and other functional modules, allowing a more general overview of protein functions in a genome or metagenome. iPath2.0′s main interface is an interactive Flash-based viewer, which allows users to easily navigate and explore the complex pathway maps. In addition to the default pre-computed overview maps, iPath offers several data mapping tools. Users can upload various types of data and completely customize all nodes and edges of iPath2.0′s maps. These customized maps give users an intuitive overview of their own data, guiding the analysis of various genomics and metagenomics projects.
INTRODUCTION
Genomes contain a variety of genes of high functional divergence. Interpretation of these huge datasets often requires an overview of various functional traits encoded by the genes (1). Metagenomics or recent pan-genomics projects (2) increase those demands even further. The KEGG database provides hand-curated pathway diagrams, and recently made their overview pathway diagram for metabolism publicly available in SVG (scalable vector graphics) format (3,4). Several tools have been developed since to utilize this diagram and the underlying database (5,6).
In our previous study, we have developed a web-based tool that provides a simple interface to navigate and customize those overview pathways, iPath (7). Here, we report iPath version 2.0 (hereafter iPath2.0) with a considerably expanded amount of underlying data, numerous changes to its data mapping capabilities and a completely overhauled interactive user interface. The underlying global pathway map, which was originally constructed using ∼120 KEGG metabolic pathways in the previous version, has been greatly extended in the current version. iPath2.0 gives overviews of (i) the complete central metabolism in biological systems, (ii) secondary metabolite biosynthesis pathways and (iii) regulatory pathways and functional modules. In total, the three overview pathway diagrams currently cover 172 pathways or functional modules. Nodes in the map correspond to various chemical compounds and edges represent series of enzymatic reactions or protein complexes. This upgrade considerably extends its usefulness in various genome, metagenome, transcriptome or proteome analysis projects.
FEATURES
iPath2.0 is an online tool, accessible using any modern web browser. Pathway diagrams are displayed through an interactive environment developed in Adobe Flex (http://www.adobe.com/products/flex/). Data mapping and customization of various maps are performed on our web server, using a set of Perl scripts and a PostgreSQL-based relational database, thus considerably reducing the local CPU needs of users.
USER INTERFACE AND BASIC FUNCTIONS
iPath2.0′s main interface allows users to easily navigate and explore the complex pathway maps. The viewer provides zooming and panning controls, with different levels of map details corresponding to various zoom levels. Clicking on nodes and edges in the map displays a popup window with detailed information about the associated data, such as enzymes, reactions and compounds involved. Names and identifiers of these associated data can be searched using the built in keyword search engine, allowing users to quickly identify map elements of interest.
UNDERLYING DATA SET
iPath contains 172 pathways or functional modules and 3733 protein orthologous groups defined in KEGG (KOs) based on sequence similarity and manual curation, which are mapped to the respective 4392 clusters of orthologous groups (COGs) (8) and other ortholgous groups derived from the eggNOG database for cross reference (9). These orthologous groups represent enzymes in metabolic or other pathways, parts protein complexes or other components of functional modules.
The content of iPath2.0 is summarized in three separate overview maps. The first one represents the central metabolism, composed of 145 pathways (2130 reactions); for example, glycolysis or amino acid metabolism. The second map gives an overview of 58 metabolic pathways mainly for secondary metabolite biosynthesis (53 pathways are shared with central metabolism; however, 5 unique pathways contains 656 reactions for this overview pathway diagram), such as polyketide biosynthesis, which has high evolutionary diversity. The third one contains 22 regulatory pathways or functional modules such as ribosome or transport systems. In addition to the default overview maps, iPath offers species-specific pathways for 933 fully sequenced genomes derived from their orthologous protein information defined in KEGG.
In the current version, iPath’s maps do not cover all genes of an organism. For example, Escherichia coli has 2549 annotated COGs/NOGs in the eggNOG database corresponding to 4493 of its genes, with only 970 currently covered in iPath as many genes are functionally ill defined or without functional context (859 COGs for 1149 genes in E. coli are classified into ‘poorly characterized’ category), thus the vast majority of well-understood function is already in the map.
PATHWAY CUSTOMIZATION
In addition to the default pre-computed overview maps, iPath2.0 offers a number of useful data mapping tools and extensive customization options. Users can upload various types of data associated with genes, proteins or compounds to generate custom representations of any overview or species-specific pathways map (for examples see below).
INPUT USER DATA
Users can upload query data in plain text, and can define colors, opacity and width for various nodes and edges in the map. Detailed explanation of parameters and example customizations are available in the iPath online help pages. The following types of data can be used to specify parts of the map to customize: KEGG pathways, KEGG compounds, KEGG KOs, KEGG proteins, enzyme EC numbers, COGs, eggNOG orthologous groups, KEGG modules (3) and STRING proteins (10). iPath also contains species information as described in the section on the underlying data sets, and, using a NCBI taxonomy ID or three-letter KEGG organism code, allows users to display only customized versions of species-specific pathways. Correct taxonomy IDs can be selected using the built-in species search engine. iPath can store map customizations within its database, allowing users to simply reload them in future visits.
DATA EXPORT
Customized maps are displayed in the interactive viewer by default, providing the same functionality available for default maps. However, customized maps can also be exported into several graphical formats, both vector and bitmap, for direct inclusion into publications or other documents. Currently supported formats are Scalable Vector Graphics (svg), Portable Network Graphics (png), Encapsulated Postscript (eps), Postscript (ps) and Portable Document Format (pdf).
ILLUSTRATIVE EXAMPLES
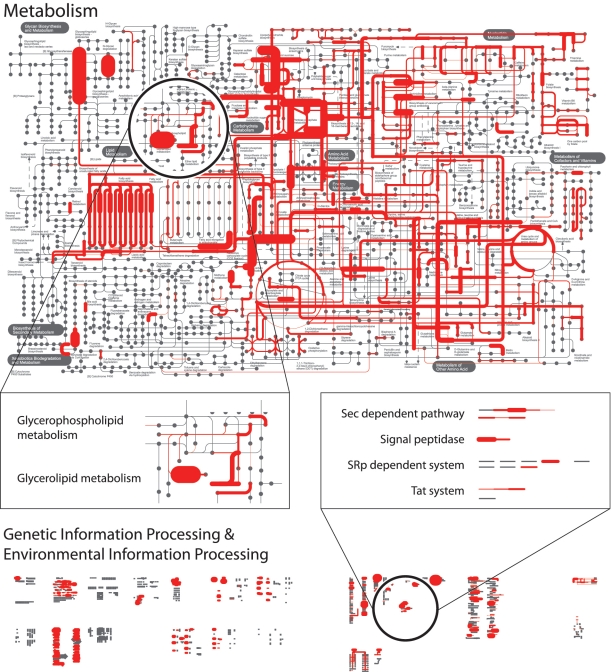
Customized maps generated by iPath allow users to digest their own data in the context of genomic or metagenomic projects. Here, we show an example of the mapping describing the enzymatic activity in human gut microbiota (Figure 1). Using the SmashCommunity pipeline for phylogenetic and functional annotation (11), metagenomic sequence reads from fecal samples of 13 Japanese individuals (12) were mapped to orthologous groups (KOs) defined in the KEGG database via the STRING database (10), and the abundance of those KOs in each sample was calculated. As highly abundant gene orthologous groups across samples might encode crucial functions for human gut microbiome (13), we computed the average abundance of each KOs in 13 metagenomic samples and projected them as widths of edges in the overview pathway diagram (Figure 1). Several pathways, such as Glycerolipid metabolism or the Sec-dependent pathway, show overrepresentation in comparison to the other pathways detected in the data set. Further analysis is required to reveal the detailed associations between abundant pathways and human gut, however, iPath easily provides a general overview of the functionality.
Figure 1.
Enzymatic activity in the human gut microbiome: the nodes colored in red represent enzymes or proteins detected in metagenomic data derived from human fecal samples (12). Users can easily get an overview of complete metabolic and other pathways. In this example, enzymes in glycerolipid metabolism (center left) or proteins in protein export systems (center right) are overrepresented compared to other pathways in the data set. Line widths in the map are proportional to the level of overrepresentation.
In addition to visualizing the metabolic capacity of the human microbiome, iPath2.0 has already been proven as a useful tool in a number of other genomic or metagenomic analyses (1,14–16).
COMPARISON WITH OTHER TOOLS
In order to quantify functionality of iPath2.0, we listed four functional categories [(i) User interface, (ii) Customization capability, (iii) Data mapping and (iv) Functional coverage] and subdivided them into 15 detailed features. Using these features, we compared iPath2.0 with KEGG Atlas (4), Pathway Projector (6) and the previous version of iPath (7) (see Supplementary Table). All tools provide integrated pathways and zooming/panning capability. Compared to the original version, keyword search and mouse over popups have been implemented in iPath2.0. iPath2.0 enables users to map COG/eggNOG, UniProt and STRING IDs directly into pathway diagrams in addition to KEGG IDs, strengthening its advantage in customization capabilities and data mapping. In addition, iPath2.0 is the only tool that provides an overview for regulatory pathways, which is a vital point for functional coverage. Taken together, iPath2.0 has the advantage in making customized maps. Although multiple conditional data is not covered by iPath2.0 yet, it will be provided in the next version.
FUTURE DIRECTIONS
The current version of iPath provides powerful visualization and customization of cellular pathway diagrams. However, a significant amount of manual intervention and analysis is still required to identify interesting pathways in a particular data set. To simplify this process, we are planning to develop an API which will enable programmatic access to iPath by end users and other software packages. Combined with other tools developed by our group, such as iTOL(17) and SmashCommunity (11), iPath will become an integral part of a Pathway-analysis-suite and will also contain more functionality, for example, to display differences between two data sets.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R, Li Y, Zheng H, Luo R, Zhu H, Li Q, Qian W, Ren Y, Tian G, Li J, et al. Building the sequence map of the human pan-genome. Nat. Biotechnol. 2010;28:57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- 3.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kono N, Arakawa K, Ogawa R, Kido N, Oshita K, Ikegami K, Tamaki S, Tomita M. Pathway projector: web-based zoomable pathway browser using KEGG atlas and Google Maps API. PLoS ONE. 2009;4:e7710. doi: 10.1371/journal.pone.0007710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letunic I, Yamada T, Kanehisa M, Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 2008;33:101–103. doi: 10.1016/j.tibs.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arumugam M, Harrington ED, Foerstner KU, Raes J, Bork P. SmashCommunity: a metagenomic annotation and analysis tool. Bioinformatics. 2010;26:2977–2978. doi: 10.1093/bioinformatics/btq536. [DOI] [PubMed] [Google Scholar]
- 12.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Beukeboom LW, Desplan C, Elsik CG, Grimmelikhuijzen CJ, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gianoulis TA, Raes J, Patel PV, Bjornson R, Korbel JO, Letunic I, Yamada T, Paccanaro A, Jensen LJ, Snyder M, et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc. Natl Acad. Sci. USA. 2009;106:1374–1379. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]