Abstract
Plant endogenous non-coding short small RNAs (20–24 nt), including microRNAs (miRNAs) and a subset of small interfering RNAs (ta-siRNAs), play important role in gene expression regulatory networks (GRNs). For example, many transcription factors and development-related genes have been reported as targets of these regulatory small RNAs. Although a number of miRNA target prediction algorithms and programs have been developed, most of them were designed for animal miRNAs which are significantly different from plant miRNAs in the target recognition process. These differences demand the development of separate plant miRNA (and ta-siRNA) target analysis tool(s). We present psRNATarget, a plant small RNA target analysis server, which features two important analysis functions: (i) reverse complementary matching between small RNA and target transcript using a proven scoring schema, and (ii) target-site accessibility evaluation by calculating unpaired energy (UPE) required to ‘open’ secondary structure around small RNA’s target site on mRNA. The psRNATarget incorporates recent discoveries in plant miRNA target recognition, e.g. it distinguishes translational and post-transcriptional inhibition, and it reports the number of small RNA/target site pairs that may affect small RNA binding activity to target transcript. The psRNATarget server is designed for high-throughput analysis of next-generation data with an efficient distributed computing back-end pipeline that runs on a Linux cluster. The server front-end integrates three simplified user-friendly interfaces to accept user-submitted or preloaded small RNAs and transcript sequences; and outputs a comprehensive list of small RNA/target pairs along with the online tools for batch downloading, key word searching and results sorting. The psRNATarget server is freely available at http://plantgrn.noble.org/psRNATarget/.
INTRODUCTION
Plant endogenous non-coding short small RNAs (20–24 nt), including microRNAs (miRNAs) and a subset of small interfering RNAs (ta-siRNAs), are derived from the cleavage products of double-strand RNAs (ds-RNAs) by DICER-like enzymes (1–4). These regulatory small RNAs (mainly include miRNAs and ta-siRNAs, sic passim) negatively regulate gene expression at post-transcriptional level by directing the cleavage of target transcript (mRNA) (5). Several transcription factors and development-related genes have been reported as targets of these regulatory small RNAs, and together they play key roles in gene expression regulatory network controlling plant growth and development (6). These discoveries have aroused wide interest and urgent demand for genome-wide analysis of small RNAs and dissect of their functions, for example, identifying their regulatory target genes in plants.
A number of algorithms and programs have been introduced to search target genes for miRNAs (7–9). However, most of them were developed for animal miRNAs which are significantly different from plant miRNAs in the target recognition process (9,10). For example, an animal miRNA generally requires loose complementarity in about first eight nucleotides of the miRNA, while a plant miRNA demands the whole miRNA mature sequence to be near perfectly aligned with its mRNA target. Secondly, an animal miRNA tends to inhibit target gene’s expression at the translational level, whereas a plant miRNA directly cleaves its target transcript. In addition, an animal miRNA inclines to recognize 3′-UTR region of its target mRNA and a plant miRNA usually has no preference in terms of position. Recent discoveries suggest that plant miRNA may inhibit gene expression at the translational level (11), though it seems to utilize a different recognition pattern compared with a typical animal miRNA’s action (see the next section). These differences therefore demand the development of separate plant small RNA target analysis tools.
A regular expression (a computer language that describes matching strings in text) like pattern matching program, PatScan (12), was adopted in identifying the miRNA targets in rice and Arabidopsis (13,14). In order to utilize PatScan, users need first prepare regular expression-style pattern sets for an input miRNA sequence, then search target sequences with matching pattern(s) in a candidate mRNA sequence dataset. However, the PatScan program was not designed for the end users such as biologists; extra training and programming skills are usually required in order to successfully install this UNIX-style program and generate comprehensive pattern sets for large-scale systematical miRNA target analysis. Xie et al. (15) developed a BLAST-based target search program, miRNAassist. More recently, Fahlgren and Carrington (16) described a pipeline for plant miRNA target prediction using the FASTA program and Perl scripts for matching and scoring. These programs require local installation on a standalone computer and are not designed for high-throughput computing; therefore are not suitable for genome-scale analysis. In addition, the performance of BLAST-like programs are controversial; for example, our study indicated that NCBI BLASTN may miss up to 70% potential targets, because these programs were designed to align long sequences, such as the Expressed Sequence Tags (ESTs) and genomic sequences (8).
Zhang (19) introduced an online analysis tool, miRU, for plant miRNA target analysis. The miRU adopted Smith–Waterman algorithm to search the optimal alignment; it also provides a simple user-friendly web interface and outputs an easily understandable list of matching results, which makes miRU a popular plant miRNA target analysis tool (17,18). However, miRU only accepts one sequence at a time for analysis (lacks high-throughput analysis capability) and the user can only search target candidates in the preloaded libraries. These shortcomings have limited its application in genomic studies, such as analyzing the popular next-generation sequences (19).
The above-mentioned plant miRNA target analysis tools generally focus on the complementarity between small RNA and target transcript. The accessibility of target site on mRNA to a small RNA, determined by secondary structure of mRNA around the target site, has been proved to be an important factor in target recognition (20–23). Incorporating such target-site accessibility evaluation to small RNA target analysis was reported to significantly improve the prediction accuracy (6).
Brodersen et al. (11) reported that miRNA translation inhibition might widely spread in plants. However to date, there is no reported plant small RNA target prediction tool that is capable of distinguishing the newly discovered mechanism from the well-accepted post-transcriptional inhibitions.
Here, we present a new plant small RNA target analysis web server, psRNATarget (http://plantgrn.noble.org/psRNATarget/). The psRNATarget integrates two important analysis functions: (i) reverse complementary matching between small RNA and target transcript using a proven scoring schema, and (ii) target-site accessibility evaluation by calculating unpaired energy (UPE) required to ‘open’ secondary structure around a small RNA target site on the mRNA. This server incorporates recent discoveries in plant small RNA target recognition, e.g. it distinguishes translational inhibition and post-transcriptional inhibition (11), and it reports the number of small RNA/target site pairs that are reportedly associated with small RNA recognition activity to the target transcript. The psRNATarget is designed for high-throughput analysis of next-generation data by implementing a distributed computing pipeline which runs on a Linux cluster at back-end. The server front-end integrates three user-friendly interfaces to accept user-submitted or preloaded small RNAs and transcript sequences and outputs a comprehensive list of small RNAs and matching target sites on candidate transcripts along with built-in online tools for batch downloading, key word searching and the results filtering.
PRINCIPLES AND psRNATarget BACK-END PIPELINE IMPLEMENTATION
Complementarity
psRNATarget evaluates complementarity between small RNA and target gene transcript using the scoring schema originally applied by miRU (19). Instead of using the NCBI BLAST program, we employed a popular Smith–Waterman (24) implementation, ssearch (Version 36.x) (25), in our back-end pipeline since the latter warrants finding the most alignments between very short small RNA sequences and the mRNA sequences.
Multiplicity of target sites
Another good reason for adopting the ssearch program is that it has capability of returning multiple alignments (ssearch versions 36.x and latter releases) for each small RNA/target transcript pair, unlike most of other Smith–Waterman implementations that only return the optimal alignment. Returning multiple optimal alignments enable the reporting of multiple target sites for each small RNA/target transcript pair. This so-called multiplicity of target sites is especially relevant to the biogenesis of siRNA because existence of dual target sites of a miRNA on a specific target transcript has been reported to be an effective trigger of ta-siRNA biogenesis from a TAS (transacting-siRNA) precursor gene (21,26).
Target site accessibility
The psRNATarget server analyzes target accessibility using the RNAup program in Vienna Package (27). RNAup calculates UPE which is the energy required to ‘open’ secondary structure around target site on mRNA. Less energy represents higher possibility to be an effective target site because the secondary structures may prevent small RNA and target site from contacting. Considering the bulk of RNA-induced silencing complex (RISC), both 5′- and 3′-flanks of target site are included for target accessibility evaluation (22).
Translational inhibition
In plants, it has been observed that mismatches occurred around the center of miRNA/mRNA complementary region tend to disable the cleavage activity of RISC; however, the binding of mRNA to RISC still can block gene expression at the translational level (11). The psRNATarget server reports translational inhibition potential when a mismatch is detected in the central complementary region of the small RNA sequence.
Parallel computing analysis pipeline
Both the Smith–Waterman-based alignment using the search program and the UPE calculation using the RNAup program are very computational intensive. Although these programs produce much more accurate results, they significantly impact the analysis throughput.
We greatly enhanced the analysis throughput by developing an efficient back-end pipeline on the basis of an in-house developed distributed computing platform, namely BioGrid. Upon user submission, the master node of BioGrid system divides the submitted miRNA datasets into multiple subsets and transfers these subsets as well as specified target transcript library to remote computing nodes in a Linux Cluster. Next, the master node remotely calls and monitors analytic programs in these computing nodes, and finally the master node collects outputs when these analysis jobs are completed. The communications between master node and computing nodes are mainly through the Linux SSH (Secure Shell) channel. The psRNATarget back-end system, including the BioGrid platform, was written in Java and Groovy languages.
WEB INTERFACES OF psRNATarget
Input
The server front-end integrates three simplified user-friendly interfaces to accept user-submitted sequences and selection of a preloaded miRNAs and transcript sequences for analysis, i.e. (i) searching user-submitted small RNAs against preloaded transcripts; (ii) searching preloaded small RNAs (miRNAs by species) against user-submitted transcripts; and (iii) searching user-submitted small RNAs against the user-submitted transcripts.
In each input interface, default parameters are suggested and preloaded based on our literature analysis; however, users may adjust the behavior of back-end pipeline by changing the parameters. Briefly, maximum expectation for complementarity and UPE (maximum energy to unpair the target site) for target accessibility analysis may be decreased to retrieve more stringent prediction results; more potential target sequences might be missed, though (Figure 1, arrows A and C). The current default values of both parameters were suggested based on our benchmark test (see ‘Performance’ section). The hspsize is the length of scoring region for complementary analysis; users are advised to reduce it to the shortest length of small RNAs if the submitted small RNAs are shorter than default hspsize value (20 nt). Otherwise, those small RNAs shorter than hspsize will be skipped in target analysis (Figure 1, arrow B). The two flanking sizes (lengths of the left and right flanking sequences of the target site) are also adjustable in the target accessibility analysis (Figure 1, arrow D). The users may also adjust the range of central region in which any detected mismatch will be considered as a trigger of translational inhibition (Figure 1, arrow E).
Figure 1.
A set of parameters for adjusting the behavior of back-end pipeline of psRNATarget server.
Output
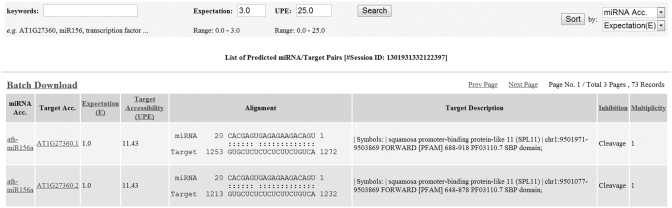
After each successful submission, the end user will be provided a unique URL to trace the analysis progress or check final results at any time. Once the submitted analysis is completed, the psRNATarget server lists the details of the potential small RNA/target site pairs page by page with a comprehensive query and sort tools on the top of each output page for user to easily browse through the results (Figure 2). In addition, psRNATarget allows users to download the entire results in a tab-delimited text file, which is very critical for large-scale data analysis.
Figure 2.
A list of comprehensive miRNA/target site pairs along with query and sort tools on top of each output page.
PERFORMANCE
The psRNATarget searches target gene based on both complementarity scoring analysis and secondary structure analysis. We demonstrate its performance by predicting target genes of 10 published Arabidopsis thaliana miRNAs (Supplementary Data 1) and comparing those predicted target genes to the experimentally validated targets reported in literature. With the default parameters (Expectation ≤ 3, a slightly relaxed threshold), psRNATarget found 92 target candidates (Supplementary Data 2) in TAIR9 cDNA library for the 10 miRNAs, which includes all of 46 validated target gene (100% coverage rate) at 50.0% potential false positive prediction rate. With a more stringent cut-off threshold (Expectation ≤ 2), psRNATarget detected 52 target candidates. Of them, 38 genes have been reportedly validated by 5′-RACE technology, which covers 82.6% of validated target genes at 26.9% potential false positive prediction rate. These results indicate that psRNATarget is able to systematically identify target transcripts; and users may trade their preference on higher prediction coverage or lower false positive prediction rate using different thresholds.
One of the popular applications for psRNATarget is to search target genes in transcript library for small RNAs sequenced by the next-generation technology (28,29). In a performance test, the published small RNA dataset from Arabidopsis Small RNA Project (http://asrp.cgrb.oregonstate.edu/), which consists of around 206 000 small RNAs, was submitted to search against the Arabidopsis TAIR9 transcript database (http://www.arabidopsis.org/). The psRNATarget took 1 h 54 min to complete the whole analytic procedure running on a Linux cluster equipped with 264 cores (52 AMD opteron processors) and generated around 2.5 million small RNA/target site pairs using default parameter values except that the expectation cutoff value was set ≤4 to generate large number of matching pairs. This benchmark results indicate that psRNATarget is well capable of performing high-throughput analysis for large-scale datasets, such as the next-generation sequencing (NGS) data.
DISCUSSION
Complementarity and target-site accessibility have been proven to be the two key factors in the plant regulatory small RNA (miRNA and ta-siRNA) target recognition mechanism (19–21). Both factors have been incorporated to improve the analysis of miRNA target genes in Arabidopsis (6); however, there is no published tool that is able to evaluate these factors for plant regulatory small RNA target analysis for general purpose. The psRNATarget server successfully integrates two well proven approaches (19,27) to evaluate the above two factors; both approaches have been widely applied and well validated by experiments, which warrant the analytic quality of the psRNATarget server.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online
FUNDING
Funding for open access charge: National Science Foundation (Grant No. ABI-0960897); Oklahoma Center for the Advancement of Science and Technology (OCAST Project No. PSB09-32 and PSB11-004); the Samuel Roberts Noble Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Ms Zhaohong Zhuang for assisting in literature analysis, Dr Rakesh Kaundal and Dr Frioz Ahmed for critical reading of the manuscript and providing valuable comments.
REFERENCES
- 1.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Chen H-M, Chen L-T, Patel K, Li Y-H, Baulcombe DC, Wu S-H. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl Acad. Sci. USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell Online. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Zhang Y-Z. Computational detection of microRNAs targeting transcription factor genes in Arabidopsis thaliana. Comput. Biol. Chem. 2005;29:360–367. doi: 10.1016/j.compbiolchem.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri K, Chatterjee R. MicroRNA detection and target prediction: integration of computational and experimental approaches. DNA Cell Biol. 2007;26:321–337. doi: 10.1089/dna.2006.0549. [DOI] [PubMed] [Google Scholar]
- 8.Dai X, Zhuang Z, Zhao PX. Computational analysis of miRNA targets in plants: current status and challenges. Brief. Bioinformatics. 2010;12:115–121. doi: 10.1093/bib/bbq065. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Xu J, Yang D, Tan X, Wang H. Computational approaches for microRNA studies: a review. Mamm. Genome. 2010;21:12. doi: 10.1007/s00335-009-9241-2. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 12.Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 13.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 15.Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett. 2007;581:1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- 16.Fahlgren N, Carrington JC. miRNA target prediction in plants. Methods Mol. Biol. 2010;592:51–57. doi: 10.1007/978-1-60327-005-2_4. [DOI] [PubMed] [Google Scholar]
- 17.Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;57:313–321. doi: 10.1111/j.1365-313X.2008.03690.x. [DOI] [PubMed] [Google Scholar]
- 18.Meng Y, Shao C, Chen M. Toward microRNA-mediated gene regulatory networks in plants. Brief. Bioinformatics. 2011 doi: 10.1093/bib/bbq091. doi:10.1093/bib/bbq091. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hausser J, Landthaler M, Jaskiewicz L, Gaidatzis D, Zavolan M. Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C-miRNA complexes and the degradation of miRNA targets. Genome Res. 2009;19:2009–2020. doi: 10.1101/gr.091181.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 22.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 23.Marin RM, Vanicek J. Efficient use of accessibility in microRNA target prediction. Nucleic Acids Res. 2011;39:19–29. doi: 10.1093/nar/gkq768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 25.Pearson WR. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 26.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 27.Muckstein U, Tafer H, Hackermuller J, Bernhart SH, Stadler PF, Hofacker IL. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 28.Ruan M-B, Zhao Y-T, Meng Z-H, Wang X-J, Yang W-C. Conserved miRNA analysis in Gossypium hirsutum through small RNA sequencing. Genomics. 2009;94:263–268. doi: 10.1016/j.ygeno.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber A, Shi B-J, Huang C-Y, Langridge P, Baumann U. Discovery of barley miRNAs through deep sequencing of short reads. BMC Genomics. 2011;12:129. doi: 10.1186/1471-2164-12-129. [DOI] [PMC free article] [PubMed] [Google Scholar]