Abstract
ChemMine Tools is an online service for small molecule data analysis. It provides a web interface to a set of cheminformatics and data mining tools that are useful for various analysis routines performed in chemical genomics and drug discovery. The service also offers programmable access options via the R library ChemmineR. The primary functionalities of ChemMine Tools fall into five major application areas: data visualization, structure comparisons, similarity searching, compound clustering and prediction of chemical properties. First, users can upload compound data sets to the online Compound Workbench. Numerous utilities are provided for compound viewing, structure drawing and format interconversion. Second, pairwise structural similarities among compounds can be quantified. Third, interfaces to ultra-fast structure similarity search algorithms are available to efficiently mine the chemical space in the public domain. These include fingerprint and embedding/indexing algorithms. Fourth, the service includes a Clustering Toolbox that integrates cheminformatic algorithms with data mining utilities to enable systematic structure and activity based analyses of custom compound sets. Fifth, physicochemical property descriptors of custom compound sets can be calculated. These descriptors are important for assessing the bioactivity profile of compounds in silico and quantitative structure—activity relationship (QSAR) analyses. ChemMine Tools is available at: http://chemmine.ucr.edu.
INTRODUCTION
Cheminformatics tools for analyzing small molecule screening data play an important role in many fields including chemical biology, chemical genomics, drug discovery and agrochemical research (1–3). Informatics resources in these areas are essential for exploring the structure, properties and bioactivity of biologically relevant molecules. To provide these capabilities, software tools are required for analyzing the structural similarities, physicochemical properties and bioactivity profiles of natural and synthetic compounds to gain insight into their modes of action in biological systems. This information is important for the development of effective small molecule probes for studying the functions of protein and cellular networks in chemical genomics and drug discovery research (4). In addition, similar informatics resources are required for identifying the structural and physicochemical relationships among compounds from metabolic or signaling pathways (5–7). The rapidly growing relevance of chemical genomics approaches for modern biology research has significantly increased demand for small molecule mining systems in academia (8).
Currently, the structures of over 30 million distinct small molecules are available in open-access databases, including PubChem, ChemBank and many others (9–15). In addition, preliminary bioactivity data from hundreds of high-throughput screening (HTS) experiments against a wide spectrum of target sites have become available for almost one million compounds in the bioassay sections of various public databases (see below; 9,10,15,16). To efficiently analyze these resources, the development of novel compound data mining and cheminformatic web services is essential.
While there has been extensive development of public domain small molecule databases in recent years (6,9–11, 13–24), the number of open access web services for analyzing public or custom small molecule data is extremely limited at this point (25,26). Thus far, most development has been focused on standalone software applications targeted toward computational rather than experimental scientists. These include Open Babel (27,28), the Chemistry Development Kit (29,30), the Chemical Descriptors Library (31) and JOELib (32). Examples of software designed for non-expert users in this field are Chembench (33) for online quantitative structure—activity relationship (QSAR) modeling and KNIME (34) for designing data analysis pipelines.
Here, we present ChemMine Tools as an online portal to a variety of cheminformatics, visualization, search and clustering tools for small molecule data. The utilities provided by this service are useful for various analysis and data mining routines of small molecule screening experiments in chemical genomics and related areas. An easy to use web interface makes these tools accessible to experimental scientists without an extensive computational background.
METHODS
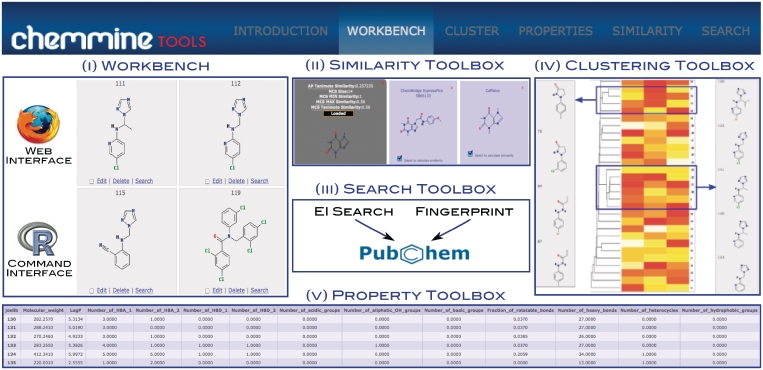
Conceptually, the ChemMine Tools online service is divided into five application domains (Figure 1 and Table 1): (i) a Compound workbench for data imports and result management; (ii) a Structure Similarity toolbox to quantify the similarities among compounds; (iii) a Search toolbox for retrieving similar compounds from PubChem; (iv) a Clustering toolbox for accessing clustering and data visualization tools; and (v) a Property toolbox for predicting physicochemical properties of compounds. To construct robust data analysis workflows, the back-end of the server employs a modular design architecture with object-oriented methods and container classes assuring compatible input/output flows and parameter settings among the different data processing units. Currently, the server integrates over 30 cheminformatics and data mining tools that were developed by this or related open source projects. The modular organization of the ChemMine Tools service has several advantages. For instance, it maximizes the transparency and maintainability of the system, and simplifies the addition of new features and analysis methods upon user request. The web interface of ChemMine Tools is written in Python using the object-oriented and highly scalable Django web framework. Modern JavaScript/Ajax utilities are embedded to generate interactive and customizable high-content web pages. Moreover, the ChemMine Tools project is dedicated to an open access and resource sharing policy. All of its online services and downloadable software components are freely available without restrictions. The following subsections give a detailed description of the underlying algorithms and software tools used by the individual ChemMine Tools services.
Figure 1.
Illustration of the functionalities provided by ChemMine Tools. The utilities of the five application domains (i–v) are listed in more detail in Table 1.
Table 1.
List of services provided by ChemMine Tools
| Functions | Program | Input | Output | Comments |
|---|---|---|---|---|
| (i) Compound workbench | ||||
| Structure import/export | Open Babel | Mouse clicks | SMILES/SDF | One or many compounds |
| Format interconversions | Open Babel | SDF/SMILES | SMILES/SDF | One or many compounds |
| Bioactivity data import | JavaScript/Ajax | Tabular data | Table/heat map | SAR table |
| Structure depictions | CACTVS | SMILES/SDF | Image file (GIF) | One or many compounds |
| Structure drawing | JME Molecular Editor | Mouse clicks | SMILES/SDF | Single compound |
| Database import | SOAP | XML/SDF | SMILES/SDF | PubChem |
| Scriptable access from R | ChemmineRa | SDF, tabular data | Online viewing | SAR table |
| (ii) Similarity toolbox | ||||
| Fragment-based similarity | Atom Pairsa | SDF/SMILES | Similarity coefficients | Pairwise comparisons |
| Maximum common substructure | MCSa | SDF/SMILES | MCS (SDF), similarity coefficient | Pairwise comparisons |
| (iii) Search toolbox | ||||
| Embedding and indexing | EI Searcha | Mouse clicks, SDF/SMILES | Ranked compound list | Database search |
| Fingerprint search | PubChem PUG | Mouse clicks, SDF/SMILES | Ranked compound list | Database search |
| (iv) Clustering toolbox | ||||
| Binning clustering | cmp.clustera | SDF/SMILES, custom table | Cluster table | |
| Hierarchical clustering | hclust | SDF/SMILES, custom table | Tree, distance matrix | Optional heat map |
| Multidimensional scaling | cmdscale | SDF/SMILES, custom table | Scatter plot | Interactive |
| (v) Property toolbox | ||||
| Physicochemical descriptors | JOELib | SDF/SMILES | Property table | 38 descriptors |
The names of software tools, libraries and environments are italicized.
aPrograms developed by the ChemMine Tools project. Acronyms defined in text.
DISCUSSION OF SERVICES
Compound workbench
A central feature of ChemMine Tools is its Compound workbench. It provides a flexible online workspace to upload, manage and visualize small molecule data. Compounds can be imported by reading them from local files, copy and paste, PubChem queries (see Search toolbox) or by interacting with the service through the ChemmineR library (35) within the statistical programming environment R. The latter is an extension of the ChemMine Tools project to provide a programmable interface to more advanced users. Alternatively, compounds can be drawn online with the JME Molecular Editor (36) and then added to the Compound workbench. Currently, the import utility supports the structure data format (SDF) and simplified molecular input line entry system (SMILES). After the import, one can organize and annotate the compounds or view their structure images in single or batch modes. These images are generated in real time from the underlying structure definition data using the structure depiction tool of the CACTVS software suite (11) which runs on the server side. To revisit instances of compound sets, users can save their workbench for later use by downloading the compounds to local files. The compound download function also serves as a format conversion tool to interconvert structure representations between SDF and SMILES formats using utilities from the Open Babel project (27,28). Once the user has populated the Compound workbench with structures, it serves as a central submission system to all downstream analysis services.
Similarity toolbox
In many small molecule screening data analysis routines it is important to compute objective similarity measures among compounds as a means to compare and prioritize structurally related lead compounds. To provide this functionality, ChemMine Tools has implemented two algorithms for computing similarity coefficients among compound structures. The first employs atom pairs as structural descriptors (37) and the widely used Tanimoto coefficient as a similarity measure (see below for more details). Alternatively, users can choose other similarity coefficients, such as Tversky or Dice (38). The second algorithm identifies the maximum common substructure (MCS) shared among compound pairs (39). Subsequently, the size of both compounds and the size of their shared MCS is used to calculate the available similarity coefficients. The underlying MCS algorithm often provides the most accurate and sensitive similarity measure, especially for compounds with large size differences (40,41).
Search toolbox
To efficiently mine much of the chemical structure and bioactivity space available in the public domain, the ChemMine Tools service provides text and structure similarity search methods that interface with the PubChem database (15) via its SOAP-based Power User Gateway (PUG) data exchange feature. During an analysis session, instantaneous search functionality is often important for retrieval of detailed property and annotation information for compounds of interest, or to identify related structures. In ChemMine Tools, structural similarity searches can be performed with PubChem's fingerprint search engine or via the EI Search method. The latter was developed in house as part of this project to provide ultra-fast structure similarity search functionality using an embedding/indexing (EI) algorithm (42). When the fingerprint method is chosen, the query is sent to PubChem, where the structure search is performed and the results are returned to the compound workbench. In contrast to this, EI Search is specific to the ChemMine Tools project and thus, runs locally on its servers. These two tools possess complementary strengths and weaknesses in identifying weak similarities among compounds (42).
Clustering toolbox
Clustering of compounds by structural or property similarity can be a powerful approach to correlating compound features with biological activity. Clustering tools are also widely utilized for diversity analyses to identify structural redundancies and other biases in compound libraries. ChemMine Tools' clustering workbench provides an online interface to three clustering algorithms which include hierarchical clustering, multidimensional scaling (MDS) and binning clustering (35). The following provides a short overview of these tools, while a more detailed outline of the underlying theory and clustering schemes is available in the online tutorial. When clustering by structural similarity, the required similarity measures are computed by first generating the atom pair descriptors (features) for each compound which are then used to calculate a similarity matrix based on the common and unique features observed among all compound pairs using the Tanimoto coefficient. The Tanimoto coefficient has a range from 0 to 1 with higher values indicating greater similarity than lower ones. For the subsequent clustering steps, the similarity matrix is converted into a distance matrix by subtracting the similarity values from 1. The hierarchical and MDS clustering methods provided by ChemMine Tools are based on the R programs hclust and cmdscale, respectively; the third method utilizes an internally developed C++ implementation. These three programs complement one another with respect to their data outputs and visualization options. Hierarchical clustering organizes compounds by similarity in a tree with branch lengths proportional to the item-to-item (compound-to-compound) similarities, while the MDS output encodes this information in a scatter plot. These two methods do not directly provide assignments of compounds to discrete similarity groups; assignments are generated downstream of the actual clustering process using various post-processing methods, such as tree cutting approaches. The binning clustering output provides these groupings directly for a user-definable similarity cutoff. For instance, if a Tanimoto coefficient of 0.6 is chosen then compounds will be joined into groups that share a similarity of this value or greater using a ‘single linkage’ rule for cluster joining. Final results are presented as interactive visualization pages to simplify the interpretation of the (often complex) clustering results. The hierarchical clustering result page uses the Google Maps API to generate zoom- and click-able trees aligned with molecular structure images. Moreover, heat maps of user uploaded data containing compound property, activity or other information can be viewed alongside the tree. A similar system is used to present the MDS results as click-able scatter plots with cursor-over viewing of compound structures. The binning clustering results are presented in a table view containing (among other information) the cluster identifiers and the corresponding compound depictions.
Property toolbox
Predictions of small molecule physicochemical properties are important for assessing their ‘druglikeness’ and ‘leadlikeness’ in silico (43,44). They are also useful for enriching compound collections with desirable properties. For instance, the famous ‘Lipinski Rule of Five’ (45) is often applied to enrich compound collections with druglike candidates. This rule filters for compounds with ≤5 hydrogen bond donors, ≤10 hydrogen acceptors, a molecular weight ≤500 daltons and an octanol-water partition coefficient log P ≤ 5. Physicochemical property data are essential for predicting bioactive and other properties of small molecules using modern machine learning approaches. These data are fundamental to the development of QSAR models (25). ChemMine Tools provides an online interface to the property prediction module of the JOELib package (32). This service can calculate 38 physicochemical property values, including Lipinski descriptors for custom compound sets. The resulting property tables can be downloaded or further processed on ChemMine Tools by sending them to the Clustering toolbox. There, they can be used to cluster compounds by similar property profiles, as described above, or the data can be visualized as a heat map next to the hierarchical clustering trees.
CONCLUSION AND FUTURE DEVELOPMENT
ChemMine Tools is an online service for compound analysis in the chemical genomics field. The service is unique in that it integrates a large number of cheminformatic programs with clustering and visualization functionalities. Additional outstanding features of ChemMine Tools include: (i) its commitment to publicly developed open source software throughout its infrastructure; (ii) its strong dedication to the development of new cheminformatic tools and their free distribution in the community; and (iii) the integration of its many components into a unified online and downloadable software infrastructure which maximizes their utility for diverse tasks with different levels of complexity and customization needs. An intuitive web interface makes these tools accessible to scientists with limited computational background, while simultaneously providing a programmable interface for advanced users. To the best of our knowledge, there are currently no related online services available that provide a comparable suite of functionalities. Overlaps exist, however they are limited to isolated functionalities. For instance, ChemDB and VCCLab (13,43) can be used for property predictions and structure format interconversions of single compound queries; and PubChem supports structure-based clustering for compounds retrieved from its own database.
In the future, many additional utilities will be added to the ChemMine Tools service including the addition of MCS-based search functionality within the Similarity toolbox to support more complex graph-based search strategies against custom compound sets imported into the Compound workbench. Existing functionalities for analyzing bioactivity data will also be expanded by adding a Bioactivity toolbox that will contain regression, machine learning and QSAR modeling tools.
FUNDING
National Science Foundation (grant numbers ABI-0957099, 2010-0520325 and IGERT-0504249). Funding for open access charge: National Science Foundation (grant number: ABI-0957099).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the community projects—Open Babel, JOELib, CACTVS and R—for providing excellent software and data resources that are used by ChemMine Tools. We also thank Peter Ertl for providing the JME Molecular Editor. TG acknowledges support from the core facilities at the Institute for Integrative Genome Biology (IIGB) at UC Riverside. Additionally, we thank our systems administrator Aleksandr Levchuk for assistance in debugging these tools, and expertly maintaining the necessary computational resources.
REFERENCES
- 1.Strausberg RL, Schreiber SL. From knowing to controlling: a path from genomics to drugs using small molecule probes. Science. 2003;300:294–295. doi: 10.1126/science.1083395. [DOI] [PubMed] [Google Scholar]
- 2.Haggarty SJ. The principle of complementarity: chemical versus biological space. Curr. Opin. Chem. Biol. 2005;9:296–303. doi: 10.1016/j.cbpa.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Oprea TI, Tropsha A, Faulon JL, Rintoul MD. Systems chemical biology. Nat. Chem. Biol. 2007;3:447–450. doi: 10.1038/nchembio0807-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- 5.Hattori M, Okuno YY, Goto S, Kanehisa M. Heuristics for chemical compound matching. Genome Inform. 2003;14:144–153. [PubMed] [Google Scholar]
- 6.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahman SA, Bashton M, Holliday GL, Schrader R, Thornton JM. Small molecule subgraph detector (SMSD) toolkitl. J. Cheminform. 2009;1:12. doi: 10.1186/1758-2946-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olah MM, Bologa CG, Oprea TI. Strategies for compound selection. Curr. Drug. Discov. Technol. 2004;1:211–220. doi: 10.2174/1570163043334965. [DOI] [PubMed] [Google Scholar]
- 9.Austin CP, Brady LS, Insel TR, Collins FS. NIH molecular libraries initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 10.Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, et al. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–D359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ihlenfeldt WD, Voigt JH, Bienfait B, Oellien F, Nicklaus MC. Enhanced CACTVS browser of the open NCI database. J. Chem. Inf. Comput. Sci. 2002;42:46–57. doi: 10.1021/ci010056s. [DOI] [PubMed] [Google Scholar]
- 12.Girke T, Cheng LC, Raikhel N. ChemMine. A compound mining database for chemical genomics. Plant Physiol. 2005;138:573–577. doi: 10.1104/pp.105.062687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JH, Linstead E, Swamidass SJ, Wang D, Baldi P. ChemDB update–full-text search and virtual chemical space. Bioinformatics. 2007;23:2348–2351. doi: 10.1093/bioinformatics/btm341. [DOI] [PubMed] [Google Scholar]
- 14.Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Cheng T, Wang Y, Bryant SH. PubChem as a public resource for drug discovery. Drug Discov. Today. 2010;15:1052–1057. doi: 10.1016/j.drudis.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voigt JH, Bienfait B, Wang S, Nicklaus MC. Comparison of the NCI open database with seven large chemical structural databases. J. Chem. Inf. Comput. Sci. 2001;41:702–712. doi: 10.1021/ci000150t. [DOI] [PubMed] [Google Scholar]
- 18.Couzin J. Molecular medicine. NIH dives into drug discovery. Science. 2003;302:218–221. doi: 10.1126/science.302.5643.218. [DOI] [PubMed] [Google Scholar]
- 19.Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 20.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Block P, Sotriffer CA, Dramburg I, Klebe G. AffinDB: a freely accessible database of affinities for protein-ligand complexes from the PDB. Nucleic Acids Res. 2006;34:D522–D526. doi: 10.1093/nar/gkj039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goede A, Dunkel M, Mester N, Frommel C, Preissner R. SuperDrug: a conformational drug database. Bioinformatics. 2005;21:1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- 25.Spjuth O, Willighagen EL, Guha R, Eklund M, Wikberg JE. Towards interoperable and reproducible QSAR analyses: Exchange of datasets. J. Cheminform. 2010;2:5. doi: 10.1186/1758-2946-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Q, Lajiness MS, Ding Y, Wild DJ. WENDI: a tool for finding non-obvious relationships between compounds and biological properties, genes, diseases and scholarly publications. J. Cheminform. 2010;2:6. doi: 10.1186/1758-2946-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. The blue obelisk-interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Boyle NM, Morley C, Hutchison GR. Pybel: a Python wrapper for the OpenBabel cheminformatics toolkit. Chem. Cent. J. 2008;2:5. doi: 10.1186/1752-153X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL. Recent developments of the chemistry development kit (CDK) - an open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 30.Guha R. Chemical Informatics functionality in R. J. Stat. Softw. 2007;18:1–16. [Google Scholar]
- 31.Sykora VJ, Leahy DE. Chemical descriptors library (CDL): a generic, open source software library for chemical informatics. J. Chem. Inf. Model. 2008;48:1931–1942. doi: 10.1021/ci800135h. [DOI] [PubMed] [Google Scholar]
- 32.Wegner JK, Fröhlich H, Zell A. Feature selection for descriptor based classification models. 2. Human intestinal absorption (HIA) J. Chem. Inf. Comput. Sci. 2004;44:931–939. doi: 10.1021/ci034233w. [DOI] [PubMed] [Google Scholar]
- 33.Walker T, Grulke CM, Pozefsky D, Tropsha A. Chembench: a cheminformatics workbench. Bioinformatics. 2010;26:3000–3001. doi: 10.1093/bioinformatics/btq556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berthold MR, Cebron N, Dill F, Gabriel TR, Kotter T, Meinl T, Ohl P, Sieb C, Thiel K, Wiswedel B. KNIME: The Konstanz Information Miner. New York: Springer; 2007. [Google Scholar]
- 35.Cao Y, Charisi A, Cheng LC, Jiang T, Girke T. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24:1733–1734. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ertl P. Molecular structure input on the web. J. Cheminform. 2010;2:1. doi: 10.1186/1758-2946-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Reynolds CH. Performance of similarity measures in 2D fragment-based similarity searching: comparison of structural descriptors and similarity coefficients. J. Chem. Inf. Comput. Sci. 2002;42:1407–1414. doi: 10.1021/ci025531g. [DOI] [PubMed] [Google Scholar]
- 38.Holliday JD, Salim N, Whittle M, Willett P. Analysis and display of the size dependence of chemical similarity coefficients. J. Chem. Inf. Comput. Sci. 2003;43:819–828. doi: 10.1021/ci034001x. [DOI] [PubMed] [Google Scholar]
- 39.Cao Y, Jiang T, Girke T. A maximum common substructure-based algorithm for searching and predicting drug-like compounds. Bioinformatics. 2008;24:366–374. doi: 10.1093/bioinformatics/btn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raymond JW, Willett P. Maximum common subgraph isomorphism algorithms for the matching of chemical structures. J. Comput. Aided Mol. Des. 2002;16:521–533. doi: 10.1023/a:1021271615909. [DOI] [PubMed] [Google Scholar]
- 41.Hattori M, Okuno Y, Goto S, Kanehisa M. Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J. Am. Chem. Soc. 2003;125:11853–11865. doi: 10.1021/ja036030u. [DOI] [PubMed] [Google Scholar]
- 42.Cao Y, Jiang T, Girke T. Accelerated similarity searching and clustering of large compound sets by geometric embedding and locality sensitive hashing. Bioinformatics. 2010;26:953–959. doi: 10.1093/bioinformatics/btq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, Palyulin VA, Radchenko EV, Zefirov NS, Makarenko AS, et al. Virtual computational chemistry laboratory–design and description. J. Comput. Aided Mol. Des. 2005;19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- 44.Monge A, Arrault A, Marot C, Morin-Allory L. Managing, profiling and analyzing a library of 2.6 million compounds gathered from 32 chemical providers. Mol. Divers. 2006;10:389–403. doi: 10.1007/s11030-006-9033-5. [DOI] [PubMed] [Google Scholar]
- 45.Lipinski CA, Lombardo F, Dominy BW, J FP. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliver. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]