Abstract
The massively parallel sequencing technologies have recently flourished and dramatically cut the cost to sequence personal human genomes. Haplotype assembly from personal genomes sequenced using the massively parallel sequencing technologies is becoming a cost-effective and promising tool for human disease study. Computational assembly of haplotypes has been proved to be very accurate, but obviously contains errors. Here we present a tool, HapEdit, to assess the accuracy of assembled haplotypes and edit them manually. Using this tool, a user can break erroneous haplotype segments into smaller segments, or concatenate haplotype segments if the concatenated haplotype segments are sufficiently supported. A user can also edit bases with low-quality scores. HapEdit displays haplotype assemblies so that a user can easily navigate and pinpoint a region of interest. As inputs, HapEdit currently takes reads from the Polonator, Illumina, SOLiD, 454 and Sanger sequencing technologies.
INTRODUCTION
In transcriptome sequencing, epigenomics, targeted sequencing and whole-genome resequencing, the use of massively parallel sequencing technologies is widespread, some notable examples of which are sequencing-by-synthesis platform (Illumina) (1), sequencing-by-ligation platforms (Polonator; ABI SOLiD) (2), pyrosequencing platform (Roche 454) (3) and single-molecule sequencing platforms (Helicos Heliscope) (4) (Pacific Biosciences SMRT) (5). The massively parallel sequencing technologies continue to extend read length, increase throughput and shorten run time. Along with this, the massively parallel sequencing technologies are becoming indispensable in genomic variation detection and clinical diagnosis.
Haplotype assembly is a useful tool for genome analysis. One example is to characterize the causal relationship between cis variation and gene expression. As genome-wide association studies have progressed, it is now essential to understand how cis variations are correlated with phenotypes. To advance this study, haplotype assembly is necessary to determine the phases between cis-regulatory regions and coding regions. The Sanger sequencing (6) or Illumina sequencing technology has been used to assemble personal human haplotypes (7,8). It is anticipated that whole-genome resequencing using the massively parallel sequencing technologies will become routine as the sequencing cost for a personal genome drops under $1000 within several years. If personal genomes can be sequenced at that low cost, haplotypes will be more frequently assembled for clinical use.
It has been a common practice to infer a haploid consensus sequence from a genome assembly even when reads were generated from two haplotypes in eukaryotes. In haploid assembly, inferring a haploid consensus sequence only requires a simple statistical method (9). To computationally assemble haplotypes from sequenced reads, however, it is necessary to disentangle reads from two haplotypes and infer two consensus sequences. The complexity of haplotype assembly is known to be NP-hard (10). Several computational methods have been developed to assemble haplotypes, which are based on Markov chain Monte Carlo approaches (11,12), heuristic approaches (7,13), and a combinatorial approach (14).
The assembly viewer Consed was originally developed to assess and edit haploid genome assemblies from reads obtained by Sanger sequencing, but now also supports reads obtained from massively parallel sequencing methods. (15). Recently, EagleView was developed to view genome assemblies by massively parallel sequencing technologies (16). However, none of these was designed to view, assess, and edit haplotype assemblies.
HapEdit was designed to assess assembled haplotypes and edit misassembled haplotypes, supporting reads sequenced by the five massively parallel sequencing technology platforms (Illumina, Polonator, ABI SOLiD, Roche 454, and Helicos) and the Sanger sequencing technology.
WORKFLOW
Software package
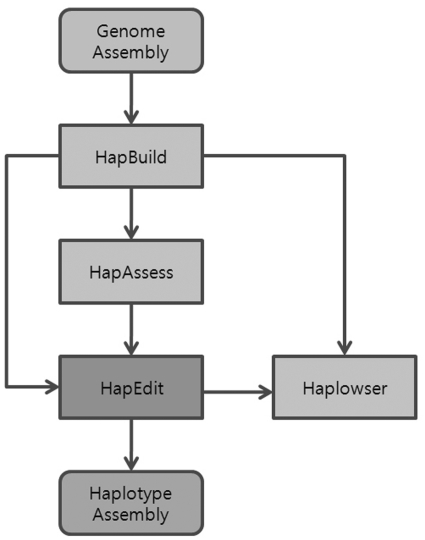
Figure 1 shows the flowchart of haplotype assembly. HapEdit imports a haplotype assembly from HapBuild (11). Optionally, HapEdit can import and display quality scores for assembled haplotypes, which are calculated by HapAssess (17). A user can compare haplotypes from different individuals, using a comparative browser, Haplowser (18). HapEdit is provided as a component in a software package for haplotype assembly.
Figure 1.
Workflow of haplotype assembly. The input is a sequence assembly, taken by HapBuid as an input. The final output is a haplotype assembly. For the description of each component software, see the main text.
Web start and standalone program
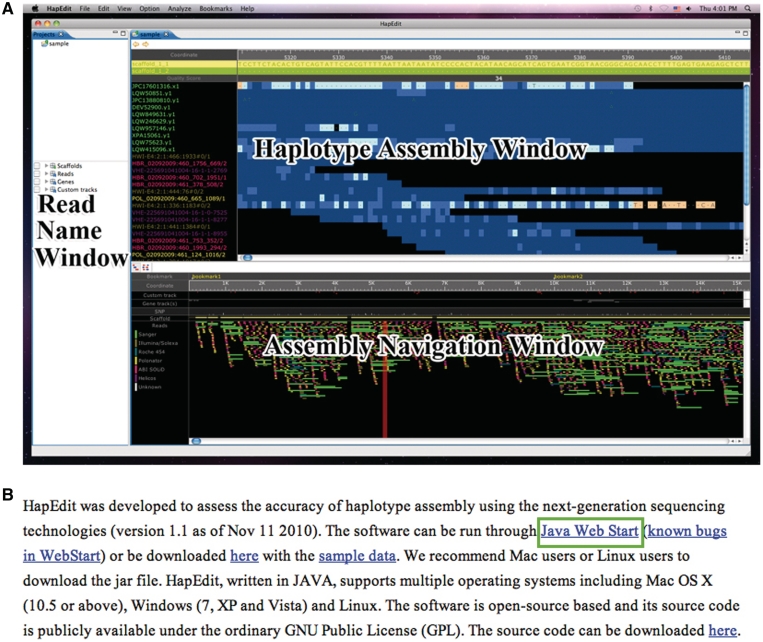
A user can download the binary files compiled for the three operating systems (MacOSX, Windows and Linux). Alternatively, a user can run HapEdit directly on the web site through Java web start (Figure 2B).
Figure 2.
(A) Screenshot of HapEdit main window. At the top of the HA Window, haplotype sequences with the genomic coordinates are displayed, where SNPs are highlighted in red. Quality scores for assembled haplotype sequences are displayed below the haplotype sequences. A haplotype assembly is located below quality scores, where read names are colored according to the sequencing technologies used. (B) HapEdit web site. HapEdit can be run simply by pressing the ‘Java Web Start’ link.
IMPLEMENTATION
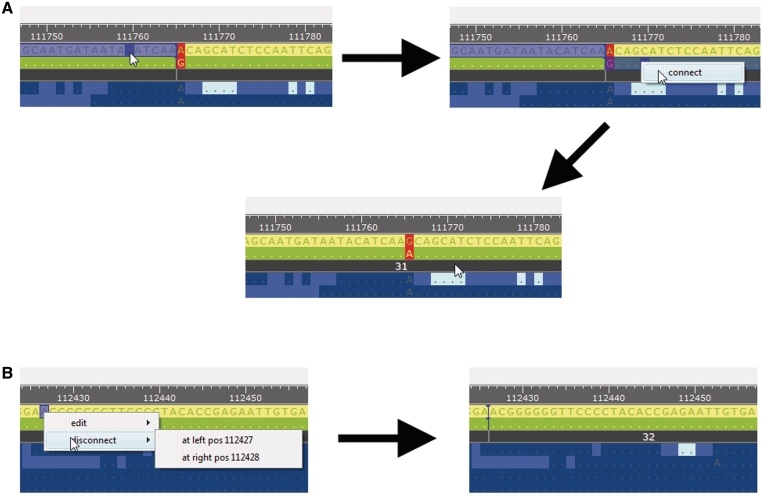
HapEdit provides different views of a haplotype assembly through three windows [Read Name Window (RN), Haplotype Assembly Window (HA) and Assembly Navigation Window (AN)]; see Figure 2A. In the Haplotype Assembly Window (HA), a detailed view of a haplotype assembly is displayed with zooming function, where haplotype sequences and alignments with quality scores are also shown. The name of each read in the alignments is differentially colored based on the sequencing technology used. Similarly, each base-call is colored based on its quality score. In this manner, a user can easily identify low-quality bases and the sequencing technology used for the bases. At the top of the HA window, a user can manually edit haplotype sequences in three ways. First, erroneous bases can be fixed by directly modifying the bases. Second, a user can connect haplotype segments if the connection is judged to be significantly supported by any read (Figure 3A). Third, a user can consider the quality scores for assembled haplotype segments, and break haplotype segments potentially containing phasing errors into pieces (Figure 3B).
Figure 3.
(A) Using a combination of a mouse and key operation, a user can connect haplotype segments. Haplotype segments to be connected are selected by pressing the left-mouse button while holding the shift key. Then, the connection menu pops up with a mouse right-click (control key + a mouse click on MacOSX). Haplotype segments are connected by clicking the ‘connect’ menu. (B) A user can choose any region of haplotypes to be disconnected by pressing the right-mouse button. Haplotype segments are disconnected by selecting the position to be disconnected.
The AN Window is synchronized with the HA window to depict a global view of a haplotype assembly. In the AN window, the sequencing technology used for a read is indicated by the color of the read. A user can navigate any region of a haplotype assembly in a mouse click. The region selected by the mouse click in the AN Window is synchronously displayed in the HA Window. Conversely, The region shown in the HA window is traced and marked by a red bar in the AN window. The gene annotation [in GFF, UCSC or SG (Simple Gene) format] and custom track information (in BLAT or BLAST format) can be imported, and displayed in two optional panes (Gene Pane and Custom Track Pane) of the AN window. The SNP information obtained from haplotype sequences is also displayed in an optional pane (SNP Pane).
The RN Window enumerates the names and genomic coordinates of all the reads included in the haplotype assembly in the HA window. A user can move to the starting point (or ending point) of a read of interest by right-clicking the name of the read and selecting the pop-up menu. The names of gene names and custom tracks shown in the AN window are also enumerated in the RN window.
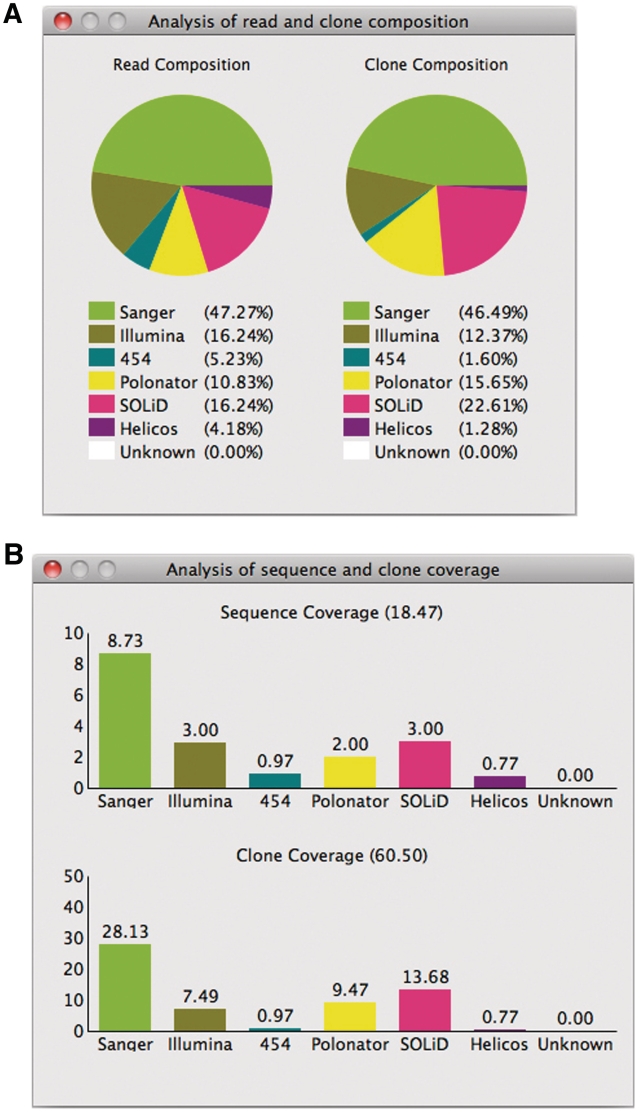
Integrating different sequencing technologies to detect structural variants in a cost-effective way has been recently explored through a simulation study (19). However, finding the optimal composition rate of each sequencing technology between cost and N50 haplotype length in assembling haplotypes is yet to be explored. To fit the composition rates into the ideal composition rates (or the rates that a user initially planned), HapEdit facilitates a user to assess the deviation from those; the composition rates of reads and clone among the entire reads and clones are summarized in pie charts (Figure 4A). Similarly, the sequence coverage and clone coverage of each sequencing technology are analyzed and summarized in bar charts (Figure 4B).
Figure 4.
(A) The composition rates of reads (left) and clones (right) sequenced by the different technologies are displayed in pie charts. The different colors represent the different sequencing technologies. (B) The sequence coverage (upper) and clone coverage (lower) are calculated and presented in a form of bar chart. Each bar indicates the coverage by a specific sequencing technology.
CONCLUSION
HapEdit is an accuracy assessment tool to view haplotype assemblies by massively parallel sequencing technologies and edit misassembled haplotypes. It offers a graphical user interface to navigate haplotype assemblies and helps a user to fit the composition rates of the reads sequenced by the (up to) six different sequencing technologies to the ideal composition rates.
FUNDING
The National Research Foundation of Korea (2009-0083311 to J.H.K., W.K. and S.P.; 2011-0004382 to S.P.); The National Institutes of Health, USA (HG002790 to L.M.L.). Funding for open access charge: The National Research Foundation of Korea [2009-0083311].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Michael Sismour and John Aach for helpful comments.
REFERENCES
- 1.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate Multiplex Polony Sequencing of an Evolved Bacterial Genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 3.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braveman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nature Biotech. 2009;17:847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 6.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The Diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchill GA, Waterman MS. The accuracy of DNA sequence: Estimating sequence quality. Genomics. 1992;14:89–98. doi: 10.1016/s0888-7543(05)80288-5. [DOI] [PubMed] [Google Scholar]
- 10.Lippert R, Schwartz R, Lancia G, Istrail S. Algorithmic strategies for the single nucleotide polymorphism haplotype assembly problem. Brief. Bioinform. 2002;3:23–31. doi: 10.1093/bib/3.1.23. [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, Waterman MS, Li LM. Diploid genome reconstruction of Ciona intestinalis and comparative analysis with Ciona savignyi. Genome Res. 2007;17:1101–1110. doi: 10.1101/gr.5894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bansal V, Halpern AL, Axelrod N, Bafna V. An MCMC algorithm for haplotype assembly from whole-genome sequence data. Genome Res. 2008;18:1336–1346. doi: 10.1101/gr.077065.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long Q, MacArthur D, Ning Z, Tyler-Smith C. HI: haplotype improver using paired-end short reads. Bioinformatics. 2009;25:2436–2437. doi: 10.1093/bioinformatics/btp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bansal V, Bafna V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008;24:i153–i159. doi: 10.1093/bioinformatics/btn298. [DOI] [PubMed] [Google Scholar]
- 15.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequencing finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 16.Huang W, Marth G. EagleView: A genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–1543. doi: 10.1101/gr.076067.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JH, Waterman MS, Li LM. Accuracy assessment of diploid consensus sequences. IEEE Trans. Comput. Biol. and Bioinfo. 2007;4:88–97. doi: 10.1109/TCBB.2007.1007. [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Kim WC, Waterman MS, Park S, Li LM. HAPLOWSER: whole-genome haplotype browser for personal genome and metagenome. Bioinformatics. 2009;25:2430–2431. doi: 10.1093/bioinformatics/btp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du J, Bjornson RD, Zhang ZD, Kong Y, Snyder M, Gerstein MB. Integrating sequencing technologies in personal genomics: optimal low cost reconstruction of structural variants. PLoS Comput. Biol. 2009;5:e1000432. doi: 10.1371/journal.pcbi.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]