Abstract
Transcription factors are important gene regulators with distinctive roles in development, cell signaling and cell cycling, and they have been associated with many diseases. The ConTra v2 web server allows easy visualization and exploration of predicted transcription factor binding sites in any genomic region surrounding coding or non-coding genes. In this new version, users can choose from nine reference organisms ranging from human to yeast. ConTra v2 can analyze promoter regions, 5′-UTRs, 3′-UTRs and introns or any other genomic region of interest. Hundreds of position weight matrices are available to choose from, but the user can also upload any other matrices for detecting specific binding sites. A typical analysis is run in four simple steps of choosing the gene, the transcript, the region of interest and then selecting one or more transcription factor binding sites. The ConTra v2 web server is freely available at http://bioit.dmbr.ugent.be/contrav2/index.php.
INTRODUCTION
Both transcription factors (TFs) and microRNAs (miRNAs) are key players in gene regulation in multicellular organisms (1). Based on pairing between miRNAs and mRNAs, miRNA targets are predicted by searching for matches with the miRNA seed regions (2). On the other hand, the use of a position weight matrix (PWM) is the leading model for detection of TF binding sites (TFBSs). A PWM represents the sequence motif and depicts the DNA binding preferences of the TF. It is constructed using a set of known binding sequences.
Traditionally, regulation of genes by TFs is predicted by analyzing promoter regions and determined experimentally by DNAse-foot-printing assays or electrophoretic mobility shift assays (EMSA). Nowadays, functional protein–DNA binding sites are increasingly studied on a genomic scale by using ChIP-seq. These studies indicate that only some of the functional TFBS are located in promoter regions; introns and untranslated regions (UTRs) also contain a substantial number of functional sites (3–5). For example, regulatory sites in the first intron might interact with sites in the promoter region due to DNA looping (6,7).
Of the estimated 2000 human TFs, ∼300 are thought to bind to the core promoter and to play a role in the general transcription machinery, whereas the rest bind more specifically and regulate a fraction of genes (8). The latter TFs are expressed in almost all tissues or only in a few tissues, depending on whether their function is broad or more specific. Over half of the human genes are believed to have alternative promoters (9) and consequently one should investigate the promoters, UTRs and intronic regions of each individual transcript.
In this update, we describe the new features and expansions of the ConTra webserver. In this tool, for any genomic region TF binding sites can be detected and visualized of the known transcripts of a gene of interest. Starting from one of nine reference organisms, a scientist can easily investigate regulation at the transcription level using the latest UCSC multiz alignments, which are accessible through the ConTra interface. Alternatively, sequence files and PWMs can be uploaded for analysis of the user's own data. Similar web tools with their pros and cons compared to ConTra v2 are listed in Supplementary Table S1.
NEW FEATURES
The first version of ConTra provided users with a flexible way to analyze promoter alignments (10). Users were able to visualize or explore TFBSs in the promoter region of a gene of interest. PWM libraries from the JASPAR CORE database and TRANSFAC database were used to identify TFBSs in a multi-species alignment with human as reference species. Even though the human genome is one of the most widely used reference genomes, the lack of other reference species and alignments was regarded as one of the most important shortcomings in the first version of ConTra. Furthermore, only the promoter region could be analyzed for TFBSs.
The 2011 update of ConTra adds the following features. In addition to the promoter region, users can now look for TFBSs in 5′-UTR, 3′-UTR and introns. Evidence is rising that these regions are at least as important in transcriptional regulation as the promoter region itself (3–5,11). Mokry et al. (3) demonstrated that many (35–40%) of the TCF4 binding sites are intronic. Furthermore, considerable fractions of ZNF-263-, CTCF-, NRSF- and STAT1 binding sites are located in 5′-UTR, 3′-UTR and intronic regions. A detailed overview of the relative importance of the aforementioned genomic regions is given in Supplementary Table S2.
In the first edition of ConTra, searching for TFBSs was only possible in multiple alignments in relation to the human genome, which left many users empty handed. In ConTra v2, multiple alignments with mouse, chicken, cow, frog, zebrafish, fruitfly, worm and yeast as reference species have been added. A detailed overview of the different genome assemblies, genes and multiz alignments available in ConTra v2 is presented in Table 1. Although the human genome is the most widely studied genome, other model organisms should not be ignored. The importance of the different model organisms is illustrated in Supplementary Figure S1, in which the popularity of the different organisms is compared in terms of PubMed hits.
Table 1.
Summary of the number of genes, non-coding genes and transcripts for each reference organism that can be analyzed in ConTra v2
| Reference species | Common name | Assembly | Genes | RefSeq transcripts | Coding (NM_) (%) | Non-coding (NR_) (%) | Ensembl transcripts | Multiple sequence alignment |
|---|---|---|---|---|---|---|---|---|
| Homo sapiens | human | hg19 | 22 167 | 37 474 | 86.3 | 13.7 | 151 222 | multiz46way of 46 vertebrate genomes (hg19) |
| Mus musculus | mouse | mm9 | 21 786 | 27 621 | 93.3 | 6.7 | 88 186 | multiz30way of 30 vertebrate genomes |
| Bos taurus | cow | bosTau4 | 11 559 | 12 427 | 97.7 | 2.3 | 31 598 | multiz5way: cow, dog, human, mouse, platypus |
| Gallus gallus | chicken | galGal3 | 4905 | 5176 | 90.1 | 9.9 | 23 392 | multiz7way: chicken, human, mouse, rat, opossum, frog, zebrafish |
| Xenopus tropicalis | frog | xenTro2 | 8358 | 9695 | 99.8 | 0.2 | 28 937 | multiz7way: frog, chicken, opossum, human, mouse, rat, zebrafish |
| Danio rerio | zebrafish | danRer6 | 13 812 | 15 776 | 95.6 | 4.4 | 32 992 | multiz6way: zebrafish, tetraodon, stickleback, frog, mouse, human |
| Drosophila melanogaster | fruit fly | dm3 | 14 230 | 23 550 | 94.1 | 5.9 | 23 017 | multiz15way of 15 insects |
| Caenorhabditis elegans | worm | ce6 | 19 903 | 24 892 | 97.1 | 2.9 | 35 019 | multiz6way of 6 worms |
| Saccharomyces cerevisiae | yeast | sacCer2 | 7130 | na | na | na | 7130 | multiz7way of 7 yeast species |
For each species, a specific UCSC multiz alignment is used.
In ConTra v2, transcripts can be searched for using the official HGNC gene name, HGNC symbol, alias, Ensembl gene ID (ENSG), the Entrez Gene ID, the RefSeq mRNA ID (NM_/NR_) or the Ensembl transcript ID (ENST). For every species, the most recent alignments are then automatically fetched from UCSC and processed.
Users can select binding motifs from different sources, including the latest versions of the TRANSFAC database (update 2010.4) (12), the JASPAR core database update 2010 (13), the phyloFACTS database (14) and a collection of homeodomain TF PWMs derived from a protein binding microarray (PBM) (15). Furthermore, PWMs can be constructed by the user using the web interface. Creating a custom PWM is as easy as uploading a fasta file containing aligned sequences. The ConTra v2 web interface automatically converts the data into the right format.
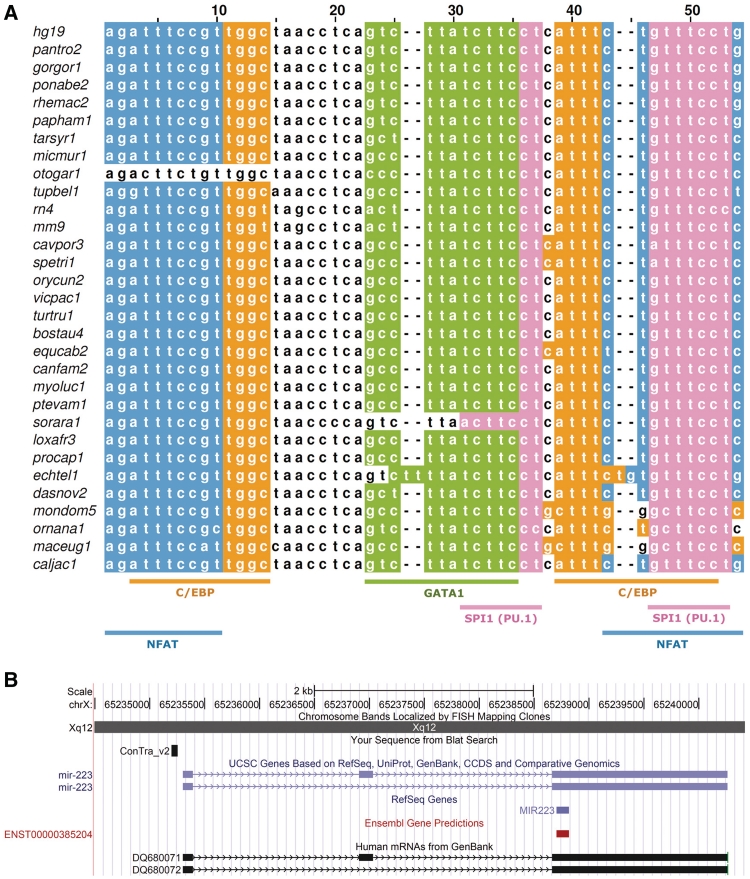
In ConTra v2, non-coding genes are no longer excluded from the analysis. TFs and miRNAs often work together in what is termed a feed-forward loop (FFL). These FFLs regulate many important biological processes, such as those in development and tumor formation (16). Non-coding transcripts are treated as regular transcripts in ConTra, and they can be analyzed in the same way. To verify whether the results on non-coding genes are meaningful, we looked for binding sites in the promoter region of miRNA-223 (hsa-miR-223 or MIR223) with RefSeq accession number NR_029637. Fukao et al. (17) have shown that MIR233 is regulated by a wide range of TFs, such as NFAT, C/EBP, GATA1 and PU.1. Analysis in ConTra v2 not only supports the presence of the binding sites for these TFs but also shows that they have been strongly conserved during evolution (Figure 1).
Figure 1.
Visualization of the evolutionarily conserved mechanism for miRNA-223 regulation in the promoter region, as described by Fukao et al. (17). (A) Multiz alignment showing the conserved binding sites. In orange, the C/EBP TF, predicted using the Jaspar positional weight matrix MA0102.2; in blue, the NFAT TF (TRANSFAC M00935); in green; the GATA1 TF (Jaspar MA0035.2); and in pink, the PU.1 TF (Jaspar MA0080.2). The figure was created with the free multiple alignment editor Jalview using the ConTra fasta and fc file on the results page. (B) Region of (A) was mapped using BLAT on the mir-223 promoter in the UCSC genome browser (black box). Blue box represents the miRNA location.
A wide variety of examples on the use of ConTra v2 can be found in online Supplementary Data. Supplementary Figures S2–S6 show results of ConTra v2 analyses on different genomic regions, using the UCSC multiz46way alignment based on the human hg19 reference sequence and illustrating experimentally validated binding sites from literature. Supplementary Figure S7 depicts an evolutionarily conserved binding site in the second intron of the Mus musculus nestin gene, as described by Jin et al. (18). In Supplementary Figure S8, two sine oculis (SO) binding sites are conserved in the second intron of the Drosophila Lz gene, which confirms the study of Yan et al. (19). Finally, the promoter of the S. cerevisiae PHD1 (FLO11) gene in Supplementary Figure S9 shows two conserved TEA TFBSs, which supports the regulatory mechanism proposed by Heise et al. (20).
If the genomic region of interest, for example, from another reference organism or for a new transcript, is not available in ConTra, alignment files in either the UCSC multiple alignment format (MAF), in multi-fasta format or in clustal format can be uploaded. On the help page of the web site are demos showing how to obtain such a MAF file in the UCSC genome browser, how to upload and analyze this file, and how to use the feature color (fc) file and fasta file on the result page to produce publication-quality figures similar to those in the online Supplementary Data of this article. If a PWM model for a particular TF is not present in the available collections, uploading one's own PWM is also possible. This can be either in the PWM format, but less experienced users can simply upload an alignment file in multi-fasta format. ConTra automatically detects the input format and subsequently builds the PWM.
TECHNICAL DETAILS AND FOUR-STEP ANALYSIS PROCESS
ConTra v2 runs on a CentOS 5 server configured with an apache web server (version 2.2.3), MySQL server (5.0.77), PHP 5.1.6 and perl 5.8.8. The interface is programmed in PHP, and alignments are fetched from UCSC using perl scripts. TFBS hits for a user-defined motif are calculated using the Match algorithm. An overview picture of these hits, created with Jalview, is embedded in the overview page with the help of the Highslide thumbnail viewer (http://www.highslide.com). Different TFs on the result page are visualized dynamically using Javascript. For each alignment block, both a file with PWM scores and a file containing a phylogenetic conservation score for each TF is provided (see File 1 in Supplementary Data for more details). Scores in the ConTra v2 exploration part are calculated in the same way as in the previous version of ConTra, with the exception that due to the inclusion of other genomic regions, we no longer take into account the distance to the transcription start site.
The ConTra v2 analysis consists of four steps. First, users have to choose whether they want to visualize or explore a gene of interest. In this step, it is also necessary to indicate the reference species and the gene of interest. The second step lists a group of available transcripts for genes matching the search terms, from which one can be selected. For every gene, all possible RefSeq and Ensembl transcript variants are listed with a link to the genomic location in the respective genome browser. This way, genes with alternative promoters, UTRs or alternative intronic regions can be analyzed for regulatory differences. In step three, different genomic regions of the selected transcript can be chosen (upstream, introns, 5′-UTR and 3′-UTR). The final step offers users an extensive choice of PWM motifs: up to 20 PWM motifs can be simultaneously taken into account for analysis.
For the visualization part, results are split into alignment blocks (Supplementary Figure S10). These blocks consist of local alignments produced by the TBA program (threaded blockset aligner) (21). In the exploration part, a list of PWMs is given, ranked according to the prediction score.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agency for Innovation through Science and Technology in Flanders (grant number 091213). Funding for open access charge: Department for Molecular Biomedical Research, VIB, Ghent, Belgium.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Amin Bredan for critical reading and editing of the article.
REFERENCES
- 1.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mokry M, Hatzis P, de Bruijn E, Koster J, Versteeg R, Schuijers J, van de Wetering M, Guryev V, Clevers H, Cuppen E. Efficient double fragmentation ChIP-seq provides nucleotide resolution protein-DNA binding profiles. PLoS One. 2010;5:e15092. doi: 10.1371/journal.pone.0015092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frietze S, Lan X, Jin VX, Farnham PJ. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J. Biol. Chem. 2010;285:1393–1403. doi: 10.1074/jbc.M109.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin H, van't Hof RJ, Albagha OM, Ralston SH. Promoter and intron 1 polymorphisms of COL1A1 interact to regulate transcription and susceptibility to osteoporosis. Hum. Mol. Genet. 2009;18:2729–2738. doi: 10.1093/hmg/ddp205. [DOI] [PubMed] [Google Scholar]
- 7.Magklara A, Smith CL. A composite intronic element directs dynamic binding of the progesterone receptor and GATA-2. Mol. Endocrinol. 2009;23:61–73. doi: 10.1210/me.2008-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooghe B, Hulpiau P, van Roy F, De Bleser P. ConTra: a promoter alignment analysis tool for identification of transcription factor binding sites across species. Nucleic Acids Res. 2008;36:W128–W132. doi: 10.1093/nar/gkn195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 12.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su N, Wang Y, Qian M, Deng M. Combinatorial regulation of transcription factors and microRNAs. BMC Syst. Biol. 2010;4:150. doi: 10.1186/1752-0509-4-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T, Tanabe M. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 18.Jin ZG, Liu L, Zhong H, Zhang KJ, Chen YF, Bian W, Cheng LP, Jing NH. Second intron of mouse nestin gene directs its expression in pluripotent embryonic carcinoma cells through POU factor binding site. Acta. Biochim. Biophys. Sin (Shanghai) 2006;38:207–212. doi: 10.1111/j.1745-7270.2006.00149.x. [DOI] [PubMed] [Google Scholar]
- 19.Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev. Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Heise B, van der Felden J, Kern S, Malcher M, Bruckner S, Mosch HU. The TEA transcription factor Tec1 confers promoter-specific gene regulation by Ste12-dependent and -independent mechanisms. Eukaryot. Cell. 2010;9:514–531. doi: 10.1128/EC.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]