Abstract
RNA molecules can achieve a broad range of regulatory functions through specific structures that are in turn determined by their sequence. The prediction of mutations changing the structural properties of RNA sequences (a.k.a. deleterious mutations) is therefore useful for conducting mutagenesis experiments and synthetic biology applications. While brute force approaches can be used to analyze single-point mutations, this strategy does not scale well to multiple mutations. In this article, we present corRna a web server for predicting the multiple-point deleterious mutations in structural RNAs. corRna uses our RNAmutants framework to efficiently explore the RNA mutational landscape. It also enables users to apply search heuristics to improve the quality of the predictions. We show that corRna predictions correlate with mutagenesis experiments on the hepatitis C virus cis-acting replication element as well as match the accuracy of previous approaches on a large test-set in a much lower execution time. We illustrate these new perspectives offered by corRna by predicting five-point deleterious mutations—an insight that could not be achieved by previous methods. corRna is available at: http://corrna.cs.mcgill.ca.
INTRODUCTION
RNA molecules can achieve a broad range of regulatory functions through specific self-folding structures that are in turn determined by their nucleotide sequence. Any modification in this sequence may result in a change in its structure and a loss of function. These deleterious mutations (1) can be the origin of metabolic disorders. For example, Halvorsen et al. (2) recently reported finding several mutations associated with diseases that were indeed deleterious. Since the role played by RNA molecules in various diseases is becoming evident (3), the development of tools for predicting deleterious mutations could be helpful to predict pathogenic mutations especially in the absence of comparative genomic data.
Geneticists could also benefit from such a predictor. Indeed, to understand the importance of specific nucleotides, mutagenesis experiments proceed by point-wise mutations in order to reveal modifications in the molecule's function. When this function is carried by the structure, these mutations can be associated with a structural change. These experiments, however, are time consuming and have a substantial cost. Since the number of possible mutations grows exponentially with the size of the sequence, exhaustive experimental studies are not feasible. It follows that the choice of which mutations to test is critical. An efficient prediction method that returns a small list of deleterious mutation candidates could help direct these experiments and generate better results.
The prediction of deleterious mutations is also important in synthetic biology. Many recent models use RNA molecules as nano devices and require sequences designed to fold into specific shapes (4–7). To be functional, the best candidate sequences should be robust to both thermodynamic and genetic perturbations. In this case, a deleterious mutation predictor can be used to filter out sequences which are too sensitive to nucleotide substitutions.
In the last 4 years, three methods have been developed to predict deleterious mutations (8–10). RDMAS (8) and RNAmute (9) have been designed to predict single deleterious mutations. However, in general, the structural instability carried by a single mutation is limited and may not produce significant changes. To address this challenge, Churkin and Barash extended their method and developed MultiRNAMute—a method searching for multiple-point mutations that greatly improves the scope and significance of the predicted deleterious candidates (10). To date, MultiRNAMute is available as a stand-alone application and only RDMAS offers a web interface.
All these previous methods combine a brute force exploration of the mutational landscape with a systematic usage of single sequence secondary structure predictors (11). This approach is unfortunately computationally limiting as the algorithm must generate and individually fold a large number of mutants that grows exponentially with the length of the sequence and the number of mutations allowed. Efforts to circumvent this problem have led to heuristics using the structural properties of the wildtype to restrict the number of candidates considered. Unfortunately, even with these techniques, the search depth is very limited and the state-of-the-art approach (i.e. MultiRNAMute) cannot efficiently predict simultaneous deleterious mutations with more than three mutations.
We have recently shown that we can simultaneously explore both the mutational and secondary structure landscape of an RNA sequence in both polynomial time and space complexity (12–14). The resulting software, RNAmutants, has been implemented as a web server (15) for general RNA mutational analysis. Although straightforward applications of RNAmutants can be used to predict deleterious mutations (14), the accuracy of these results is limited as RNAmutants does not implement any strategy to bias the search toward deleterious mutations, neither does it provide an evaluation function for quantifying the deleterious effect of the predicted mutations. Nevertheless, as noted in a recent review by Barash and Churkin (16), our statistical sampling algorithms provide the best perspectives for a time-efficient multiple-point deleterious mutation analysis.
In this article, we describe corRna, a method for predicting multiple-point deleterious mutations in RNA sequences using our RNAmutants framework. Our approach enables us to predict deleterious mutations with a large number of substitution sites, while preserving the accuracy of a brute force approach. To achieve these results, we combined RNAmutants with the structural heuristic search introduced in Ref. (10), thus producing similar quality predictions in a much shorter time. In addition, we propose a novel mutational heuristic search and show that it also improves the accuracy of the mutation predictions.
This article is organized as follows. First, we describe the web server input parameters and the prediction output provided by corRna. Then, in the ‘Definitions and methods’ section, we describe the algorithms and the search heuristics which have been used to improve the accuracy of the results. Finally, in the ‘Results’ section, we evaluate the performance of our methods. In particular, we (i) show that corRna predictions correlate with mutagenesis experiments (17), (ii) estimate the impact of various heuristics on the quality of the predictions, and (iii) compare our methods with previous approaches on a newly created test set extracted from the Rfam database (18). We also illustrate the new perspectives offered by corRna by predicting five-point deleterious mutations—an insight that could not be achieved by any previous methods. corRna is the first web server that enables the prediction of deleterious multiple-point mutations for an RNA sequence.
WEB SERVER
Hardware and compatibility
The web server (http://corrna.cs.mcgill.ca) runs Ubuntu-Server 10.04 on a Dell PE T610 2x Intel Quad core X5570 Xeon Processor, 2.93 GHz 8M Cache, 64 GB Memory (8 × 8 GB), 1333 MHz Dual Ranked RDIMMs for 2 Processor, Advanced ECC. The web server has been tested and is functional in Internet Explorer, Firefox and Google Chrome.
Input
The input form of corRna is shown in Figure 1. First, the user inputs an RNA sequence and an optional email address. Then, the user can choose between a ‘Structure’ (default) and a ‘Mutation’ heuristic to guide the mutational landscape exploration, or to simply decide to perform an unbiased search without using any heuristics. The structural heuristic explores mutations that favor alternate structures present in the suboptimal structural ensemble. The mutation heuristic performs successive searches while limiting the location at which mutations can occur along the RNA sequence. Details on these heuristics will be discussed in the ‘Definitions and methods’ section.
Figure 1.
corRna Input Form.
corRna also enables the user to choose between two methods for probing the mutational landscape. By default, it uses the the original RNAmutants algorithm (14). However, if no search heuristic is selected, the user may also use a novel extension of RNAmutants called fixedCGSampling, which enables us to compute multiple mutations while preserving the G + C content of the input sequence (19). In both cases, the user can define the maximum number of k-point mutations allowed in the input sequence, using the field called ‘Mutation depth’.
Finally, users are able to refine their search by modifying extra options, depending on the heuristic chosen. With the structure heuristic, the user can define the number of suboptimal base pairings that corRna will use. In the mutation heuristic, the user can define how many successive searches will be performed, as well as restrict results to mutation sequences that fall below a defined threshold. The user can also modify the dangling end energy setting used when running RNAfold to obtain the base pairing probability distribution.
Output
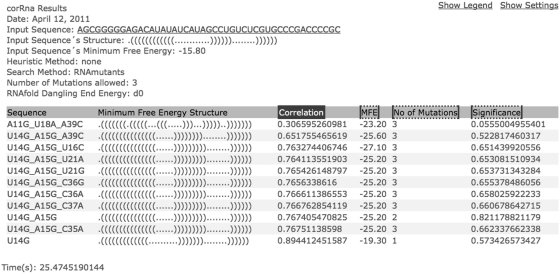
Upon submission of the input form, a link to the results page is posted at the top of the web site. If the user provided an email address, this link and the results will also be sent by email. Before the results are generated, the page will refresh every 5 s and display the status of the query, whether it be ‘Waiting in Queue’ or ‘Processing’. A sample of the results page for the HCV cis-acting replication element (5BSL3.2) is shown in Figure 2. The page consists of a table that displays all deleterious mutations predicted by corRna. The additional columns include the minimum free energy (MFE) secondary structure, the base pair correlation (i.e. a measure of the deleteriousness of the mutations described in the ‘Definitions and methods’ section), the MFE value of the mutant, the number of mutations and the significance of the candidate (i.e. an estimate of how likely this mutant can be found by chance. See ‘Definitions and methods’ section). If the structural heuristic is used, then the base pair constraint and its break number are also included. By clicking on the header of each column, the user can sort the results according to the value stored in this column.
Figure 2.
corRna Results Page—results for the HCV cis-acting replication element (5BSL3.2) without heuristics and allowing up to three-point mutations. Note that although corRna calculates the correlation based on the whole structural ensemble, only the MFE structure is displayed.
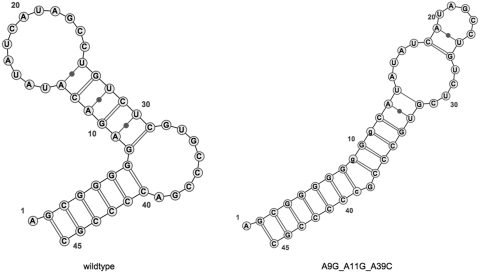
Moreover, if a user clicks on a sequence in the table, the server will display a graphical representation of the associated secondary structure using the java applet VARNA 3.7 (20). This functionality is illustrated in Figure 3 and is useful to quickly compare the structural differences between the wildtype and the mutation candidate.
Figure 3.
Secondary structure candidate comparison of the 5BSL3.2 wildtype and the A9C_A11G_A39C mutation candidate through the use of VARNA 3.7.
DEFINITIONS AND METHODS
The core component of corRna is RNAmutants, an efficient mutational analysis tool that explores the complete mutational landscape of a given RNA sequence. Given an RNA sequence, RNAmutants uses a dynamic programming algorithm to compute, for each integer k, the minimum free energy MFE(k) and Boltzmann partition function Z(k) of all sequences with k mutations over all secondary structures (14). Then, RNAmutants uses a stochastic backtracking procedure to sample mutants and secondary structures.
corRna works in two steps. First, it uses RNAmutants to compute a sample set of candidate deleterious mutations. This search can be aided either by a structural or mutation heuristic to prune the RNA mutational landscape. Then, corRna ranks the samples by the strength of their deleterious effect.
Structural heuristic
The structural heuristic uses structural constraints on the base pairings allowed in the sequence to guide corRna in the exploration of the mutational landscape. corRna will first use the base pairing probability matrix generated by Vienna's RNAFold to find base pairing locations with significant probabilities that are not used in the MFE secondary structure. Then, it calculates the break number of each base pair, defined by the number of base pairs that must be removed from the wild-type sequence in order to insert the target base pair. Finally, corRna runs RNAmutants while constraining the search to mutations which preserve these identified base pairs. This strategy was inspired by and implemented from the method used in MultiRNAMute (10).
Mutation heuristic
The mutation heuristic uses constraints on the allowed mutation locations to guide corRna. In RNAmutants, the mutants with the lowest MFE are more likely to be sampled than other sequences. Thus, deleterious mutations that do not improve the free energy of the input sequence can be missed. To find other mutations, corRna performs successive runs of RNAmutants and progressively removes from the sample set, mutation locations that were explored in the previous runs (i.e. we constrain RNAmutants to not mutate the positions used in previous runs). This novel heuristic provides a way to explore the mutation space at locations that would otherwise be obscured by the more probable candidates provided by RNAmutants. This strategy thoroughly differs from the structural heuristic and enable us to explore regions of the mutational landscape that could have been otherwise missed.
Measurement of ‘deleterious-ness’
We quantify the “deleterious-ness” or destabilizing effect of a candidate mutation with a base pair correlation measure that compares the structural ensemble of the mutation sequence to that of the wildtype (i.e. the input sequence). Briefly, this correlation method computes the base pairing probabilities of the wild-type and a sampled mutant using RNAfold (11). Then, it calculates the Pearson's correlation coefficient between the two distributions to estimate the deleterious effect of the mutation(s). This correlation value ranges between −1 and 1 and quantifies the deleterious effect of a mutation. Values close to 1 denote non-deleterious mutations, values close to −1 stand for highly deleterious mutations. This method was first proposed by Halvorsen et al. (2), who demonstrated that a comparison between ensembles of base pair probabilities more accurately predicts structural changes than a single point comparison between MFE structures. The implementation of this correlation method in corRna gives us an important analytical advantage over MultiRNAMute, which only uses the base pair or Hamming distance to quantify the “deleterious-ness” of a mutation (10).
Bootstrap significance
We use a bootstrap method to estimate the significance of a candidate sequence compared with a set of randomly generated sequences. Briefly, for each number of mutations k, we sample 1000 k-mutants of the input sequence uniformly. Then, we calculate the base pair correlation for each of these samples with the wildtype, and derive a distribution of correlation values for the whole set. Finally, corRna returns the percentile (between 0 and 1) of each candidate sequence by where it is ranked in this correlation distribution. A sequence with a significance value close to 0 would indicate that the candidate sequence has a low base pair correlation to wildtype that is significantly separated from a random sample of mutation sequences. It is worth noting that even if some rare random mutations may have a lower correlation value than RNAmutants samples, the latter have much more thermodynamically stable structures and thus provide better deleterious mutation candidates.
RESULTS
Comparison with mutagenesis experiments
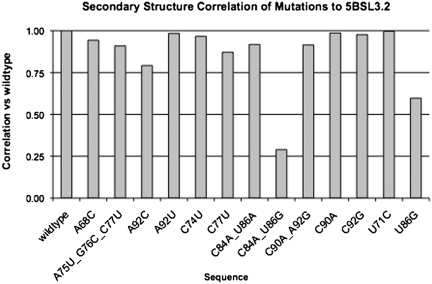
To validate the accuracy in which this correlation method can predict mutation-based structural changes, we used a benchmark of mutations used by You et al. (17) on the Hepatitis C virus cis-acting replication element (5BSL3.2). These mutations were analyzed with our correlation method. Our results, shown in Figure 4, found the C84A_U86G mutation to have the lowest correlation (0.290) with respect to wildtype. This result is consistent with the findings in Ref. (17), where the authors found that the most deleterious mutation was the C84A_U86G mutant and confirmed that the loss in viability was due to the disruption of the upper helix of the RNA secondary structure. This benchmark shows that the correlation calculation can accurately identify the most deleterious mutation among a set of candidate sequences that have been analyzed experimentally.
Figure 4.
Correlation values generated by corRna on the sequences used in the mutational analysis described in You et al. (17).
Predictive power
To test the predictive power of corRna, we compared the predictive performance of corRna against MultiRNAMute over a benchmark set of 30 sequences obtained from the Rfam database (18). Since the accuracy of our predictions is necessarily determined by the performance of the nearest neighbor energy model (21), we selected sequences on which the energy model performs well. This set was generated by first taking all the sequences in the Rfam database with a size <100 nt. Then, for each sequence we computed the MFE structure together with its probability in the Boltzmann low energy ensemble. RNA sequences were selected if their MFE was equal to that of the consensus structure. If two sequences belonged to the same family, the more stable one (the structure with the highest probability in the ensemble) was selected. The lengths of the selected benchmark set ranged from 19 to 98 nt. This benchmark set is freely available on our web site and we encourage any future research on mutational analysis to include this benchmark set as a comparison between different methods.
The sequences in the benchmark set were run with both MultiRNAMute and corRna. The parameters of MultiRNAMute, were set to: dist1 to 15, dist2 to 15, e range to 15, mutations to 3 and distance to ‘Hamming, method = Fast, stabilizing and destabilizing’. corRna was run using no heuristic, the structural heuristic and the mutation heuristic. We first predicted up to three-point mutations. However, to demonstrate the advantage offered by the efficient methods underlying corRna's algorithm, we ran these sequences to predict up to five-point mutations. These five-point mutations could not be run in a reasonable time frame with MultiRNAMute. Once the candidate sequences were generated, the correlation values were computed for each candidate mutation sequence. The number of candidates predicted (including duplicates), average correlation to wildtype (excluding duplicates) and best candidate (defined by the lowest correlation) were then averaged across all the 30 sequences. During any trial, if no sequences were predicted, the number of candidates was set to 0 and the trial was given an average and minimum correlation of 1. Average results over all sequences in the set are shown in Table 1. Detailed results are available on the web site.
Table 1.
Benchmark results of corRna methods versus MultiRNAMute
| Method | m | Avg. cand. | Avg. corr. | Min corr. |
|---|---|---|---|---|
| corRna - structural heuristic | 3 | 236 | 0.575 | 0.025 |
| corRna - mutation heuristic | 3 | 230 | 0.683 | 0.244 |
| corRna - no heuristic | 3 | 17 | 0.668 | 0.479 |
| corRna - structural heuristic | 5 | 243 | 0.425 | −0.098 |
| corRna - mutation heuristic | 5 | 246 | 0.570 | 0.011 |
| corRna - no heuristic | 5 | 21 | 0.551 | 0.312 |
| MultiRNAMute | 3 | 16982 | 0.366 | −0.007 |
Benchmark tests were based on a test set of 30 sequences pulled from the Rfam database. ‘m’ indicates the number of mutations allowed in the method. ‘Avg. cand’ indicates the average number of candidates presented for each test set including any duplicates. ‘Avg. corr.’ indicates the global correlation average of all sequences excluding any duplicates generated over all test sets of the method. ‘Min corr.’ indicates the average of each test set's minimum correlation candidate.
The ‘Avg. cand.’ column indicates the average number of candidates generated by each method over all benchmark sequences. MultiRNAMute generated a large and varied number of candidates with an average of 16 982 and a range of 0–258 240 sequences. In addition, MultiRNAMute failed to find any predictions for four of the sequences. The number of candidates generated by any corRna method was both smaller and less varied. When calculating up to three-point mutations (m = 3), corRna with no heuristic had an average of 17 candidates and a range of 2–23. The structure heuristic had an average of 236 and a range of 94–489. Finally, the mutation heuristic had an average of 230 candidates with a range of 199–238. Similar results were obtained when calculating up to five-point mutations (m = 5).
Compared to MultiRNAMute, the lower number of candidates returned by corRna presents some advantages. From a user standpoint, it provides a simpler set of candidate sequences for consideration in mutagenesis experiments.
The ‘Avg. corr.’ column indicates the average correlation of candidates given by each method over all sequences. At m = 3, the corRna structural, mutation and no heuristic methods obtained an average correlation of 0.575, 0.683 and 0.668, respectively. At m = 5, these values improved to 0.425, 0.570 and 0.551. The average correlation of MultiRNAMute was 0.366.
Finally, the ‘Min. corr’ column indicates the average of the most deleterious mutation found for each sequence by each method. At m = 3 the corRna structural, mutation and no heuristic methods obtained an average correlation of 0.025, 0.244 and 0.479, respectively. At m = 5, these values improved to −0.098, 0.011 and 0.312. The average minimum correlation of MultiRNAMute was −0.007.
These results indicate that both the structural and mutational heuristic improves the basic corRna method. Furthermore, the ability to search to higher k-point mutants improved the average correlation and min correlation. Overall, the structural heuristic performed better than the mutational heuristic. However, the performance of the mutational heuristic significantly improved when allowing up to five-point mutations. Indeed, there were some cases in the five-point mutation case where the mutation heuristic would find sequences with a markedly lower correlation than either the corRna structural heuristic or MultiRNAMute (data not shown).
When comparing the results from corRna and MultiRNAMute, MultiRNAMute provided a lower average correlation. However, corRna matched the average minimum correlations found when using the structural heuristic at m = 3 and when using either heuristic at m = 5. In addition, corRna managed to predict deleterious mutations even when MultiRNAMute failed to find any. Although corRna had a slightly higher average correlation of sequences predicted, corRna guaranteed results and predicted at a similar accuracy the more interesting mutations – those mutations that were most likely to be deleterious.
Running time
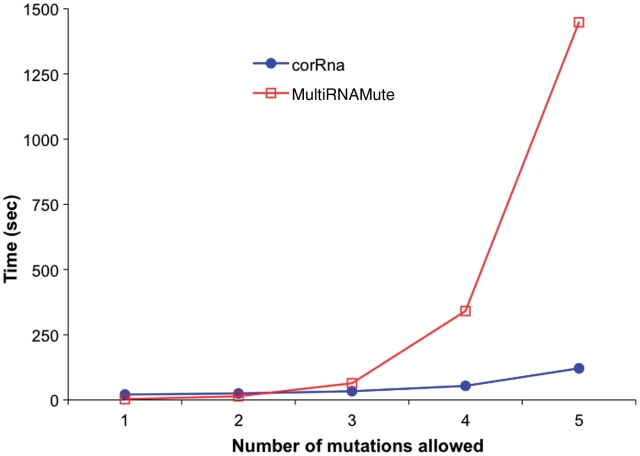
The efficient algorithm used in RNAmutants gives corRna a runtime advantage over other mutational analysis applications such as MultiRNAMute. A running time comparison between RNAmutants and MultiRNAMute conducted by Barash and Churkin (16) showed that RNAmutants has a better scaling factor that becomes advantageous when extending searches to four-point and five-point mutations. This advantage becomes especially important when implementing a web server which would be expected to give prompt results. To illustrate this point, we plot in Figure 5 the execution time of corRna and multiRNAmute on a sequence of size 40 used in Ref. (10) as a time benchmark. As expected, our results show that while the running time of multiRNAmute increases exponentially with the number of mutations allowed, corRna only requires an amount of time proportional to the square of the number of mutations. Here, this advantage becomes highly beneficial at mutation depth of 4. This phenomenon is amplified on longer sequences (data not shown).
Figure 5.
Running time comparison between corRna (in blue) and multiMultiRNAMute (in red) on a sequence of 40 nucleotides. The x-axis indicates the number of mutations allowed in the input sequence, and the y-axis gives the execution time in seconds.
CONCLUSION
In conclusion, corRna provides (and guarantees) a smaller candidate mutation set than MultiRNAMute, while still maintaining predictive power. More importantly, these results come with a significant reduction of the computational complexity, which allows corRna to extend the mutational analysis to larger numbers of k-point mutations. Finally, corRna also implements a correlation method which gives corRna an analytical advantage over MFE structure comparison methods used by MultiRNAMute.
corRna is the first web server that predicts multiple-point mutations and analyzes their deleterious nature using a correlation of structural changes compared with the wildtype. One of the interesting implications of corRna is that it is possible for corRna to predict mutations that would cause greater structural changes than any mutation found experimentally. These predictions are accessible through our web server (http://corrna.cs.mcgill.ca). We hope that corRna provides an avenue for new experimental research to test the deleterious nature of RNA mutations in vitro and in vivo.
FUNDING
Natural Sciences and Engineering Research Council of Canada discovery program. Funding for open access: Natural Sciences and Engineering Research Council of Canada Discovery Program discovery grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barash D. Deleterious mutation prediction in the secondary structure of RNAs. Nucleic Acids Res. 2003;31:6578–6584. doi: 10.1093/nar/gkg872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum. Mol. Genet. 2006 doi: 10.1093/hmg/ddl181. 15 Spec No 2, R162–169. [DOI] [PubMed] [Google Scholar]
- 4.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011;11:878–887. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 8.Shu W, Bo X, Liu R, Zhao D, Zheng Z, Wang S. RDMAS: a web server for RNA deleterious mutation analysis. BMC Bioinformatics. 2006;7:404. doi: 10.1186/1471-2105-7-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churkin A, Barash D. RNAmute: RNA secondary structure mutation analysis tool. BMC Bioinformatics. 2006;7:221. doi: 10.1186/1471-2105-7-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Churkin A, Barash D. An efficient method for the prediction of deleterious multiple-point mutations in the secondary structure of RNAs using suboptimal folding solutions. BMC Bioinformatics. 2008;9:222. doi: 10.1186/1471-2105-9-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofacker IL. RNA secondary structure analysis using the vienna RNA package. Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi1202s26. Chapter 12. Unit 12.2. [DOI] [PubMed] [Google Scholar]
- 12.Waldispühl J, Behzadi B, Steyaert J-M. An approximate matching algorithm for finding (sub-)optimal sequences in S-attributed grammars. Bioinformatics. 2002;18(Suppl 2):S250–S259. doi: 10.1093/bioinformatics/18.suppl_2.s250. [DOI] [PubMed] [Google Scholar]
- 13.Clote P, Waldispühl J, Behzadi B, Steyaert J-M. Energy landscape of k-point mutants of an RNA molecule. Bioinformatics. 2005;21:4140–4147. doi: 10.1093/bioinformatics/bti669. [DOI] [PubMed] [Google Scholar]
- 14.Waldispühl J, Devadas S, Berger B, Clote P. Efficient algorithms for probing the RNA mutation landscape. PLoS Comput. Biol. 2008;4:e1000124. doi: 10.1371/journal.pcbi.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waldispühl J, Devadas S, Berger B, Clote P. RNAmutants: a web server to explore the mutational landscape of RNA secondary structures. Nucleic Acids Res. 2009;37:W281–W286. doi: 10.1093/nar/gkp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barash D, Churkin A. Mutational analysis in RNAs: comparing programs for RNA deleterious mutation prediction. Brief. Bioinformatics. 2010;12:104–114. doi: 10.1093/bib/bbq059. [DOI] [PubMed] [Google Scholar]
- 17.You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 2004;78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldispühl J, Ponty Y. In Proceedings of the 15th Annual International Conference on Research in Computational Molecular Biology (RECOMB 2011), Lecture Notes in Computer Science, Vol. 6577/2011, 501-515, 2011. Heidelberg: Springer Berlin; An unbiased adaptive sampling algorithm for the exploration of RNA mutational landscapes under evolutionary pressure. [DOI] [PubMed] [Google Scholar]
- 20.Darty K, Denise A, Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner DH, Mathews DH. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010;38:D280–D282. doi: 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]