Abstract
Cellular response to stimuli is typically complex and involves both regulatory and metabolic processes. Large-scale experimental efforts to identify components of these processes often comprise of genetic screening and transcriptomic profiling assays. We previously established that in yeast genetic screens tend to identify response regulators, while transcriptomic profiling assays tend to identify components of metabolic processes. ResponseNet is a network-optimization approach that integrates the results from these assays with data of known molecular interactions. Specifically, ResponseNet identifies a high-probability sub-network, composed of signaling and regulatory molecular interaction paths, through which putative response regulators may lead to the measured transcriptomic changes. Computationally, this is achieved by formulating a minimum-cost flow optimization problem and solving it efficiently using linear programming tools. The ResponseNet web server offers a simple interface for applying ResponseNet. Users can upload weighted lists of proteins and genes and obtain a sparse, weighted, molecular interaction sub-network connecting their data. The predicted sub-network and its gene ontology enrichment analysis are presented graphically or as text. Consequently, the ResponseNet web server enables researchers that were previously limited to separate analysis of their distinct, large-scale experiments, to meaningfully integrate their data and substantially expand their understanding of the underlying cellular response. ResponseNet is available at http://bioinfo.bgu.ac.il/respnet.
INTRODUCTION
Cells live in a dynamic environment in which they encounter environmental changes, toxins and mutations. Cellular response to these stimuli is typically complex and involves regulatory, proteomic and metabolic changes. Identification and comprehensive understanding of these changes is very important: it may reveal the mechanism of the stimulus, such as the mode of action of a potent drug [e.g. (1)], and identify proteins and processes that can be used to manipulate the cellular response [e.g. (2,3)], potentially opening new avenues for therapeutic intervention (4).
In an effort to accomplish these goals, large-scale genetic screens and transcriptomic profiling assays are applied extensively in current research [e.g. (5–7)]. Genetic screens identify proteins that when mutated significantly alter the phenotype of stimulated cells. In the past few years, genetic screens using gene inactivation or overexpression libraries identified scores of proteins whose association with specific stimuli were previously unknown [e.g. (6)]. However, while providing strong evidence that these proteins were functionally related to the underlying cellular processes, these relationships were often indirect and hard to decipher, leaving many of the identified associations unexplained. A different approach to illuminate the cellular response is provided by transcriptomic profiling assays. Using microarrays or RNA-Seq technologies, these assays reveal genes whose expression levels are significantly altered in stimulated cells. Transcriptomic profiling assays were extensively applied to identify the processes that are activated or inhibited in response to stimuli [e.g. (5,8,9)], yet, similarly to genetic screens, the series of events that led to the transcriptomic change often remained hidden.
It had been previously noted in few instances that in the yeast Saccharomices cerevisiae, genetic screens do not identify the same genes as transcriptomic profiling assays conducted in the same conditions [e.g. (10–15)]. By systematically comparing the results of the two types of screens in over 150 distinct conditions, we recently showed that this discrepancy is, in fact, a general rule (16). Importantly, we found that genetic screens tend to identify response regulators, whereas transcriptomic profiling assays tend to identify metabolic responses.
In order to make the most of the distinct data provided by genomic screens and transcriptomic profiling assays, we developed ResponseNet (16). ResponseNet is an integrative approach that uses known molecular interactions to bridge the gap between these data and reveal a broader image of the cellular response. Specifically, ResponseNet identifies signaling and regulatory pathways, by which the proteins detected by genetic screens may lead to the measured transcriptomic response (Figure 1). The novelty of ResponseNet lies in its formulation as a minimum-cost flow optimization problem that is solved efficiently using linear programming solvers (17). We applied ResponseNet to reveal cellular pathways that respond to the toxicity of α-synuclein, a small human protein implicated in several neurodegenerative disorders including Parkinson’s disease (16). We conducted a genetic screen of a well-established yeast model for α-synuclein toxicity and identified 77 proteins that when overexpressed altered α-synuclein toxicity. We also profiled the transcritpome of the yeast model and identified 612 differentially expressed genes (16,18). We then used ResponseNet to search the yeast molecular interaction network (interactome), which related over 11 000 proteins and genes via over 57 000 molecular interactions, for a high-probability sub-network connecting the identified proteins and genes. ResponseNet successfully mapped previously unknown as well as recognized pathways responding to α-synuclein toxicity. We experimentally validated four de novo predictions suggested by ResponseNet analysis, including the presence of nitrosative stress, the involvement of the TOR pathway, the disturbance of the sterol biosynthesis pathway and the mode-of-action of the genetic suppressor Gip2 in the response to α-synuclein toxicity (16).
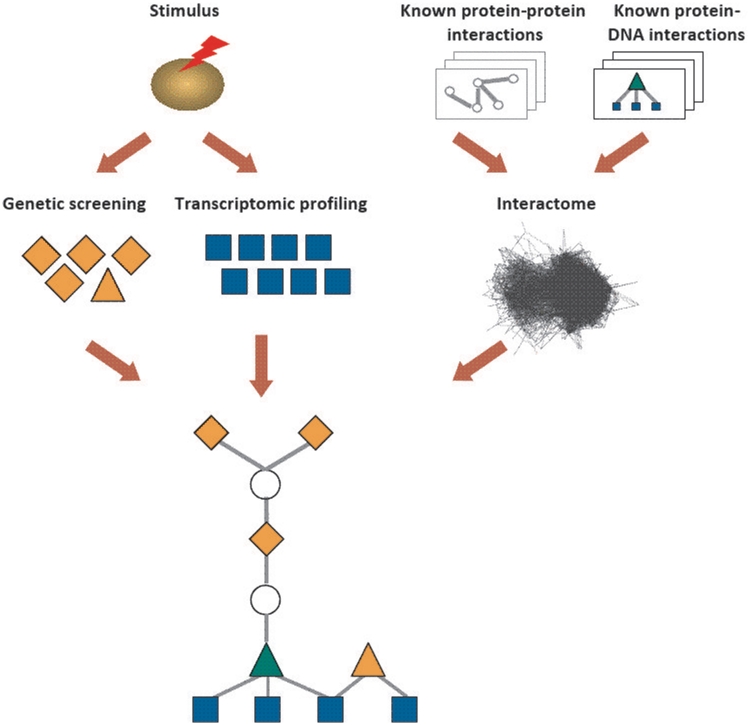
Figure 1.
The signaling and regulatory sub-network, by which stimulus-related proteins detected by genetic screens may lead to the measured transcriptomic response. ResponseNet integrates the identified stimulus-related proteins and genes with known molecular interactions to find molecular interaction paths, through which a subset of the proteins may regulate the transcription of a subset of the genes. The regulation may be direct, e.g. when the stimulus-related protein is the transcriptional regulator of a stimulus-related gene, or indirect via intermediate proteins and transcriptional regulators. Stimulus-related proteins appear as orange nodes; stimulus-related genes appear as blue nodes; intermediary proteins appear as white nodes. Known protein–protein and protein–DNA interactions appear as gray edges. Transcriptional regulators appear as triangles.
In recent years, several other methodologies for identifying interactome pathways that bridge the gap between potential causal proteins and affected genes have been developed [e.g. (19–25)]. These methodologies were based on a variety of computational techniques including maximum likelihood (19), integer programming (21), Steiner trees (23,24), electric circuits (22,26) and Bayesian networks (2,27). Notably, many of these techniques were computationally intensive and therefore required the use of approximation schemes [e.g. (24)] or arbitrary constraints over the topology of the output network [e.g. (21,23)]. Unfortunately, this rich set of sophisticated techniques is not readily available for use. In fact, only in few cases downloadable code was provided (21,27).
Here, we present the ResponseNet web server for predicting the signaling and regulatory network connecting stimulus-related proteins and genes. This web server accepts lists of proteins and genes from the user, and then searches within a given interactome for a high-probability sub-network connecting these proteins and genes. The web server presents the identified signaling and regulatory sub-network as a graphical network image in Cytoscape format (28). It also reports the gene ontology (GO) enrichment of the connected input lists and of the output sub-network, further illuminating the underlying cellular response to the stimulus. By default, ResponseNet analysis is carried in yeast using a recently complied version of the yeast interactome that is stored in our database. However, users may upload their own interactome data, opening the way to apply ResponseNet to analyze cellular responses in other organisms, including human. The ResponseNet web server is free and open to all users with no login requirement.
ResponseNet METHODOLOGY
Prediction of signaling and regulatory response pathways in the yeast S. cerevisiae is extremely challenging. For only a handful of stimuli, like the mating pheromone or high osmolarity, these pathways were fully characterized, and efforts to map the yeast signaling network are yet ongoing (29–31). Moreover, due to the vast number of known interactions, a search for all interaction paths connecting stimulus-related proteins to genes typically results in a ‘hairball’ sub-network that is very hard to interpret. To overcome these limitations and identify meaningful signaling and regulatory sub-networks, ResponseNet is designed as a network-optimization approach that uses a graphical model in which: (i) proteins and genes are represented as separate network nodes; (ii) a directed edge leads from a protein node to a gene node only if they correspond to a transcription factor and its target gene; and (iii) each network edge is associated with a probability that reflects its likelihood (16).
We implemented ResponseNet as a ‘flow algorithm’, a well-known computational approach for efficiently finding connectivity in graphs (17). Flow algorithms deliver an abstract flow from a source node (S) to a sink node (T) through the edges of a network, which, similarly to pipes, are associated with a capacity that limits the flow and with a cost. (As a loose analogy, this resembles water finding the path of least resistance through a complex landscape.) Because S and T are the two endpoints for the flow, by linking S to the stimulus-related proteins and the stimulus-related genes to T, the flow is forced to find paths that connect the stimulus-related proteins and genes through protein–protein and protein–DNA interactions. To give preference to connecting paths of high probability, we formulated ResponseNet as a minimum-cost flow optimization problem (17). This formulation aims to maximize the flow between S and T while minimizing the cost of the connecting paths. Hence, by setting the cost of an edge to the negative log of its probability, a sparse, high-probability connecting sub-network is obtained.
Importantly, the minimum-cost flow problem is solved efficiently using linear programming tools [LOQO (32)]. A typical optimal solution connects a subset of the stimulus-related proteins to a subset of the stimulus-related genes through known interactions and intermediary proteins; these interactions and proteins are weighted by the amount of flow they pass, thus illuminating core versus peripheral components of the response. A detailed description of ResponseNet formulation is given in Yeger-Lotem et al. (16).
As described in Yeger-Lotem et al. (16), we tested ResponseNet on the well-characterized pheromone and DNA damage responses by applying it to proteins and genes identified by genetic and transcriptomic profiling assays, respectively. In both cases, ResponseNet revealed a connecting sub-network that was highly enriched for the relevant response pathway and exposed core pathway members. To thoroughly test ResponseNet despite the lack of known response pathways, we applied it to a set of 101 distinct, less-characterized stimuli, which corresponded to gene deletions. For each stimulus, we created a randomized solution set composed of 100 sub-networks that were obtained by applying ResponseNet to partially randomized inputs (either the stimulus-related proteins or the genes were randomly chosen, while all other inputs were fixed). We considered as success solutions that were significantly enriched for the stimulus, or ranked stimulus-related proteins significantly high, relative to the randomized solution set (P ≤ 0.05). Although in 86 of the 101 test cases screening data were very limited, ResponseNet displayed 41% sensitivity, 98% specificity and 30% precision. Moreover, when applied to α-synuclein toxicity screening data ResponseNet successfully revealed meaningful response pathways that we validated experimentally (16).
ResponseNet INPUT AND OUTPUT
The ResponseNet web server is designed for predicting stimulus-related signaling and regulatory pathways in the yeast S. cerevisiae. The basic input to ResponseNet includes a source set comprising a list of stimulus-related proteins, preferably identified through genetic screening, and a target set comprising a list of stimulus-related differentially expressed genes. An advanced form of input specifies each set as a weighted list, with weights being positive real numbers. Higher weights increase the probability that the corresponding proteins or genes will appear in the output network, and may reflect, e.g. the strength of the effect of the mutated protein or the fold-change in the gene’s expression level. If no weights are given, they are considered to be uniform. Upon submitting the data to the web server the user may choose to perform a GO enrichment analysis of the input sets and the output sub-network, to identify processes that are significantly associated with the cellular response. This analysis is carried using the GOrilla tool (33).
The ResponseNet web server also offers three advanced features. By default, given the sets of proteins and genes, ResponseNet searches an up-to-date version of the yeast interactome that we compiled as described in Yeger-Lotem et al. (16). Using the advanced options the user can append or replace the interactome data: user-provided protein–protein interactions are considered bidirectional, unless specified otherwise by the user; user-provided protein–DNA interactions are directed from a protein to its target gene. Notably, by replacing the interactome data the user can apply the server to data of other organisms. The two other advanced features relate to the free parameters of the minimum-cost flow formulation: The capping value is an upper bound on the probability of each interaction, and therefore a lower bound on its cost; lowering the capping value reduces the bias toward extensively studied interactions and may lead to sparser solutions. The gamma value controls the connectivity of the solution, with increasing values enabling identification of lower probability connecting sub-networks. More information about the input options can be found in the ResponseNet tutorial that is stored on the server.
The computation of the minimum-cost flow optimization problem typically takes a few minutes due to the need to solve a linear programming problem with thousands of variables. Once complete, the output is provided as a multi-panel window (Figure 2). The middle panel contains a graphical image of the identified signaling and regulatory sub-network that connects the input sets. This panel utilizes a Cytoscape plug-in that enables the user to manipulate the visualization of the sub-network (34). The left panel provides various statistics, such as the fraction of input proteins and genes that were actually connected by ResponseNet, and, if requested by the user, a link to the GO enrichment analysis of the input sets and the output sub-network. The right panel provides a link to a text version of the identified sub-network in a Cytoscape table format.
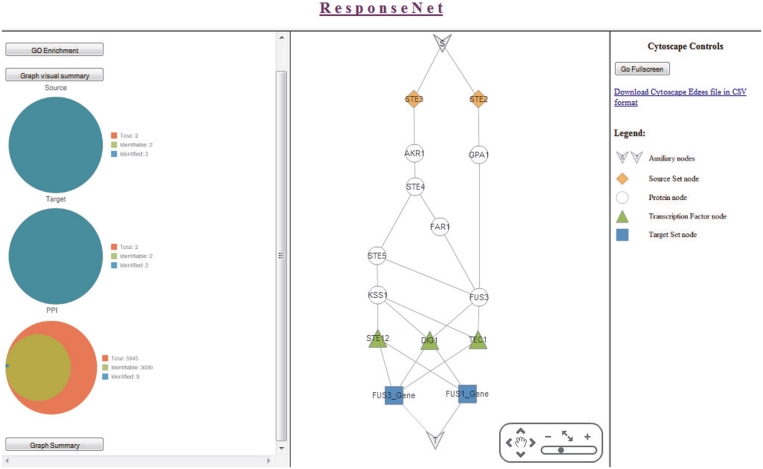
Figure 2.
The ResponseNet web server output. In this example the ResponseNet web server was applied to a protein set containing Ste2 and Ste3 and to a gene set containing Fus1 and Fus3 using default parameters. The middle panel presents the regulatory and signaling output sub-network identified by ResponseNet. The left panel presents a visual summary of the coverage of the output sub-network, as well as a link to the GO enrichment analysis of the connected input sets and the output sub-network. The right panel presents a link to a text version of the output sub-network in a Cytoscape-compatible format, and a legend for the sub-network appearing in the middle panel.
An example of ResponseNet output is shown in Figure 2. In this example, we used ResponseNet to identify the signaling and regulatory sub-network connecting the two pheromone receptor proteins, Ste2 and Ste3, to the genes Fus1 and Fus3 that are up-regulated during the pheromone response. As shown in the middle panel of Figure 2, ResponseNet indeed identified core components of the pheromone response signaling cascade. The connecting sub-network was most significantly enriched for ‘pheromone-dependent signal transduction involved in conjugation with cellular fusion’ (P-value of 9.72E-13). Notably, ResponseNet is applicable to much larger protein and gene input sets as detailed in (16).
DISCUSSION
ResponseNet is a unique web server that is designed to identify the signaling and regulatory sub-networks that connect stimulus-related proteins and genes. These stimulus-related data typically result from large-scale genetic screens and transcriptomic profiling assays. While each of these assays provides unprecedented genome-wide views into the cellular response, the function of the identified proteins in the response to the stimulus, and the signaling paths leading to transcriptomic changes, often remain hidden. By integrating these data with data of known molecular interactions, ResponseNet simultaneously illuminates the mode-of-action of some of the proteins and the signaling cascades that lead to the measured transcriptomic response.
The ResponseNet algorithm is based on the minimum-cost flow optimization problem. This formulation is advantageous over other approaches for finding explanatory paths in molecular interaction networks, since it gives preference to high-confidence interactions, allows for a simultaneous solution for the input proteins and genes, puts no a priori bounds on the structure of the output sub-network, and is solved efficiently using linear programming tools. These features of ResponseNet, together with its verified success (16) substantiate the minimum-cost flow optimization as a powerful framework to illuminate cellular pathways from multidimensional, large-scale experimental data. Importantly, it is one of the few approaches that are freely available on the web and can be readily applied by users. While being designed for integrating the results of genetic screens and transcriptomic profiling assays in yeast, ResponseNet is a general framework for identifying explanatory paths underlying other types of input. For example, it can readily be used to analyze phosphoproteomic assays (23), to reveal the paths underlying eQTLs associations (22) or to identify disease-related explanatory paths in other organisms, including human (27). The ease of use of the ResponseNet web server and its enhanced functionality make it suitable for illuminating the pathways underlying a wide-range of cellular states in a variety of organisms. We believe that many researchers that are limited today to separate analysis of their large-scale experiments can take full advantage of their valuable data by analyzing them using this unique integrative web server.
FUNDING
European Union Seventh Programme under the FP7-PEOPLE-MCA-IRG Funding scheme, grant agreement n°[256360 to E.Y.-L.]; and the United States-Israel Binational Science Foundation (BSF) [2009323 to E.Y.-L. and S.L.]. Funding for open access charge: United States-Israel Binational Science Foundation (BSF) [2009323 to E.Y.-L. and S.L.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Vered Chalifa-Caspi, Daniel Dovrat and Ruth Barshir for helpful comments on the ResponseNet web server and manuscript.
REFERENCES
- 1.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 2.Tu Z, Argmann C, Wong KK, Mitnaul LJ, Edwards S, Sach IC, Zhu J, Schadt EE. Integrating siRNA and protein-protein interaction data to identify an expanded insulin signaling network. Genome Res. 2009;19:1057–1067. doi: 10.1101/gr.087890.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat. Rev. Drug Discov. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- 5.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 6.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Wageningen S, Kemmeren P, Lijnzaad P, Margaritis T, Benschop JJ, de Castro IJ, van Leenen D, Groot Koerkamp MJ, Ko CW, Miles AJ, et al. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell. 2010;143:991–1004. doi: 10.1016/j.cell.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 11.Deutschbauer AM, Williams RM, Chu AM, Davis RW. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davis RW, Brown JM. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl Acad. Sci. USA. 2002;99:8778–8783. doi: 10.1073/pnas.132275199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haugen AC, Kelley R, Collins JB, Tucker CJ, Deng C, Afshari CA, Brown JM, Ideker T, Van Houten B. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 2004;5:R95. doi: 10.1186/gb-2004-5-12-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry RC, Begley TJ, Samson LD. Genome-wide responses to DNA-damaging agents. Annu. Rev. Microbiol. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- 15.Smith JJ, Sydorskyy Y, Marelli M, Hwang D, Bolouri H, Rachubinski RA, Aitchison JD. Expression and functional profiling reveal distinct gene classes involved in fatty acid metabolism. Mol. Syst. Biol. 2006;2:2006 0009. doi: 10.1038/msb4100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat. Genet. 2009;41:316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cormen TH, Leiserson CE, Rivest RL, Stein C. Introduction to Algorithms. Cambridge, MA: The MIT Press; 2001. [Google Scholar]
- 18.Su LJ, Auluck PK, Outeiro TF, Yeger-Lotem E, Kritzer JA, Tardiff DF, Strathearn KE, Liu F, Cao S, Hamamichi S, et al. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson’s disease models. Dis. Models Mech. 2010;3:194–208. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeang CH, Ideker T, Jaakkola T. Physical network models. J. Comput. Biol. 2004;11:243–262. doi: 10.1089/1066527041410382. [DOI] [PubMed] [Google Scholar]
- 20.Tu Z, Wang L, Arbeitman MN, Chen T, Sun F. An integrative approach for causal gene identification and gene regulatory pathway inference. Bioinformatics. 2006;22:e489–e496. doi: 10.1093/bioinformatics/btl234. [DOI] [PubMed] [Google Scholar]
- 21.Ourfali O, Shlomi T, Ideker T, Ruppin E, Sharan R. SPINE: a framework for signaling-regulatory pathway inference from cause-effect experiments. Bioinformatics. 2007;23:i359–i366. doi: 10.1093/bioinformatics/btm170. [DOI] [PubMed] [Google Scholar]
- 22.Suthram S, Beyer A, Karp RM, Eldar Y, Ideker T. eQED: an efficient method for interpreting eQTL associations using protein networks. Mol. Syst. Biol. 2008;4:162. doi: 10.1038/msb.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang SS, Fraenkel E. Integrating proteomic, transcriptional and interactome data reveals hidden components of signaling and regulatory networks. Sci. Signal. 2009;2:ra40. doi: 10.1126/scisignal.2000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yosef N, Ungar L, Zalckvar E, Kimchi A, Kupiec M, Ruppin E, Sharan R. Toward accurate reconstruction of functional protein networks. Mol. Syst. Biol. 2009;5:248. doi: 10.1038/msb.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Missiuro PV, Liu K, Zou L, Ross BC, Zhao G, Liu JS, Ge H. Information flow analysis of interactome networks. PLoS Comput. Biol. 2009;5:e1000350. doi: 10.1371/journal.pcbi.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanderbei RJ. LOQO User’s Manual–Version 3.10. Optimization Methods and Software. 1999;12:485–514. [Google Scholar]
- 33.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]