Abstract
AnnotQTL is a web tool designed to aggregate functional annotations from different prominent web sites by minimizing the redundancy of information. Although thousands of QTL regions have been identified in livestock species, most of them are large and contain many genes. This tool was therefore designed to assist the characterization of genes in a QTL interval region as a step towards selecting the best candidate genes. It localizes the gene to a specific region (using NCBI and Ensembl data) and adds the functional annotations available from other databases (Gene Ontology, Mammalian Phenotype, HGNC and Pubmed). Both human genome and mouse genome can be aligned with the studied region to detect synteny and segment conservation, which is useful for running inter-species comparisons of QTL locations. Finally, custom marker lists can be included in the results display to select the genes that are closest to your most significant markers. We use examples to demonstrate that in just a couple of hours, AnnotQTL is able to identify all the genes located in regions identified by a full genome scan, with some highlighted based on both location and function, thus considerably increasing the chances of finding good candidate genes. AnnotQTL is available at http://annotqtl.genouest.org.
INTRODUCTION
The final steps of genetic mapping research programs require close analysis of several QTL regions to select candidate genes for further studies. Despite several websites (NCBI genome browser, Ensembl Browser, UCSC Genome Browser) or web tools (Biomart, Galaxy) developed to achieve this task, the selection of candidate genes remains a laborious process. The information made available on the more prominent web sites differs slightly in terms of gene prediction and functional annotation, while other websites provide extra information that researchers may want to use (HGNC approved gene symbols, Gene Ontology (GO) Annotation or functional data, conservation of synteny with other species, etc.). It is possible to manually merge and compare this information for one QTL containing few genes, but not for many different QTL regions containing dozens of genes.
Here, we propose a web tool that, for a given region of interest, merges the list of genes available in NCBI and Ensembl, removes redundancy, adds functional annotations from different prominent web sites, and highlights the genes for which functional annotation fits the biological function or diseases of interest. The tool is dedicated to sequenced species of livestock including cattle, pig, chicken and horse as well as dog, i.e. species that have been extensively studied (with over 8000 QTLs detected; see http://www.animalgenome.org/cgi-bin/QTLdb/index). Nevertheless, because of the family designs and the low number of animals used in these species, most of the studies use linkage analysis, and the QTL regions identified remain large (containing dozens of genes). Conversely, in human and model species, most analyses now draw heavily on association studies involving large cohorts, thus providing more power and accuracy, and the web tools already available focus on these species through functional annotation of SNPs in association with the trait (1–8). Most of these tools focus on the SNP annotation itself, describing whether the SNP is located in a gene, or even in a coding sequence, and defining if it could have a functional effect. While these web tools are highly efficient in providing a good annotation for specific SNPs, they clearly cannot be used to collect information on the large regions obtained in livestock species.
METHODS
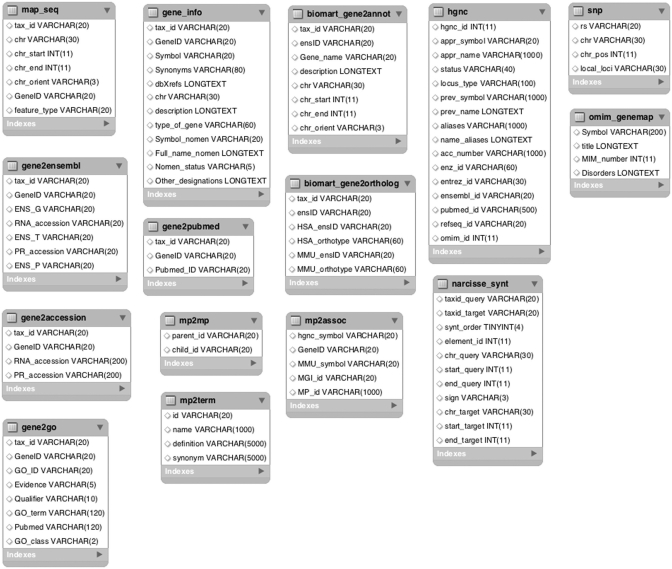
The main objective of AnnotQTL is to minimize redundancy so as to display the maximum amount of information from several sources on the genes in the region of interest. The main AnnotQTL program is implemented in PERL. The data are downloaded from several FTPs or websites (see Figure 1 for details on the data and fields used) and stored on our server for further computation. Location and annotation data from Ensembl are downloaded via BioMart (9) using MartService. AnnotQTL gives several sets of information from comparative mapping of selected species against the human and mouse genomes using a local dump of the data provided by the Narcisse web site (10) and orthologous gene information from the Ensembl comparative database. PERL scripts import the downloaded files into our SQL databases. All PERL scripts and official GO database updates are inserted into a BioMaj (11) workflow to automate the updating process. Updates are performed monthly.
Figure 1.
Schematic diagram of the database and source data files. The table map_seq is filled using file xxx_seq_gene.md.gz, where xxx is the species name, located in the NCBI FTP directory: /genomes/xxx/mapview. The tables gene_info, gene2accession, gene2go, gene2ensembl and gene2pubmed are filled using data files stored in the NCBI FTP directory: /gene/DATA. The table biomart_xxx is filled using the BioMart service for the Ensembl databases. For each species, a SQL table is created to store SNP data (here, only one is detailed). The data files are downloaded from NCBI FTP directory: /snp/organisms/xxx/chr_rpts (where xxx is species name). The tables mp2mp, mp2term and mp2assoc are filled using files HMD_HumanPhenotype.rpt and MPheno_OBO.ontology available from the MGI FTP site (ftp.informatics.jax.org/pub/reports). The hgnc table is filled using the data stored at the GeneName website together with its LWP agent (http://www.genenames.org/cgi-bin/hgnc_downloads.cgi). The table omim_genemap is filled using the data file located in the NCBI FTP directory /repository/OMIM. The table narcisse_synt is filled using the comparative data provided by the Narcisse website (http://narcisse.toulouse.inra.fr).
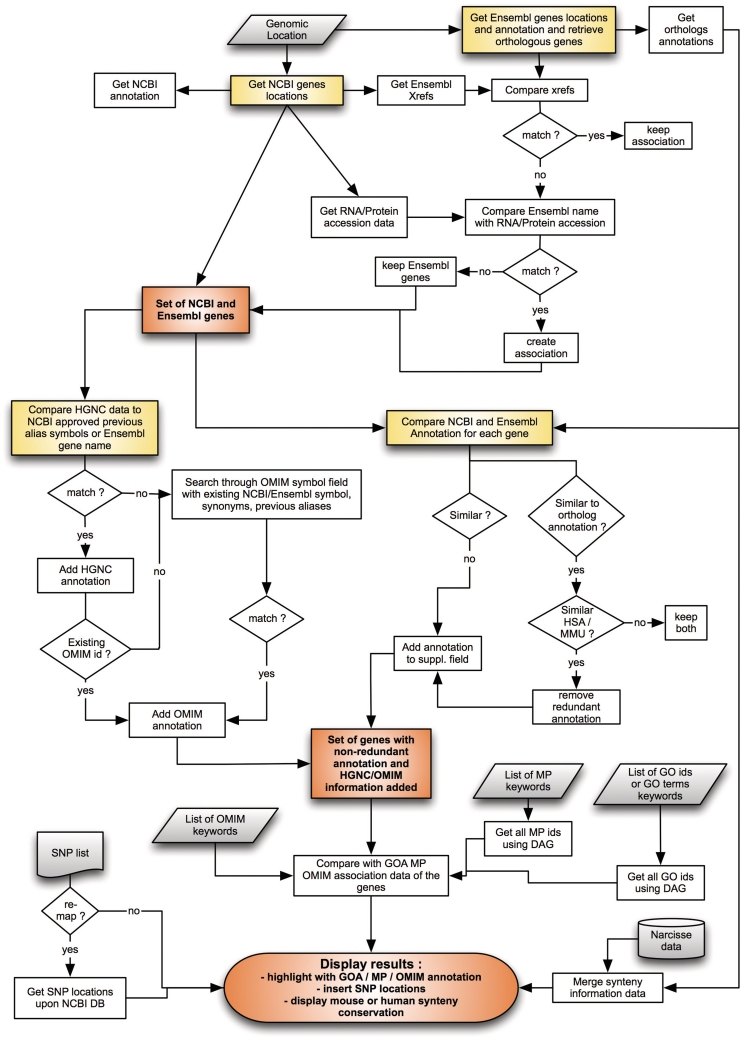
The principle and workflow of AnnotQTL is depicted in Figure 2. Starting with the genome coordinates entered by the user, the program extracts the NCBI GeneID of genes contained in the region, the corresponding annotations (name, description, symbol and so on), plus the associated cross-references (RNA accession number, protein accession number and Ensembl identifier) and the Pubmed identifier. This Pubmed identifier is specific of the requested species and does not list the publications related to this gene in other species. Using the same genome coordinates, the program then extracts Ensembl ID, gene annotation and human and mouse ortholog gene identifiers from the Ensembl database.
Figure 2.
AnnotQTL—principle and workflow. Boxes shaded in gray represent user input or database (i.e. Narcisse) input. Boxes shaded in yellow show the main processes in the AnnotQTL workflow. Boxes shaded in orange represent intermediate results. MP: Mammalian Phenotype.
The main step now is to remove the redundancy between NCBI and Ensembl data while keeping the specific annotation of both databases. As there is a slight difference in gene location between the two web sites, the filtering process cannot be based on gene location, which leaves two approach options. The first is to use the Ensembl cross-reference provided by NCBI. However, this approach is not exhaustive since few cross-references are missing even for genes annotated in both databases. A second strategy has therefore been developed based on a textual query search in the annotation fields provided by the two sites. Values for the symbol, synonyms, RNA accession and protein accession fields from NCBI are compared against the values in the Ensembl gene name field for the gene of the species of interest. When one or more of these fields match, all the information is combined under one record, thus removing duplicates and enriching the annotation (without losing the annotation specific to both sites). If available, the gene annotations of human and mouse orthologs are also included in this comparison. Each record is also filtered for potential intra-redundancy of annotation between the gene and its orthologs (i.e. the same gene description is found between the requested species and Human or mouse orthologs). This set of genes combining NCBI- and Ensembl-specific information is then compared to the HGNC database. The goal of this procedure is to retrieve the HGNC approved symbol by searching for correspondences between annotation fields and the symbol or aliases fields of the HGNC database. Then, the values found in the HGNC database (symbol and OMIM identifier, if any) are included in the final results output displayed. If the OMIM identifier is still undefined, a search through the OMIM symbol fields is performed using the HGNC symbol or aliases. Where relevant, OMIM identifier, title and related disorders are retrieved from the OMIM database. Finally, the user-specified genome region is cross-compared against the Narcisse database to fetch the human or mouse-orthologous genomic region.
To clarify the output and adapt it to the scientist's query, certain information is only available through menu options. Human and mouse orthology information from Ensembl can be used to more accurately define certain genomic regions left undefined in Narcisse data. Users can also select level of synteny (synt order, see (10) for more details) between studied species and target species (human or mouse). Another option is to upload a set of genetic markers (which can be of any type provided physical location is given) to be inserted in the final results display. User can choose to keep their own marker locations or re-map markers to NCBI genome coordinates (only available with approved marker identifiers). A fourth non-processed column is available for displaying user-defined information. Adding the markers to the results display should ease the identification of the genes that most closely match the most significant markers. Finally, AnnotQTL can highlight genes based on functional annotations provided by GO, Mammalian Phenotype (MP), or OMIM disorders. For GO or MP terms organized in a hierarchical ‘parent–children’ directory structure, user-inputted keywords provide options for selecting the corresponding terms and associated children. For OMIM, a query is performed against OMIM disorders data retrieved in the previous step with user-input keywords: if the keywords matched, then the OMIM disorders are highlighted in the display. For GO, the genes are highlighted if their GeneID matches with the GO association provided by NCBI. As they do not have a GeneID, the match-up between GO annotation and genes specific to the Ensembl database is based on their HGNC name, where available. Users can improve this ‘GOA highlight function’ by adding the GOA from human and/or mouse species from orthologs to current genes. For MP, genes are highlighted if their approved symbols match the HGNC approved symbols stored in the MP database. The aim here is to provide functional information and facilitate the identification of genes linked to the trait-of-interest (i.e. functional candidate genes).
APPLICATION
To demonstrate the utility of AnnotQTL and test the efficiency of this web tool, we present different examples using real data aimed at identifying functional and positional candidate genes.
The first example focuses on a bovine mutation controlling muscular hypertrophy. In 1995, the mutation was mapped to the extremity of the BTA2 in a 12 cM interval (12). Using AnnotQTL on this region of the bovine chromosome (0–8 Mb) retrieved the location and functional annotation of 95 genes. We then applied the ‘GO highlight function’ on this region in two separate queries, using ‘muscle’ and ‘growth’ as keywords best describing the observed phenotype. These two terms highlighted two and three genes, respectively, from these 95 genes. Both lists highlighted the MSTN (GDF8) gene, which has been demonstrated as the validated causal gene (myostatin) (13).
A second step analyzed a more extensive set of 21 QTL regions shaping abdominal fatness in chickens (14,15). Average length of these regions was 4.8 Mb. After running AnnotQTL, all the regions were enriched with genes by comparing NCBI and Ensembl information against information provided by either NCBI or Ensembl only (Table 1). For all the genomic regions, working from an initial set of 1734 genes from the NCBI database and 1902 genes from the Ensembl database, AnnotQTL retrieved a non-redundant set of 2220 genes. On this large dataset, we applied the ‘highlight function’ on each region to underline genes whose functional annotation was related to the studied phenotype. Among the 2220 genes located in these 21 QTL regions, 127 were highlighted using the GO term, ‘lipid’ and the MP term ‘adipose’ as keywords, with an average 5.4 genes highlighted per region.
Table 1.
Statistics of the QTL/eQTL regions analyzed using AnnotQTL
| Number of regions | Regions mean size (Mb) | NCBI genes | Ensembl genes | AnnotQTL genes obtained merging NCBI and Ensembl | GO and MP terms screening results |
||
|---|---|---|---|---|---|---|---|
| Genes found | Average of genes found per region | ||||||
| QTL | 21 | 4.8 | 1734 | 1902 | 2220 | 127 | 5.8 |
| eQTL | 25 | 3.4 | 1198 | 1283 | 1506 | 93 | 3.7 |
Finally, AnnotQTL can also be exploited to look at eQTL regions. Strategies combining transcriptomics and genotyping data have recently been developed to better characterize QTL regions for traits of interest by identifying co-localized eQTLs and QTLs (16–21). Whatever the context, this strategy identifies a much higher number of eQTL regions than in QTL studies, thus creating a need for tools that can efficiently find positional and functional candidate genes. Here, we focus on 25 chicken eQTL regions affecting 70 genes involved in lipid metabolism (i.e. sharing the GO term GO:0006629 ‘lipid metabolic process’). Average length of these regions is 3.4 Mb. Running AnnotQTL found similar results to those obtained for the QTL regions. All the regions were enriched with genes by comparing NCBI and Ensembl information against information provided by either NCBI or Ensembl only (Table 1): working from an initial set of 1,198 genes from the NCBI database and 1283 genes from the Ensembl database, AnnotQTL retrieved a non-redundant set of 1506 genes. Again, in order to select possible candidate genes, we used the ‘highlight function’ to pinpoint the genes related to the studied phenotype. Among these 1506 genes, and using the same GO term ‘lipid’ and MP term ‘adipose’ as keywords, a total of 93 genes were identified, with an average 3.7 genes highlighted per region.
These examples corresponding to two different contexts (QTL and eQTL analyses) clearly demonstrate how in just a couple of hours, AnnotQTL can accurately analyze the gene content of numerous regions identified by a full genome scan and go on to highlight some of these genes based on both their location and function, whereas in the same time period, a manually run procedure would only have been able to analyze one single region.
CONCLUSION
AnnotQTL is a web tool designed to gather the functional annotation of different prominent web sites while minimizing redundant information. Using all known information substantially accelerates the gene analysis of QTL regions for livestock species traits and improves the selection of candidate genes.
FUNDING
INRA, Agrocampus Ouest, the Regional Council of Brittany; French Ministry in charge of Agriculture (DGER). Funding for open access charge: INRA.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank A.T.T. scientific editing services for proofreading the article and all the beta testers for their help on debugging AnnotQTL.
REFERENCES
- 1.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166–166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodswen S, Gondro C, Watson-Haigh N, Kadarmideen H. FunctSNP: an R package to link SNPs to functional knowledge and dbAutoMaker: a suite of Perl scripts to build SNP databases. BMC Bioinformatics. 2010;11:311. doi: 10.1186/1471-2105-11-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P, Dai M, Xuan W, McEachin RC, Jackson AU, Scott LJ, Athey B, Watson SJ, Meng F. SNP Function Portal: a web database for exploring the function implication of SNP alleles. Bioinformatics. 2006;22:e523–e529. doi: 10.1093/bioinformatics/btl241. [DOI] [PubMed] [Google Scholar]
- 4.Shen TH, Carlson CS, Tarczy-Hornoch P. SNPit: a federated data integration system for the purpose of functional SNP annotation. Comp. Methods Prog. Biomedicine. 2009;95:181–189. doi: 10.1016/j.cmpb.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25:1431–1432. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riva A, Kohane IS. A SNP-centric database for the investigation of the human genome. BMC Bioinformatics. 2004;5:33. doi: 10.1186/1471-2105-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reumers J, Maurer-Stroh S, Schymkowitz J, Rousseau F. SNPeffect v2.0: a new step in investigating the molecular phenotypic effects of human non-synonymous SNPs. Bioinformatics. 2006;22:2183–2185. doi: 10.1093/bioinformatics/btl348. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Ma L, Li H, Vang S, Hu Y, Bolund L, Wang J. Snap: an integrated SNP annotation platform. Nucleic Acids Res. 2007;35:D707–D710. doi: 10.1093/nar/gkl969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal–unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courcelle E, Beausse Y, Letort S, Stahl O, Fremez R, Ngom-Bru C, Gouzy J, Faraut T. Narcisse: a mirror view of conserved syntenies. Nucleic Acids Res. 2008;36:D485–D490. doi: 10.1093/nar/gkm805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filangi O, Beausse Y, Assi A, Legrand L, Larre JM, Martin V, Collin O, Caron C, Leroy H, Allouche D. BioMAJ: a flexible framework for databanks synchronization and processing. Bioinformatics. 2008;24:1823–1825. doi: 10.1093/bioinformatics/btn325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlier C, Coppieters W, Farnir F, Grobet L, Leroy PL, Michaux C, Mni M, Schwers A, Vanmanshoven P, Hanset R, et al. The mh gene causing double-muscling in cattle maps to bovine Chromosome 2. Mamm. Genome. 1995;6:788–792. doi: 10.1007/BF00539005. [DOI] [PubMed] [Google Scholar]
- 13.Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 14.Lagarrigue S, Pitel F, Carre W, Abasht B, Le Roy P, Neau A, Amigues Y, Sourdioux M, Simon J, Cogburn L, et al. Mapping quantitative trait loci affecting fatness and breast muscle weight in meat-type chicken lines divergently selected on abdominal fatness. Genet. Sel. Evol. 2006;38:85–97. doi: 10.1186/1297-9686-38-1-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Mignon G, Pitel F, Gilbert H, Le Bihan-Duval E, Vignoles F, Demeure O, Lagarrigue S, Simon J, Cogburn LA, Aggrey SE, et al. A comprehensive analysis of QTL for abdominal fat and breast muscle weights on chicken chromosome 5 using a multivariate approach. Anim. Genet. 2009;40:157–164. doi: 10.1111/j.1365-2052.2008.01817.x. [DOI] [PubMed] [Google Scholar]
- 16.Blum Y, Le Mignon G, Lagarrigue S, Causeur D. A factor model to analyze heterogeneity in gene expression. BMC Bioinformatics. 2010;11:368. doi: 10.1186/1471-2105-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Mignon G, Desert C, Pitel F, Leroux S, Demeure O, Guernec G, Abasht B, Douaire M, Le Roy P, Lagarrigue S. Using transcriptome profiling to characterize QTL regions on chicken chromosome 5. BMC Genomics. 2009;10:575. doi: 10.1186/1471-2164-10-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponsuksili S, Jonas E, Murani E, Phatsara C, Srikanchai T, Walz C, Schwerin M, Schellander K, Wimmers K. Trait correlated expression combined with expression QTL analysis reveals biological pathways and candidate genes affecting water holding capacity of muscle. BMC Genomics. 2008;9:367. doi: 10.1186/1471-2164-9-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babak T, Garrett-Engele P, Armour CD, Raymond CK, Keller MP, Chen R, Rohl CA, Johnson JM, Attie AD, Fraser HB, et al. Genetic validation of whole-transcriptome sequencing for mapping expression affected by cis-regulatory variation. BMC Genomics. 2010;11:473. doi: 10.1186/1471-2164-11-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drost DR, Benedict CI, Berg A, Novaes E, Novaes CR, Yu Q, Dervinis C, Maia JM, Yap J, Miles B, et al. Diversification in the genetic architecture of gene expression and transcriptional networks in organ differentiation of Populus. Proc. Natl Acad. Sci. USA. 2010;107:8492–8497. doi: 10.1073/pnas.0914709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Nas A, Ingram-Drake L, Sinsheimer JS, Wang SS, Schadt EE, Drake T, Lusis AJ. Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics. 2010;185:1059–1068. doi: 10.1534/genetics.110.116087. [DOI] [PMC free article] [PubMed] [Google Scholar]