Abstract
Most life science processes involve, at the atomic scale, recognition between two molecules. The prediction of such interactions at the molecular level, by so-called docking software, is a non-trivial task. Docking programs have a wide range of applications ranging from protein engineering to drug design. This article presents SwissDock, a web server dedicated to the docking of small molecules on target proteins. It is based on the EADock DSS engine, combined with setup scripts for curating common problems and for preparing both the target protein and the ligand input files. An efficient Ajax/HTML interface was designed and implemented so that scientists can easily submit dockings and retrieve the predicted complexes. For automated docking tasks, a programmatic SOAP interface has been set up and template programs can be downloaded in Perl, Python and PHP. The web site also provides an access to a database of manually curated complexes, based on the Ligand Protein Database. A wiki and a forum are available to the community to promote interactions between users. The SwissDock web site is available online at http://www.swissdock.ch. We believe it constitutes a step toward generalizing the use of docking tools beyond the traditional molecular modeling community.
INTRODUCTION
Molecular docking
Back in the 19th century, Emil Fischer introduced the lock-and-key model to explain enzyme specificity. Nowadays, it is well known that most of the processes in life sciences involve, at the atomic scale, complex interactions between at least two molecules. The prediction of such interactions, by so-called docking software, is a non-trivial task. Docking programs intrinsically have a wide range of applications that go far beyond the creation of simple visual illustrations. In the context of protein engineering, for instance, the prediction of the detailed molecular interactions between a protein and one of its interacting partners paves the way for the rational selection of amino acids that could be mutated to promote or disrupt this interaction (1,2). Conversely, a small molecule interacting with a protein can be modified in order to change its affinity and, in fine, its biological activity to obtain new molecular probes or drugs (3–9). In addition, by docking the same compound into several protein targets, one can gain insights into the underlying molecular mechanisms of selectivity (10). The properties of the predicted complex can also be investigated by further calculations, so that complex recognition mechanisms might be unveiled. For instance, a recent MD simulation study allowed a better understanding of the molecular switch involving the helix 12 of the peroxisome proliferator-activated receptor alpha (11). Lastly, docking predictions can be post-processed using free energy calculations. For example, several β-secretase (BACE) and HIV-1 protease (HIV-1 PR) inhibitors have been identified by such a combination (12,13).
Current limitations to widespread use
While docking programs have already been very helpful in drug design (14), their wide and general usage remains impaired by the steep learning curve still required before being able to setup, carry out and analyze a docking assay. This technical barrier is due to the following caveats:
Structure setup
First, a docking assay requires the structure of a target protein. Of total, 87% of those available in the most comprehensive resource, the Protein Databank [(15) as of November 2010], have been obtained by X-ray crystallography. Despite the methodological improvements in this field, most of these structures usually do not meet the quality criteria required for a modeling study. For instance, some atoms in the flexible part of the protein are not resolved and the corresponding spatial coordinates omitted in the PDB file. The orientation of asparagine, glutamine and histidine side chains, as well as the position of water molecules should be considered with caution since their determinations remain challenging for X-ray crystallography (16). Crystal packing may also lead to constrained structures, and the crystallization conditions (solvent, pH and concentration) might differ considerably from in vivo conditions (16). A careful investigation of all these issues is required before carrying out the docking assay, and a molecular modeling background is required to reconstruct and correct the defective parts of the structure (16). Similarly, the structure of the ligand must also be carefully inspected to make sure that the topology of the molecule is correct, as well as its protonation state and tautomeric form. Both the setup of the protein structure and the ligand require a sufficient understanding of biophysics as well as organic and/or medicinal chemistry, which often prevents docking software from being used by a wide audience beyond the limits of molecular modeling experts.
Software's specific use
Additionally, once the two structures have been setup, several docking parameters must be defined, such as the region of space to which the docking search space will be limited, the exhaustiveness of the conformational search or the maximum duration of the docking assay. Most docking programs are complex computational machineries and require specific additional sampling or scoring parameters to which they might be very sensitive. While convenient default values are often proposed, a good understanding of the user manual and/or of the original article is always required in order to achieve meaningful predictions. Therefore, extensive practice is generally essential to perform relevant and reliable dockings.
Once all parameters have been defined, calculations are usually started from a command line interface that might discourage non-specialists, and may require computing power beyond that of common personal computers.
Embedding predictions in an existing research pipeline
Expectations regarding docking predictions range from illustrating a hypothesis up to prioritizing compounds according to their putative biological activity. While docking software has proven to be a valuable tool for drug development (3–9,17), the visualization, classification and rational analysis of their predictions often remain so challenging that only a tiny fraction of them can be properly incorporated into existing research pipelines.
The SwissDock web server
A large interest for docking web servers has emerged recently, as can be seen through the growing list of similar web services currently available, such as DockingServer (http://www.dockingserver.com/web), Blaster (18), DockingAtUTMB (http://docking.utmb.edu/), Pardock (http://www.scfbio-iitd.res.in/dock/pardock.jsp), PatchDock (http://bioinfo3d.cs.tau.ac.il/PatchDock/), MetaDock (http://dock.bioinfo.pl/), PPDock (http://140.112.135.49/ppdock/index.html) and MEDock (http://medock.ee.ncku.edu.tw/) (a comprehensive list is being kept up to date on the click2drug web portal, http://www.click2drug.org).
With the SwissDock web site, we aim at extending the use of protein-small molecule docking software far beyond experts in the field by providing convenient answers to many of the difficulties mentioned above. First, manually curated protein structures can be downloaded from the web site, and original PDB files can be prepared through ad hoc scripts (19,20). Second, the docking software is easily accessible through either a web browser or a programmatic interface. Third, predicted binding modes (BMs) can be viewed online with a simple embedded applet or analyzed in more details thanks to a seamless integration with the UCSF Chimera molecular viewer (21), with the help of the online documentation and the user community.
SWISSDOCK
SwissDock is a docking web server that addresses the limitations described above. The structure of the target protein, as well as that of the ligand, can be automatically prepared for docking. In addition, the cumbersome syntax of the docking engine is hidden behind a clean web interface providing reasonable alternative sets of parameters as well as sample input files. All calculations are performed on the server side, so that docking runs do not require any computational power from the user. The interpretation of docking results and their integration into existing research pipelines is greatly facilitated by the seamless visualization of docking predictions in the UCSF Chimera molecular viewer, which can be launched directly from the web browser.
Web interface
Inputs
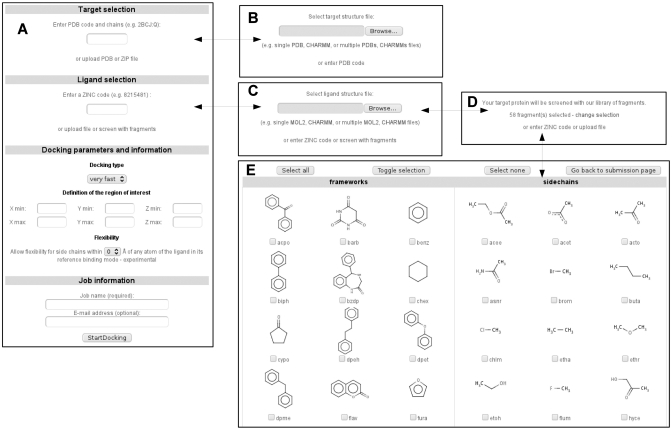
Only three steps are required to start a docking assay through the web interface of SwissDock: users must define a protein structure, one or several putative ligands and docking parameters (Figure 1). They are guided throughout this short and simple submission process by a comprehensive contextual help. As mentioned above, several sample files are supplied to users and can be directly uploaded into the form simply by clicking on a link. The corresponding sample output files are also provided.
Figure 1.
(A) Screenshot of the job submission form. It is dynamically modified so that the user can replace the default target selection from PDB codes by a target upload form (B). Similarly, the default ligand selection from ZINC codes can be replaced either by a ligand upload form (C) or by a fragment selection form (D) if a screening assay is to be performed. In such a case, the fragments to dock can be manually selected (E). The contextual help is automatically adapted to submission form content.
Target selection
A target protein structure can be determined either by specifying its identifier from the Protein Data Bank (15) or by uploading structure files.
The first option allows users who are not familiar with 3D structure files to start a docking assay with only a PDB code. If several PDB records are available for the same target, those with a high resolution and a ligand similar to the one that will be docked should be considered first. When several chains are present in the PDB file, the user can specify the identifiers of those on which the docking will be performed. If the structure file has been generated by nuclear magnetic response experiment, it must be edited first to keep only one structural model.
The second option allows the uploading of user-defined or edited target structures. Since the calculations are performed in the CHARMM force field, SwissDock supports the uploading of CHARMM formatted files in addition to the commonly used PDB format: protein structures can be uploaded as a set of protein structure file (PSF), coordinate file (CRD) and extra topology (RTF) and parameter files (PAR), if needed. A high-quality library of 260 protein complexes that have been manually curated (22) can be downloaded in the CHARMM format from the SwissDock web site.
Once the target protein structure has been defined, it is immediately prepared for use with CHARMM, and the curated structure can be downloaded and reviewed prior to the docking assay if needed (see ‘Materials and methods’ section for details).
Ligand selection
A ligand can be selected either by specifying its identifier from the ZINC database (23) or by uploading structure files. The former possibility allows users who are not familiar with 3D structure files to start a docking assay with only a ZINC accession code (AC). The latter allows uploading several ligands at once or uploading ligands that are not present in the ZINC database.
As for the target protein, SwissDock not only supports the widely used Mol2 format, but also the direct upload of CHARMM input files describing the ligand: a coordinate file (PDB), extra topology (RTF) and parameter files (PAR). The ligand is immediately set up after its definition and its prepared structure can be downloaded and reviewed prior to the docking assay, if needed (see ‘Materials and methods’ section for details).
Docking parameters
Docking type
Three docking parameter presets can be selected: very fast, fast and accurate. They correspond to presets described previously (20). In brief, several docking parameters are adjusted in order to reach the desired docking time and exhaustiveness of the search: the number of sampled BMs, the number of minimization steps that are performed to relax the ligand and the number of predicted BMs. If the ligand has less than 15 rotatable bonds and/or is likely to fit exactly into a binding pocket of the target protein, the very fast and fast modes are most probably sufficient (see below).
Definition of the region of interest
When using default parameters, the whole target protein structure is considered during the docking. However, if the target protein is particularly large and/or if the putative binding pocket is already known, the docking can be restricted to an rectangular region of space to perform a local docking assay.
Outputs
After a docking assay has been submitted, it can be tracked thanks to a dedicated URL provided on the submission confirmation page. If an (optional) email address has been specified in the submission form, this URL is also sent to the user by email, as well as a link to the docking result web page once the docking is completed.
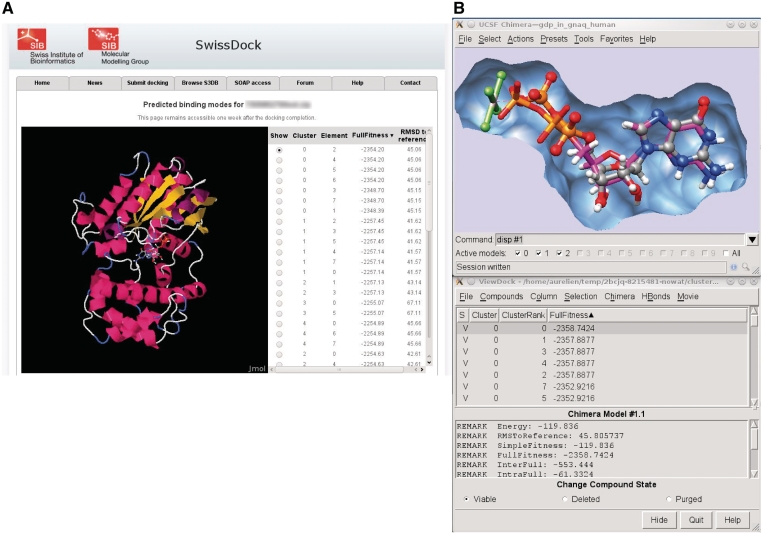
This docking result web page features a Jmol applet (24) for the visualization of the predicted BMs within the web browser (Figure 2A). For further visual investigations, UCSF Chimera can be started by a single click, and the predicted BMs are automatically loaded in its ViewDock plugin (Figure 2B). A short video illustrating this process is available on the SwissDock home page.
Figure 2.
This figure shows a typical output of SwissDock for the docking of the Guanine nucleotide-binding protein G(q) subunit alpha, a target involved in uveal melanoma (35). (A) Screenshot of the Jmol applet which renders predicted BMs within the web browser. (B) Visual investigation using the ViewDock plugin of UCSF Chimera. The predicted BM of the guanosine diphosphate (magenta sticks) is superimposed to the X-ray BM (ball and sticks). As it can be seen in the lower part of the figure, this particular predicted BM has the most favorable energy.
Predicted BMs are also available for download as a ZIP file. It contains PDB files for the complex, as well as the predicted BMs in the DOCK format. For experienced users, CHARMM PSF/CRD/RTF/PAR files are also provided for subsequent calculations.
Community
An online help is available using the form of a wiki. A searchable forum is proposed to promote communication among users, such as feedbacks, tips and tricks, questions and answers, so that the experience gained by some may be shared with others more broadly than what a mailing list would allow for.
Privacy policy
Uploaded files and docking predictions are automatically deleted after 7 days. Other parameters are kept anonymously for statistical purposes.
SOAP interface
Alongside the web interface, a programmatic access has been implemented via SOAP. The SwissDock SOAP server is implemented in the Python programming language, with the soaplib library. Sample client codes written in PHP, Perl or Python can be downloaded from the web site. They rely on the NuSOAP (http://nusoap.sourceforge.net/), SOAP::WSDL (http://search.cpan.org/~mkutter/SOAP-WSDL-2.00.10/lib/SOAP/WSDL.pm) and soaplib libraries (http://pypi.python.org/pypi/soaplib/0.8.1), respectively.
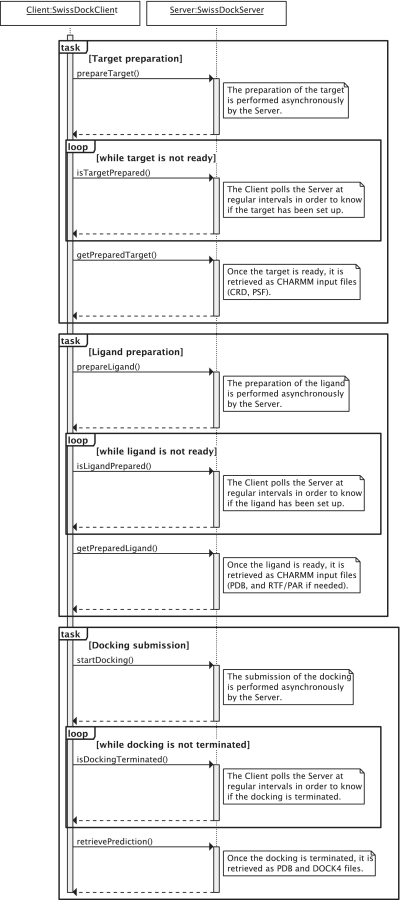
The server implements the three tasks of setting up the target protein, setting up the ligand and submitting the docking. A typical sequence is represented in Figure 3.
Figure 3.
Sequence diagram of a typical interaction between a SwissDock SOAP client and the SwissDock SOAP server. The target preparation, ligand preparation and docking submission tasks can be performed in a row or independently. Each task is divided into three steps. First, the client submits the task. Second, it polls the server at regular intervals in order to know if the task has been processed. If so, the outcome of the task is retrieved from the server.
Predictive performance assessment
The performance of the backend of SwissDock, i.e. EADock DSS, was assessed by a blind docking assay on 251 test complexes (22) taken from the Ligand Protein DataBase (LPDB) (25) with the different presets available from the web interface: fast, very fast, accurate and very accurate.
With accurate parameters, the first predicted BM was within 2 Å of the crystal structure for 54.5% of the test complexes, representing the success rate in rank 0 (SR0). This success rate increased up to 63.7% when the five most favorable predicted BMs were considered (SR5). The performance varies depending on the set of parameters and it is noteworthy that even a very short docking assay already managed to propose a correct BM for about 50% of the test complexes. The maximum performance is obtained for the very accurate parameter preset, with a SR5 slightly above 70%.
The performance of EADock DSS depends on the number of free dihedral angles of the ligand (Table 1). The vast majority (93%) of Food and Drug Administration (FDA)-approved drugs [subset of the ZINC database as of October 2010 (23)] has <10 rotatable bonds. The success rates observed for such ligands were 77% for SR0 and 86% for SR5. It is worth noting that the 251 ligands present in our validation set of complexes have 16 free dihedrals on average, making them much harder to predict. The performance observed here underestimates what can be expected in a typical drug design docking assay.
Table 1.
Influence of the flexibility of the ligand on the success rate observed with EADock DSS, the docking engine of SwissDock, in a redocking assay carried out on 251 protein–ligand complexes
| Max no. of rotatable bonds of the ligand | FDA-approved drugs (%) | SR0 (%) | SR5 (%) |
|---|---|---|---|
| 5 | 63 | 84 | 93 |
| 10 | 93 | 77 | 86 |
| 15 | 99 | 69 | 83 |
| 20 | 100 | 66 | 81 |
As can be expected, the success rate decreases as the ligand flexibility increases. Interestingly, 93% of FDA-approved drugs have 10 or less rotatable bonds. For 86% of such ligands in our data set, EADock is able to propose a binding mode within 2 Å root mean square deviation to the experimental BM within among the five most favorable predictions. See text for details.
The fraction of the surface of the ligand which becomes buried upon complexation also has a significant effect: the higher this fraction, the easier it is for SwissDock to identify the binding pocket, and therefore to dock the ligand inside (Table 2).
Table 2.
Influence of the fraction of the ligand which is buried upon complexation (%BS) on the same data set
| Min %BS | SR0 (%) | SR5 (%) |
|---|---|---|
| 95 | 82 | 95 |
| 90 | 70 | 85 |
| 85 | 66 | 80 |
| 80 | 62 | 75 |
The more buried the ligands are, the more successful their dockings are. See text for details.
MATERIALS AND METHODS
Algorithm
SwissDock is based on the docking software EADock DSS (20). Its algorithm consists of the following steps. First, a large number of BMs (typically from 5000 to 15 000) are generated, either in a user-defined box (local docking) or in the vicinity of the target cavities of the entire protein surface (blind docking). Simultaneously, their CHARMM (26) energies are estimated on a grid. Then, BMs with the most favorable energies are ranked, taking account of the solvent effect using the FACTS implicit solvation model (27), and clustered. Finally, the most favorable clusters are dumped into the result file. This unique combination of features allows accurate docking assays to be carried out within minutes.
Implementation
The SwissDock web server is implemented with open-source technologies (Apache web server, PHP). Calculations are currently carried out on 32 computing nodes with Dual Xeon E5440 2.83 GHz and 16 GB of RAM, accounting for a total of 256 CPU and 512Go of RAM, which belong to the Molecular Modeling group of the Swiss Institute of Bioinformatics and are integrated into the Vital-IT cluster.
Input files preparation
Since docking assays are carried out in the CHARMM22/27 all-hydrogen force field (26), target proteins and ligands that have been uploaded as CHARMM-formatted files can be used as is. Conversely, proteins and ligands that have been submitted in PDB or Mol2 format, respectively, have to be converted prior to the docking itself. This procedure, which generally requires a good knowledge of this software and of the physico-chemical properties of proteins and small molecules, is normally performed by experimented users. However, this step has been automatized for the SwissDock web service and is performed transparently for the user.
Preparation of the target protein
In short, the PDB structure file, either provided by the user or downloaded from the PDB if a PDB identifier has been provided, is divided into protein and non-protein parts. The former is further decomposed into CHARMM segments, which correspond to protein chains or units. These segments are identified by calculating the distance between the C and N atoms of subsequent residues in the PDB file. A new chain is defined if the distance between the C atom of a residue and the N atom of the next residue in the PDB file is larger than 1.7 Å. This value, which is significantly larger than the standard distance between two covalently linked C and N atoms in a peptide bond (i.e. 1.33 Å), was chosen in view of some abnormally long CN distances between covalently linked residues in certain PDB files. It is, however, small enough to identify residues that are not covalently linked, since the corresponding distance should not be smaller than 3.8 Å in the CHARMM force field. Similarly, disulfide bridges are automatically added when the distance between two S atoms is lower than a threshold of 2.25 Å (the standard distance between two S atoms in a S-S bond is 2.03 Å, while two non-covalently linked S atoms should not be closer than 3.95 Å). The CD1 atom of isoleucine is renamed CD in agreement with the CHARMM nomenclature. Selenomethionine residues, which are often replacing methionine residues to facilitate elucidating protein structures, are automatically replaced by methionine residues and the atom names changed accordingly. Heme residues are automatically added to the PSF when present. In that case, patches to link possible histidine or cysteine residues to the Fe atoms are used. The standard protonation state of residues at physiological pH is applied to all residues, leading to negatively charged aspartate and glutamate residues, positively charged lysine and arginine residues and neutral serine, threonine, tyrosine and cysteine residues. However, the protonation state of histidine residues is determined automatically by checking the chemical environment of the Nε and Nδ atoms of the imidazole ring. Protons are added, or not, to these two atoms to allow possible hydrogen bonds with nearby polar functions of the protein or water molecules, and to prevent unfavorable contacts between a possible NεH or NδH function and another protonated function or metal ion. When no contact is possible, the histidine is protonated on Nδ by default. The residue numbers given in the PDB file are kept during the setup, to facilitate the analysis of the docking results. Hydrogen atoms and missing side chains are added automatically using the HBUILD and IC BUILD commands of CHARMM. Molecules not belonging to the protein part of the target are added to the PSF if the corresponding compounds exist in the standard CHARMM22/27 force field. Otherwise, they are removed from the system. However, experimented users can take them into account by providing their own PSF/CRD files as mentioned below. Water molecules are removed. The whole system is minimized by 100 steps of steepest descent algorithm to remove possible clashes due to atom additions or uncertainties in the PDB file. During minimization, heavy atoms are restrained using a 5 kcal mol−1 A−2 constraint to prevent unphysical motions. The root mean square deviation between the starting and minimized conformations is generally <0.15 Å.
Preparation of the ligand
If a ZINC AC or a Mol2 file are provided for the ligand, CHARMM topology, parameters and coordinates, files are derived automatically (19) from the Merck Molecular Force Field (MMFF) (28–32). In brief, dihedral angle terms are taken as is, while only the harmonic part of the bond, angle and improper terms are retained. Charges are taken from MMFF. Van der Waals parameters are taken from the closest atom type in CHARMM22 (26). For consistency with CHARMM, atoms with identical names are renamed.
Uploaded Mol2 files must contain all hydrogens and 3D coordinates. The topology of the Mol2 files has to be correct, hence we recommend the use of the following software, working most of the time: UCSF Chimera, OpenBabel (33), MarvinSketch (http://www.chemaxon.com/) (save as MDL, then convert into Mol2 with OpenBabel) and ChemDraw (34).
CONCLUSION
The SwissDock web server aims at providing a wide scientific community with a free and user-friendly, yet state-of-the-art protein/small molecule, docking tool. The automatic setup of protein and ligand structures, the different parameter presets and the convenient visualization and analysis of docking predictions makes it accessible to a wide audience. The EADock DSS engine behind SwissDock is especially suited for drug design, with very good success rates for small and relatively rigid ligands with less than 10 flexible rotatable bonds: the most favorable predicted BM is found within 2 Å to the crystal structures for 77% of the 251 test complexes, and for 86% of them, such a correct BM is found within the five most favorable ones. This performance is even increased if the ligand can be buried in a well-defined binding site of its target protein. The SwissDock web server is available at http://www.swissdock.ch. We believe it constitutes a step toward generalizing the use of docking tools beyond the traditional molecular modeling community.
FUNDING
Swiss Institute of Bioinformatics; Swiss National Science Foundation (SCORE 3232B0-103172 and 3200B0-103173 to O.M., and 310030-130857 to O.M. and V.Z.); Oncosuisse (OCS 01381-08-2003 to O.M. and OCS 02555-02-2010 to O.M. and V.Z.). National Center of Competence in Research (NCCR) Molecular Oncology, a research instrument of the Swiss National Science Foundation; National Institutes of Health (P41 RR001081) for Molecular graphics images. Funding for open access charge: Swiss Institute of Bioinformatics; Swiss National Science Foundation (SCORE 3232B0-103172 and 3200B0-103173 to O.M., and 310030-130857 to O.M. and V.Z.); Oncosuisse (OCS 01381-08-2003 to O.M. and OCS 02555-02-2010 to O.M. and V.Z.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the VITAL-IT project of the Swiss Institute of Bioinformatics (Lausanne, Switzerland) for providing part of the computational resources. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco.
REFERENCES
- 1.Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:132–133. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 2.Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 3.Boehm HJ, Boehringer M, Bur D, Gmuender H, Huber W, Klaus W, Kostrewa D, Kuehne H, Luebbers T, Meunier-Keller N, et al. Novel inhibitors of DNA gyrase: 3D structure based biased needle screening, hit validation by biophysical methods, and 3D guided optimization. A promising alternative to random screening. J. Med. Chem. 2000;43:2664–2674. doi: 10.1021/jm000017s. [DOI] [PubMed] [Google Scholar]
- 4.Shoichet BK, McGovern SL, Wei B, Irwin JJ. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 2002;6:439–446. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- 5.Blake JF, Laird ER. Chapter 30. Recent advances in virtual ligand screening. Annu. Rep. Med. Chem. 2003;38:305–314. [Google Scholar]
- 6.Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet BK. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J. Med. Chem. 2002;45:2213–2221. doi: 10.1021/jm010548w. [DOI] [PubMed] [Google Scholar]
- 7.Paiva AM, Vanderwall DE, Blanchard JS, Kozarich JW, Williamson JM, Kelly TM. Inhibitors of dihydrodipicolinate reductase, a key enzyme of the diaminopimelate pathway of Mycobacterium tuberculosis. Biochim. Biophys. Acta. 2001;1545:67–77. doi: 10.1016/s0167-4838(00)00262-4. [DOI] [PubMed] [Google Scholar]
- 8.Röhrig UF, Awad L, Grosdidier A, Larrieu P, Stroobant V, Colau D, Cerundolo V, Simpson AJ, Vogel P, Van den Eynde BJ, et al. Rational design of indoleamine 2,3-dioxygenase inhibitors. J. Med. Chem. 2010;53:1172–1189. doi: 10.1021/jm9014718. [DOI] [PubMed] [Google Scholar]
- 9.Huang D, Lüthi U, Kolb P, Cecchini M, Barberis A, Caflisch A. In silico discovery of beta-secretase inhibitors. J. Am. Chem. Soc. 2006;128:5436–5443. doi: 10.1021/ja0573108. [DOI] [PubMed] [Google Scholar]
- 10.Cai J, Han C, Hu T, Zhang J, Wu D, Wang F, Liu Y, Ding J, Chen K, Yue J, et al. Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation. Protein Sci. 2006;15:2071–2081. doi: 10.1110/ps.062238406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalik L, Zoete V, Krey G, Grosdidier A, Gelman L, Chodanowski P, Feige JN, Desvergne B, Wahli W, Michielin O. Combined simulation and mutagenesis analyses reveal the involvement of key residues for peroxisome proliferator-activated receptor alpha helix 12 dynamic behavior. J. Biol. Chem. 2007;282:9666–9677. doi: 10.1074/jbc.M610523200. [DOI] [PubMed] [Google Scholar]
- 12.Huang D, Caflisch A. Efficient evaluation of binding free energy using continuum electrostatics solvation. J. Med. Chem. 2004;47:5791–5797. doi: 10.1021/jm049726m. [DOI] [PubMed] [Google Scholar]
- 13.Huang D, Lüthi U, Kolb P, Edler K, Cecchini M, Audetat S, Barberis A, Caflisch A. Discovery of cell-permeable non-peptide inhibitors of beta-secretase by high-throughput docking and continuum electrostatics calculations. J. Med. Chem. 2005;48:5108–5111. doi: 10.1021/jm050499d. [DOI] [PubMed] [Google Scholar]
- 14.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 15.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis AM, St Gallay SA, Kleywegt GJ. Limitations and lessons in the use of X-ray structural information in drug design. Drug Discov. Today. 2008:831–841. doi: 10.1016/j.drudis.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolb P, Irwin JJ. Docking screens: right for the right reasons? Curr. Top Med. Chem. 2009;9:755–770. doi: 10.2174/156802609789207091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irwin JJ, Shoichet BK, Mysinger† MM, Huang N, Colizzi F, Wassam P, Cao Y. Automated docking screens: a feasibility study. J. Med. Chem. 2009;52:5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoete VF, Cuendet M, Grosdidier A, Michielin O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011 doi: 10.1002/jcc.21816. May 3 (doi:10.1002/jcc.21816; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 20.Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011 doi: 10.1002/jcc.21797. May 3 (doi:10.1002/jcc.21797; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 21.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;13:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Grosdidier A, Zoete V, Michielin O. Blind docking of 260 protein-ligand complexes with EADock 2.0. J. Comput. Chem. 2009;13:2021–2030. doi: 10.1002/jcc.21202. [DOI] [PubMed] [Google Scholar]
- 23.Irwin JJ, Shoichet BK. ZINC-a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jmol: an Open-Source Java Viewer for Chemical Structures in 3D. http://www.jmol.org/ (October 2010, date last accessed) [Google Scholar]
- 25.Roche O, Kiyama R, Brooks CL., III Ligand-protein database: linking protein-ligand complex structures to binding data. J. Med. Chem. 2001;44:3592–3598. doi: 10.1021/jm000467k. [DOI] [PubMed] [Google Scholar]
- 26.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haberthür U, Caflisch A. FACTS: fast analytical continuum treatment of solvation. J. Comput. Chem. 2008;29:701–715. doi: 10.1002/jcc.20832. [DOI] [PubMed] [Google Scholar]
- 28.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996;17:490–519. [Google Scholar]
- 29.Halgren TA. Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J. Comput. Chem. 1996;17:520–552. [Google Scholar]
- 30.Halgren TA. Merck molecular force field. III. Molecular geometries and vibrational frequencies for MMFF94. J. Comput. Chem. 1996;17:553–586. [Google Scholar]
- 31.Halgren TA. Merck molecular force field. IV. Conformational energies and geometries for MMFF94. J. Comput. Chem. 1996;17:587–615. [Google Scholar]
- 32.Halgren TA. Merck molecular force field. V. Extension of MMFF94 using experimental data, additional computational data, and empirical rules. J. Comput. Chem. 1996;17:616–641. [Google Scholar]
- 33.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. The Blue Obelisk-interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mills N. ChemDraw Ultra 10.0. J. Am. Chem. Soc. 2006;128:13649–13650. [Google Scholar]
- 35.Onken MD, Worley LA, Long MD, Duan S, Council ML, Bowcock AM, Harbour JW. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest. Ophthalmol. Vis. Sci. 2008;49:5230–5234. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]