Abstract
HMMER is a software suite for protein sequence similarity searches using probabilistic methods. Previously, HMMER has mainly been available only as a computationally intensive UNIX command-line tool, restricting its use. Recent advances in the software, HMMER3, have resulted in a 100-fold speed gain relative to previous versions. It is now feasible to make efficient profile hidden Markov model (profile HMM) searches via the web. A HMMER web server (http://hmmer.janelia.org) has been designed and implemented such that most protein database searches return within a few seconds. Methods are available for searching either a single protein sequence, multiple protein sequence alignment or profile HMM against a target sequence database, and for searching a protein sequence against Pfam. The web server is designed to cater to a range of different user expertise and accepts batch uploading of multiple queries at once. All search methods are also available as RESTful web services, thereby allowing them to be readily integrated as remotely executed tasks in locally scripted workflows. We have focused on minimizing search times and the ability to rapidly display tabular results, regardless of the number of matches found, developing graphical summaries of the search results to provide quick, intuitive appraisement of them.
INTRODUCTION
The goal of the HMMER project is to make advanced probabilistic methods for sequence homology detection available in widely useful tools. The HMMER software suite has been widely used, particularly by protein family databases such as Pfam (1) and InterPro (2) and their associated search tools. HMMER 3.0, released in early 2010, includes new technology producing roughly 100-fold speed improvements relative to previous versions of HMMER (3), such that HMMER3 search times are competitive with BLASTP (4) search times. This new technology includes a combination of striped vector-parallelized alignment algorithms (5) [using single instruction, multiple data (SIMD) vector instructions called SSE on Intel-compatible platforms and Altivec/VMX on PowerPC platforms]; a new heuristic acceleration algorithm; and a ‘sparse rescaling’ method enabling the Forward and Backward profile hidden Markov model (profile HMM) algorithms to be implemented using multiply/add instructions on scaled probabilities without numerical underflow (3). HMMER3 has now been adopted by most major protein family databases (1,2,6–9). In addition to speed improvements, HMMER also now uses log-odds likelihood scores summed over alignment uncertainty (Forward scores), rather than optimal alignment (Viterbi) scores, which improves sensitivity. Forward scores are better for detecting distant homologs as there can often be several possible ways of aligning a distantly related query to a target. By summing over all possible alignments, each alternative alignment contributes to the score, sufficient to indicate the similarity. However, by taking the best alignment, as in the case of Viterbi, from a set of poor alignments is often insufficient to distinguish the remote homolog from the noise. Furthermore, posterior probabilities of alignment confidence are reported, enabling detailed and intuitive assessments of alignments on a residue-by-residue basis.
Previous versions of HMMER have largely only been available as computationally intensive UNIX command-line applications requiring local installation and local computing resources. The greatly increased speed of HMMER3 makes it feasible to address this major usability hindrance with public HMMER web services. We have developed the HMMER web site (http://hmmer.janelia.org) to not only provide downloadable HMMER binaries, documentation and source code as it has done in the past, but now also to provide an interface for performing protein sequence searches with near interactive response times.
THE HMMER WEB SERVER
The HMMER 3.0 software suite includes four database search programs for protein sequence analysis: phmmer, hmmscan, hmmsearch and jackhmmer. The web site implements the first three of these search algorithms.
phmmer
The phmmer program is analogous to BLASTP. It takes a single protein sequence, in FASTA format, as an input query and searches it against a target sequence database. To perform the search, the query sequence is converted into a profile HMM. Traditionally, profile HMMs have been thought of only as position-specific models of an input multiple sequence alignment. However, in essentially the reverse of how the original position-independent scoring model in BLASTP was generalized to the position-specific scoring model in PSI-BLAST (10), profile HMMs can be devolved to simple position-independent probabilistic scoring models as a special case. Given a single query input sequence, profile HMM residue probabilities are set by deriving the implicit probabilistic basis of a standard score matrix such as BLOSUM62 (11,12), plus empirically set insertion/deletion transition probabilities (parameters analogous to standard gap open, gap extend penalties).
The simple phmmer search submission form (http://hmmer.janelia.org/search/phmmer) allows only the query sequence to be entered, in which case default search parameters are used. Clicking the ‘Advanced’ option, found in the top-right corner of the form, reveals more expert options for modifying the way that the search is performed. The default scoring matrix and gap parameters can be modified via the ‘Advanced’ form. It is also possible to set cut-off thresholds that sequence matches must achieve in order to be displayed (or reported) and for the match to be deemed significant (inclusion). HMMER reports both bit scores and E-values (expectation values). A bit score is a log-odds ratio score (base two) comparing the likelihood of the profile HMM to the likelihood of a null hypothesis (an independent, identically distributed random sequence model, as in BLAST). An E-value is the number of hits expected to achieve this bit score or greater by ‘chance’, i.e. if the search had instead been done on an identically sized database composed only of random non-homologous sequences (13). It is also possible to turn off the bias filter, used as part of the HMMER3 acceleration pipeline. Under certain circumstances, when the query contains a lot of low complexity, tandem repeats or trans-membrane regions, the bias filter may exclude homologous target sequences. Turning off the bias composition filter can increases sensitivity, but at a high cost in speed, as more sequences have to undergo more computationally expensive analysis, hence it is on by default.
In addition to running a phmmer search, a ‘Pfam Search’ is run by default. This triggers an inexpensive hmmscan search (described in the following section) against the Pfam library of profile HMMs, in parallel to the phmmer search.
Currently, the query sequence can be searched against one of six different target sequence databases: NR (14), UniProtKB (15), SwissProt, PDB (16), UniMes and the environmental division of NR. These target sequence databases have been chosen either because they represent large, comprehensive sequence collections (NCBI NR/UniProtKB), annotated or structurally characterized sequences (SwissProt and PDB) or metagenomic sequence databases (UniMes and env NR). On the web site, the default database is selected based on where the geographical location of the IP address found in the incoming HTTP request. Users from the USA have the NCBI NR database as default, whereas UniProtKB is the default database for users in Europe.
A phmmer search via the web can also take protein accessions or identifiers, found in one of the six underlying target sequence databases, as a query, instead of a sequence. An autocompletion provides suggestions of known accessions or identifiers after the first three characters of the name have been entered.
The number of results that are displayed per page and the columns that are included in the results table can also be configured. By default, ‘Target’ (accessions and/or identifiers), ‘Description’ (functional annotations), ‘Species’ and ‘E-value’ columns are displayed, with 100 results per page. This default view allows the results to be displayed even when the browser window is narrow (typically on mobile devices). However, the amount of data can be expanded, both in terms of additional columns and the number of rows per page in the table. The results can be customized either before or after the search is performed.
hmmscan
The hmmscan program (previously called hmmpfam in HMMER2) takes a query sequence and searches it against the Pfam profile HMM library as a target database (http://hmmer.janelia.org/search/hmmscan). As with phmmer, significance and reporting thresholds can be defined either by bit score or E-value, and additionally, thresholds can be defined by the Pfam ‘gathering threshold’. Each Pfam profile HMM has a specific, curated gathering threshold that sets the inclusion bit score cut-off. Pfam defines gathering thresholds conservatively, such that no known false-positive matches are detected for that family. Any match scoring above the gathering threshold is very likely to be a true positive. These thresholds are generally most useful in fully automated searches. However, using the conservative gathering thresholds may miss borderline matches that are true hits, so when trying to establish distant relationships in more manual searches, one of the alternative thresholding methods may be more appropriate.
hmmsearch
This program takes a profile HMM and searches it against a target sequence database, with the profile HMM being built from a query multiple sequence alignment. The web search allows either a HMMER3 formatted profile HMM or a multiple sequence alignment to be submitted as the query. A variety of different multiple sequence alignment formats are permitted (Clustal, MSF, SELEX, STOCKHOLM and aligned FASTA format). Once uploaded, the multiple sequence alignment is converted to a profile HMM using hmmbuild (in its default mode). The target sequence databases available for searching and cut-off settings are as with phmmer searches. A comparison of the HMMER program to their equivalents in BLAST is shown in Table 1.
Table 1.
A comparison of HMMER and BLAST programs for protein sequence analysis
| HMMER | BLAST | Comments | |
|---|---|---|---|
| Program | phmmer | blastp | Produces similar results in terms of homolog detection. Searching with the sequence from PDB ID 2abl, chain A against PDB yields 244 matches compared with 214 matches for phmmer and blastp, respectively, using an E-value threshold of 0.01 and default search parameters. The matches were inspected for the presence of an SH3 (Src homology 3) and/or SH2 (Src homology 2) domain(s). phmmer results have the added advantage of scoring each residue in the alignment, giving users an indication of the parts of the alignment that are trustworthy. HMMER web server allows configuration of the cut-offs and provides access to all matches. |
| Query | Single sequence | ||
| Target Database | Sequence database | ||
| Program | hmmscan | rpsblast | Typically used for detection of domains on a sequence. Profile HMMs are used by the majority of protein family databases. Both are run as by default as part of the phmmer/blastp web searches. Available as separate search on the HMMER web servers. |
| Query | Single sequence | ||
| Target Database | Profile HMM database, e.g. Pfam | PSSM database, e.g. CDD | |
| Program | hmmsearch | Not applicable | There is no equivalent to hmmsearch in the BLAST suite. The web site uses hmmbuild to convert input alignments to a profile HMM. The command-line version psi-blast can be forced to perform a similar style of search by jump-starting it with a multiple sequence alignment. |
| Query | Profile HMM | ||
| Target Database | Sequence database | ||
| Program | jackhmmer | psi-blast | Both are used to iteratively search sequence databases. Subsequent iterations use the significantly scoring sequences from the previous round as input data. Jackhmmer is currently not supported on the HMMER web server. Gaps are weighted on observations for jackhmmer, rather than arbitrary open and extend penalties. |
| Query | Single sequence | ||
| Target database | Sequence database | ||
All of these HMMER methods are run on a compute farm at Janelia Farm using a new program, hmmpgmd (the hmmer program daemon). hmmpgmd is a custom IP (Internet Protocol) socket-based parallel system that establishes persistent server and worker daemons to broker search jobs across 144 cores [12 × 12 2.67 GHz Intel(R) Xeon(R) CPU] as they are received from the web servers. We chose to implement a custom IP socket communication protocol rather than using an established message-passing system such as message passing interface (MPI) in order to minimize latency. Further optimizations in hmmpgmd include the persistent caching of the database in memory upon starting the daemon and a compact binary format protocol for returning results to the client, in this instance the web server. This program will be included in the next release of the HMMER3 software to aid the deployment of HMMER on other web sites or high-throughput pipelines.
RESULTS VISUALIZATION
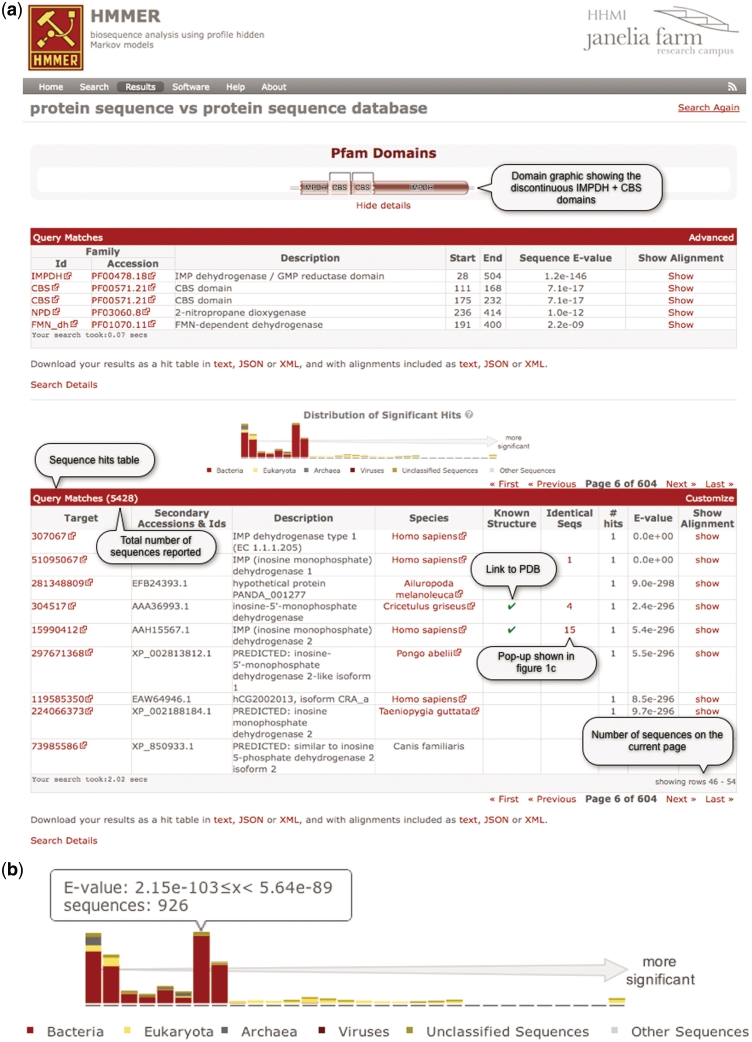
Views from a typical phmmer search result are shown in Figure 1. The top section of Figure 1a contains a graphical representation of the results from the Pfam search. A new domain graphic JavaScript library, which draws on ideas from the original Pfam graphic library, extends the Raphael JavasScript library (http://raphaeljs.com/) to produce an scalable vector graphics (SVG) image indicating the positions of Pfam domains on the query sequence. The details of any Pfam hits can be revealed by clicking a ‘show details’ link. This action renders a table of all of the Pfam domain matches, with each domain hit shown as a separate row. The domain graphic removes overlapping Pfam matches that belong to the same ‘Clan’ according to a simple ‘best match wins’ postprocessing (17). However, all significant matches are shown in the match table for completeness (Figure 1a). In the standard view, the domain details include coordinates and E-value for each domain. The positional coordinates correspond to the domain ‘envelope’, within which HMMER has determined that significant probability mass exists for an ensemble of possible alignments. Clicking on ‘Advanced’ to the top right of the Pfam domain match table toggles to a more detailed table, which includes the alignment coordinates—the region over which HMMER produced its single best estimated alignment within the envelope [using a probabilistic maximum expected accuracy (MEA) method]—and also the model positions that are matched, the bias estimate, the alignment accuracy score (the mean of the posterior probabilities) and the bit score. For each domain, there is an individual and a conditional E-value. The individual E-value is the significance of that hit as if it where the only domain/hit that had been identified. The conditional E-value is an attempt to measure the statistical significance of each domain, given that it has already been decided that the target sequence is a true homolog. It is the expected number of additional domains or hits that would be found with a score this big in the set of sequences reported in the top hits list, if those sequences consisted only of random nonhomologous sequence outside the region that sufficed to define them as homologs. The conditional E-value is evaluated against the inclusion and reporting thresholds when using E-value-based cut-offs. With either table, it is possible to ‘show’ the alignment between the query and the profile HMM. This alignment contains five rows: (i) an alignment position indicator, with every tenth position marked with ‘*’; (ii) the ‘Model’, which is the most probable sequence from the HMM that is colored according to the match; (iii) the match row indicates identical residues (letters) or similar residues (+) between the model and query, (iv) the ‘Query’ sequence aligned to the profile HMM, which is colored according to the posterior probability (below); and (v) the ‘PP’ row that is the per-position posterior probability or alignment accuracy of the residue in the query to the model.
Figure 1.
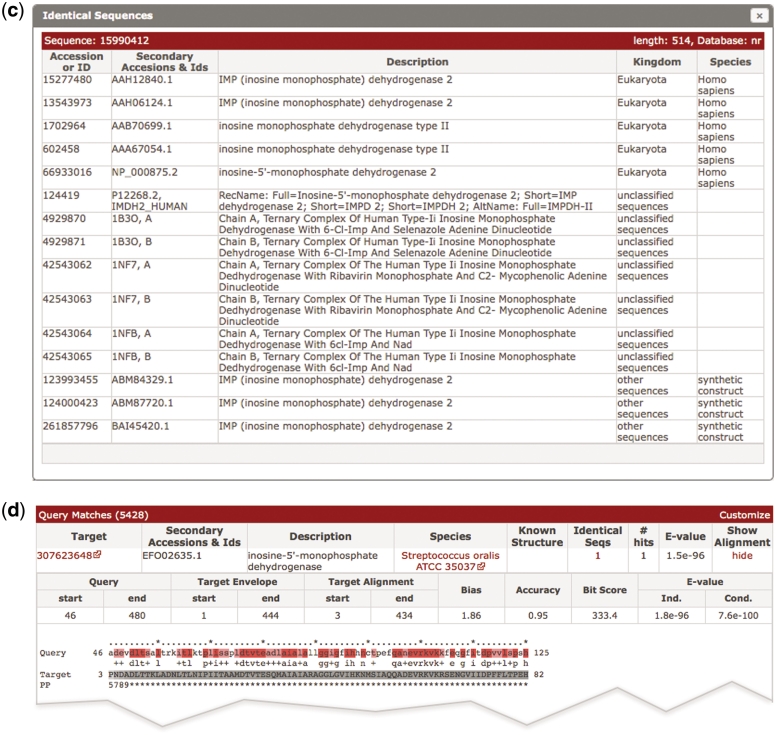
(a) A screen shot of the phmmer search results page, using the sequence IMDH1_HUMAN (UniProtKB) and default parameters. The results from the ‘Pfam search’ are shown as both a graphic and as a table, which has been revealed in this figure. Below this is a hit distribution graph and a table of the results from the phmmer search. Key features in the table are labeled and discussed in the text. This search resulted in over 5000 sequence matches, which are accessible either by going through the paginated table or can be navigated using the hit distribution histogram. (b) An enlarged version of the hit distribution graph, showing the taxonomic ranges of sequences matched in the search. The tool tip indicates the E-value range represented by the bar, and the number of sequences from the search that fall within the range.(c) As more and more sequences are being deposited, the large comprehensive sequence databases contain increasing numbers of duplicate sequences. There is no need to show an identical sequence match several times, but the annotation assigned to these duplicate sequences can differ. Thus, we indicate when we have not displayed identical sequences with a number in the results table (Figure 1a). When this number is clicked, a ‘pop-up’ displays the additional annotations for the sequence numbers. As in this example, when there are more than 20 sequences, the list is paginated. (d) Example of an alignment between a match and a query. When the show link is clicked in the results table as shown in (a), the table is expanded to show the alignments between the query and the target sequence. The query is color coded according to the match line found below it in the alignment block (identical residues are colored red similar to pink). The target sequence is colored according to the posterior probability, with lighter shades of gray indicating regions where the alignment confidence is lower. Each number in the ‘PP’ line represents the probability (or alignment accuracy) that the residue in the row above is assigned to the corresponding HMM state found in the first row of the alignment block. The posterior probability is encoded as 11 possible characters 0–9*: 0.0 ≤ P < 0.05 is coded as 0, 0.05 ≤ P < 0.15 is coded as1, and so on, 0.85 ≤ P < 0.95 is coded as 9 and 0.95 ≤ P ≤ 1.0 is coded as ‘*’.
The phmmer search results are found below the Pfam match table and horizontal rule in Figure 1a. The distribution histogram (Figure 1a, enlarged in Figure 1b) summaries two things: the distribution of hits scoring above the inclusion threshold and the taxonomic distribution of the sequence source organism. Each bar in the histogram is broken down according to the taxonomic kingdom to which the source organism belongs. This is achieved by cross-referencing each organism name that comes from the source sequence database with the NCBI taxonomy (14). In the case of the PDB, the FASTA files do not include the source organisms, so we use the SIFTS resource (18), which contains a mapping between PDB identifiers (and chains) and the source organism. When a source organism cannot be placed on the NCBI taxonomic tree (i.e. the given species name does not correspond to a known NCBI taxonomic identifier), we assign it to the unclassified sequences kingdom. Thus, it is possible to quickly determine if a query is specific to a kingdom of life, and if not, at what E-value ranges sequences from different kingdoms start to be matched. Furthermore, the bars of the graph can be used to navigate the table below it. Clicking on a bar within the histogram navigates the table to the most significant hit for the E-value range corresponding to that bar. Moving the mouse over the bars in the graph displays a tool tip that shows the E-value range of the bar and the total number of sequences that fall within that range in the search results (Figure 1b).
For example, the sequence IMDH1_HUMAN (inosine-5′-monophosphate dehydrogenase 1) was used as the query sequence for Figure 1. The hit distribution graph (Figure 1b) indicates some very similar eukaryotic sequence matches (yellow bars). Using the graph to navigate through the hits table shows that there is a very high-scoring match in mouse. Moving through the less-significant sections shows that there are homologs in other model organisms such as Drosophila melanogaster and Saccharomyces cerevisae. At lower significance values, matches to bacterial homologs are found (red bars), such as Bacillus subtilis, Aquifex aeolicus and various strains of Escherichia coli.
The table below the hit distribution graph (Figure 1a) contains the sequence matches from the target database to the query. Searches can result in many thousands of matches. Returning thousands of results across the web and rendering them as a table in the browser is often the most time-consuming part of a search. Therefore, once the search has completed on the server, with the default results configuration the first 100 matches are returned in the results table. The remaining results can be accessed using the pagination navigation tools found above the top and bottom right corners of the table, or by using the distribution graph. This strategy enables the rapid display of even the very largest result sets. The results table provides information on the target match, including: accessions and/or identifiers, functional annotations, species and E-value of the match. Using the ‘customize’ link at the top right of the table, the user can add columns to the results table depending on their needs. For example, the number of hits to the target sequence (those that score above the reporting threshold), the number of significant hits (those that score above the inclusion threshold), bit score and the kingdom that the species belongs to. An additional optional column is ‘known structure’. Where appropriate, this column indicates whether a structure has been deposited in the PDB (16) for some or all of the sequence. To do this, we use the SIFTS PDB/UniProtKB mapping (18) to first map structure links to UniProt sequences, then propagate these links to other databases based on matching identical sequences in them to the UniProtKB sequence. As most of the supported target sequence databases contain some sequence redundancy, we collapse identical sequences into a single row of the table. Note that in the distribution graph, the counts are for the hits in the table and are not multiplied by the number of redundant sequences, but we preferentially order them so that unclassified and other sequences are excluded from this navigation pane in preference to those that were assigned a kingdom. The redundant sequence information (accessions, description and species) is accessible by clicking the number found in the ‘Identical Seqs’ column (Figure 1a). This produces a pop-up table that paginates the list of redundant sequences (Figure 1c). Rows that are highlighted pink in the table indicate hits that score about the reporting thresholds, yet below the inclusion or significance thresholds.
At the end of each row in the table, there is a ‘show’ link. Clicking on this link displays the alignment between the query and the target (Figure 1d). There can be multiple hits per sequence because HMMER performs local–local searches (meaning any subsequence of the query model can align to any subsequence of the target sequence). The alignments are similar to those described previously, with the query and target labeled. The alignment view also contains the co-ordinate of the alignment boundaries. The envelope positions of the match on the target, the target bias composition score, alignment accuracy, bit score and individual and conditional E-values of the match are found above the alignment.
Below both tables (Figure 1a) there are two links, ‘Download’ and ‘Search details’. The ‘Download’ link allows the search results to be downloaded and saved in text, JSON or XML format. The ‘Search details’ reveals text that contains a 36-character unique identifier that is assigned to the job (and part of the results URL). This identifier can be used to retrieve the job results at a later date. The pop-up also contains the entire provenance of the search: the time and date that the search was performed, the HMMER command executed, and information about the version of the target database. If the query was a sequence, then that sequence is also displayed.
The hmmscan results page is essentially the same as described for ‘Pfam search’ results that are displayed as part of a default phmmer search. The only difference is that the table showing the hit details is displayed by default. Similarly, the hmmsearch results page contains the same distribution graph and table of sequence hits that is produced in a phmmer search.
Retrieval of results
Every single job is assigned a unique 36-character identifier. This identifier can be used to retrieve results at a later date via the results retrieval form, which is accessible under the ‘results’ tab. We anticipate being able to store search results for at least 1 week, after which we will delete the results as disk space becomes limiting.
Searching multiple individual sequences
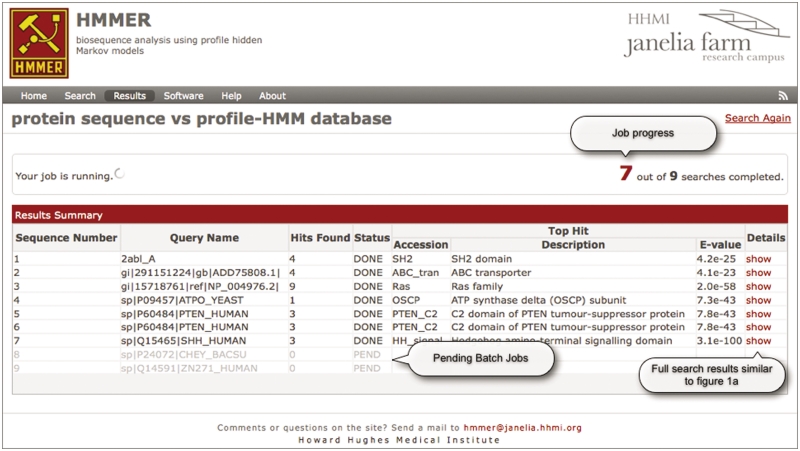
In addition to being able to submit single sequences, the advanced search submission form for phmmer and hmmscan allows ‘batch processing’ of files containing up to 500 sequences in FASTA format. The 500 sequence limit is imposed to prevent the web servers from becoming overloaded when processing and validating large uploaded query files. (Larger batch jobs should use the RESTful web services interface described in the next section.) After submitting a batch search, the user is taken to a different results page that tabulates the input file and indicates the progress of the batch job as a whole (Figure 2), and the progress of each query sequence. During the processing of the job, the batch summary table automatically updates, indicating the progress of each sequence search and providing access to the results of completed jobs. After viewing the individual sequence results, the user can either navigate back to the batch search progress page or follow the link at the top of that result page. The user can also provide an email address, which the server uses to inform them when the batch search has completed and provide a summary of results, similar to the table shown in Figure 2.
Figure 2.
After the submission of a batch job, the user is taken to a table indicating the progress of the job similar to the one shown in this figure. The top bar indicates the progress of the batch job. As sequences are successfully searched, the results are immediately viewable. Pending sequence searches are indicated as ‘grayed out’ entries in the table.
HMMER RESTful web services
In addition to the classical, user-oriented, HTML-based web server interface, we also provide all these search tools as RESTful web services, thereby allowing HMMER to be integrated as remote compute tasks in local workflows. The simple application programming interface (API) allows HMMER web services to be incorporated effectively like subroutines or functions in local scripts to enable batch processing of searches, for example the large-scale annotation of a proteome. This makes large-scale HMMER/Pfam analysis available to users who lack access to sufficient local computing resources. HMMER services have been registered with BioCatalogue (19), the registry of web services for life sciences, making them discoverable alongside other, related web services.
Almost half of our current searches are arriving via REST. There are currently two main categories of end points to the web services:
POST http://hmmer.janelia.org/search/[algo]
and
GET http://hmmer.janelia.org/results/[uuid]
The web site helps pages to contain a complete description of the API, and lists the endpoints and the parameters that they accept. For users familiar with the command-line version of HMMER, we have tried to keep the names of parameters the same as the command-line option, wherever possible. The help section also contains several examples of simple clients written in Perl, Python and Java. These can be downloaded and modified to generate more sophisticated clients according to a user’s needs. Because the RESTful searches and interactive web searches use the same pipeline, it is possible to visualize on the web site results that have been submitted via the web services. For example, an annotation pipeline may be run in batch mode, and individual selected results may later be visually inspected by a human. Retaining the job identifier is all that is necessary to link back to the HTML version of the results.
CONCLUSIONS
The focus of this initial version of the HMMER web server has been on speed and minimizing response time. Most searches take 1–2 s to search against even the largest target databases. Our long-term aim is to drive search times down even further, such that typical search times are in the 100–200 ms range, which human users perceive as a near-real-time interaction. This would permit users to interactively explore protein sequence space.
We also plan to add support for the fourth protein search algorithm, jackhmmer, in the near future. This allows iterative searches, starting from a single query sequence, analogous to PSI-BLAST (10).
We believe the most important issue to address in the future is the visualization of search results. A batch-mode tabular output was adequate in the days when most searches returned zero, one, or a few hits. Today, with the sequencing of thousands of genomes, typical searches return hundreds and thousands of hits. The most informative matches are often obscured by numerous matches to less well-annotated sequences in less accessible organisms. A principal future aim for us is the development of graphical visualizations that show results organized on phylogenetic trees. This would allow users to browse the most relevant clades and organisms while temporarily hiding other results. Such a paradigm would synergize with our goal of reducing search times by another order of magnitude, because we could organize the search and the target databases themselves along phylogenetic lines. An initial search could be conducted against a standard ‘framework’ phylogeny, which consists of a subset of better-known or characterized representative organisms, in an initial results display. Subsequent deeper searches, expanded to all sequences, could be conducted only when the user clicked to request a deeper look at some particular clade. This paradigm also provides a recipe for managing the exponential growth of the sequence databases. The framework phylogeny would be expected to be a slowly growing set of complete reference proteomes, while the exponential explosion of additional sequence data would be hidden in the smaller, higher resolution branches of that tree.
FUNDING
Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We dedicate our work to the memory of our colleague and friend Michael Farrar, the original author of hmmpgmd, who died unexpectedly in December 2010.
REFERENCES
- 1.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Farrar M. Striped Smith-Waterman speeds database searches six times over other SIMD implementations. Bioinformatics. 2007;23:156–161. doi: 10.1093/bioinformatics/btl582. [DOI] [PubMed] [Google Scholar]
- 6.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lees J, Yeats C, Redfern O, Clegg A, Orengo C. Gene3D: merging structure and function for a thousand genomes. Nucleic Acids Res. 2010;38:D296–D300. doi: 10.1093/nar/gkp987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lima Morais DA, Fang H, Rackham OJ, Wilson D, Pethica R, Chothia C, Gough J. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic Acids Res. 2011;39:D427–D434. doi: 10.1093/nar/gkq1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF. Amino acid substitution matrices from an information theoretic perspective. J. Mol. Biol. 1991;219:555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu YK, Wootton JC, Altschul SF. The compositional adjustment of amino acid substitution matrices. Proc. Natl Acad. Sci. USA. 2003;100:15688–15693. doi: 10.1073/pnas.2533904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy SR. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 2008;4:e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velankar S, McNeil P, Mittard-Runte V, Suarez A, Barrell D, Apweiler R, Henrick K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhagat J, Tanoh F, Nzuobontane E, Laurent T, Orlowski J, Roos M, Wolstencroft K, Aleksejevs S, Stevens R, Pettifer S, et al. BioCatalogue: a universal catalogue of web services for the life sciences. Nucleic Acids Res. 2010;38:W689–W694. doi: 10.1093/nar/gkq394. [DOI] [PMC free article] [PubMed] [Google Scholar]